Abstract
Epithelial–mesenchymal transition (EMT), crucial during embryogenesis for new tissue and organ formation, is also considered to be a prerequisite to cancer metastasis. We report here that the protein tyrosine phosphatase Pez is expressed transiently in discrete locations in developing brain, heart, pharyngeal arches, and somites in zebrafish embryos. We also find that Pez knock-down results in defects in these organs, indicating a crucial role in organogenesis. Overexpression of Pez in epithelial MDCK cells causes EMT, with a drastic change in cell morphology and function that is accompanied by changes in gene expression typical of EMT. Transfection of Pez induced TGFβ signaling, critical in developmental EMT with a likely role also in oncogenic EMT. In zebrafish, TGFβ3 is co- expressed with Pez in a number of tissues and its expression was lost from these tissues when Pez expression was knocked down. Together, our data suggest Pez plays a crucial role in organogenesis by inducing TGFβ and EMT.
Introduction
Epithelial–mesenchymal transition (EMT) is fundamental during early development for the reorganization of groups of progenitor cells within a preexisting epithelium into complex sets of juxtaposing tissues, thus allowing new inductive patterning events necessary for organogenesis. In addition to the formation of the three germ layers and the neural crest, EMTs give rise to, amongst others, sclerotome from ventral somites; endocardial cushions of the atrio-ventricular (A-V) canal of the heart from the endocardial epithelium; and bones, cartilage, and musculature from cells of the neural plate (for review see Savagner, 2001; Shook and Keller, 2003).
The process of EMT enables epithelial cells, which are normally constrained by cell-to-cell adherence, to delaminate and take on the invasive and migratory properties that are a feature of mesenchymal cells. Aberrant reactivation of this process in the adult can give rise to undesirable pathological states such as tumor progression, whereby delamination of the tumor cells and acquisition of invasive and migratory properties is the forebear of metastatic disease (for review see Thiery, 2002). Loss of E-cadherin expression facilitates the loss of cell-cell adhesion. This is often achieved during EMT by the induction of repressors of E-cadherin transcription, such as Snail, ZEB1/δEF1 (Grooteclaes and Frisch, 2000), ZEB2/SIP1 (Comijn et al., 2001), E47 (Perez- Moreno et al., 2001), Twist (Yang et al., 2004), and Slug/Snail2 (Bolos et al., 2003).
In vivo, the importance of E-cadherin transcriptional repressors in EMT is demonstrated by the role of Snail in EMT. Snail-null mice cannot complete gastrulation because they fail to down-regulate E-cadherin to undergo EMT (Carver et al., 2001). Evidence is also accumulating that indicates a role for Snail and other transcriptional repressors of E-cadherin in cancer progression. In malignant tumors in general, E-cadherin expression correlates with more highly differentiated cancers that exhibit cell–cell adhesion and are less invasive (Thiery, 2002). Elevated expression of Snail1, ZEB1, and Twist have been found in metastatic cancer cell lines and invasive human cancers, with concomitant loss of E-cadherin expression in these cell lines and tissues (Peinado et al., 2004; Yang et al., 2004). Although these observations clearly illustrate that regulation of E-cadherin and its transcriptional repressors are key elements in embryonal and pathological EMT, the molecular events that initiate the EMT are ill defined.
The TGFβs are pleiotrophic cytokines that regulate cellular functions ranging from embryonal development to maintaining homeostasis in the adult. The three mammalian TGFβ isoforms (TGFβ1, TGFβ2, and TGFβ3) show high sequence homology and have overlapping functions. Most cells, including epithelial cells, make TGFβs and express TGFβ receptors (Fleisch et al., 2006); hence, the TGFβs can act in a paracrine, autocrine, and/or endocrine fashion (Smith, 1996). The role of TGFβ in EMT, where it regulates cell–cell adhesion, cytoskeletal remodelling, and cell-matrix adhesion, is well documented (for review see Zavadil and Bottinger, 2005). In embryonal development, TGFβ plays a role in organogenesis in the late stages of embryogenesis. The requirement for TGFβ in EMT in the formation of the endocardial cushions during cardiac valve development (Romano and Runyan, 2000) and in EMT involved in palate fusion (Kaartinen et al., 1995) support a role for TGFβ in EMT in vivo. TGFβ has also been shown to play opposing roles in oncogenesis and metastasis (Roberts and Wakefield, 2003; Siegel et al., 2003). In the early stages of carcinogenesis, its epithelial growth suppressive properties liken its role to that of a tumor suppressor. However, in tumor cells that have progressed to evade its anti-proliferative effects, apoptosis and anoikis, TGFβ can promote oncogenic EMT leading to an increase in invasiveness and motility, the beginnings of metastasis.
Phosphorylation of proteins on tyrosine residues by tyrosine kinases and the reversal of the process by protein tyrosine phosphatases (PTPs) regulate many diverse cellular functions from proliferation to differentiation. The PTP Pez is an intracellular PTP that is located at the adherens junctions (AJ) in endothelial and epithelial cells (Wadham et al., 2000, 2003). Dominant-negative mutants of Pez induce an increase in tyrosine phosphorylation at the AJ, which together with the observation that Pez directly dephosphorylates β-catenin (Wadham et al., 2003) suggests that Pez plays a role in regulating the tyrosine phosphorylation status of the AJ. However, Pez can be localized elsewhere in the cytoplasm and in the nucleus (Wadham et al., 2000), suggesting that it may have other targets and functions apart from regulating the phosphorylation status of the AJ. Here, we show that Pez expression in zebrafish embryos is developmentally regulated and that knocking down its expression results in embryos with defective organ development. In vitro and in vivo studies revealed a role for Pez in regulating TGFβ and EMT, processes critical for the formation of new tissues and organs during embryonic development.
Results
Pez expression is transiently induced at specific stages during zebrafish embryogenesis
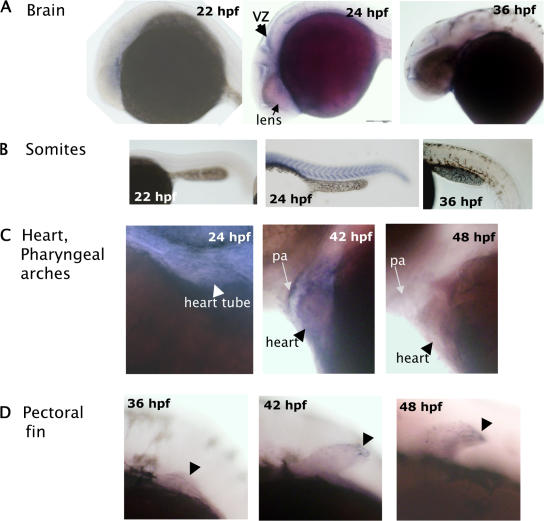
The temporal and spatial expression profiles of a gene in the context of a whole organism can shed light on its physiological function. To assess whether Pez has a developmental role, we examined its temporal and spatial expression profiles during zebrafish embryonal development up to 48 h post-fertilization (hpf) by whole-mount in situ hybridization. With the exception of maternally derived mRNA present up to 4 hpf (unpublished data), Pez expression was not detectable until 16 hpf, at which time it became faintly detectable throughout the embryo (unpublished data). Between 22–24 hpf Pez expression was strongest in the ventricular/subventricular zone of the developing brain (Fig. 1 A), a region rich in proliferating and migrating neuronal progenitors. Pez expression in the brain subsequently declined, with very low levels remaining at 36 hpf (Fig. 1 A). Pez expression was also detected in the somites at 24 hpf, but was no longer detectable by 36 hpf (Fig. 1 B; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200705035/DC1), suggesting that Pez may play a role in the later stages of somite maturation. Transient Pez expression was also observed during cardiac development. Pez expression was visible in the early developing heart tube at 24 hpf, reaching a peak between 42–44 hpf and then rapidly down-regulated, so that by 48 hpf little Pez expression remained (Fig. 1 C). Another tissue in which Pez expression was observed is the pectoral fin, where little or no Pez expression was detected in the fin bud at 36 hpf, but more sustained expression was observed, between 42 to 48 hpf, the latest time point investigated. These examples of transient Pez expression at specific stages of tissue development suggest that Pez plays a regulatory role in development.
Figure 1.
Pez expression during zebrafish embryogenesis. Zebrafish Pez mRNA was detected by whole-mount in situ hybridization. Lateral views of embryos at various stages of development are shown; dorsal to the top, anterior to the left. (A) View showing developing brain. Arrow indicates the ventricular/subventricular zone of the brain in 24-hpf embryo. (B) View of trunk showing Pez expression in somites in 24-hpf embryo. (C) View showing Pez expression in the pharyngeal arches (pa) and heart. (D) View showing pectoral fin (arrowhead) development. Embryos in B–D were cleared with BBA post-staining.
Pez expression is crucial for organogenesis
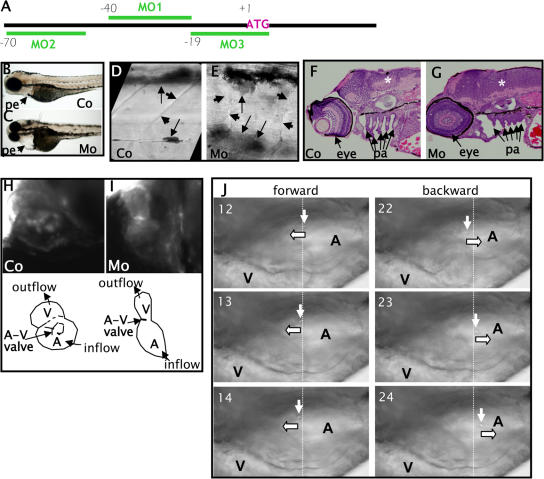
To determine whether Pez plays a crucial role in the regulation of developmental processes, three different antisense morpholino- oligonucleotides (MOs) (Fig. 2 A), designed to inhibit translation of Pez protein, were microinjected into zebrafish embryos at the one- to four-cell stage. Embryos injected with a control MO were indistinguishable from uninjected controls, whereas injection with each of the anti-Pez MOs produced identical morphant phenotypes.
Figure 2.
Phenotype of Pez knock-down zebrafish embryos using antisense morpholino oligonucleotides. (A) Schematic showing relative positions of Pez antisense morpholino oligonucleotides used for knocking-down Pez expression. B, D, F, and H are control (Co) and C, E, G, and I are Pez morphant (Mo) zebrafish embryos; B–G embryos are 96 hpf, H–J embryos are 53 hpf. Anterior to the left. (B and C) Lateral view of whole zebrafish embryos; pe, pericardium. (D and E) Lateral view of trunk showing somite boundaries (short arrow) and pigment (long arrow). (F and G) Longitudinal H&E-stained sections showing increased cellular density in the developing brain (*), and abnormal formation of pharyngeal arches (pa) in morphant embryos. (H and I) Ventro-lateral view. Fluorescence micrographs showing structure of the developing heart of live fli-GFP embryos. Schematic below shows relative positions of atrium (A), ventricle (V), and A-V valve, and arrows indicate direction of normal blood flow. Opening and closing of A-V valve in control and lack thereof in Pez morphants can be viewed in Videos 1 and 2, respectively (Videos available at http://www.jcb.org/cgi/content/full/jcb.200705035/DC1). (J) Time-lapse light micrographs showing blood flow in atrium of representative MO-injected embryo. Left panels: three sequential frames (12–14) from a time-lapse video showing a forward-moving blood cell. Right panels: three sequential frames (22–24) from the same time-lapse video showing backward-moving blood cell. Downward arrows track the blood cell in each series and block arrows indicate direction of movement of the indicated blood cell.
Pez morphants displayed a number of defects in several organs and tissues (summarized in Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200705035/DC1) that correlated with regions of Pez expression (see Fig. 1). Most notable was a severe pericardial edema obvious by 2–3 dpf (Fig. 2, B and C; Fig. S2). From in situ hybridization (Fig. 1), we had noted that Pez expression generally peaks after the rudimentary tissues and organs have formed, suggesting that Pez may play a role in the development of specific structures within the organs rather than in initiating organ development. In concordance with this hypothesis, the organs were formed in the Pez morphants, but with architectural defects in multiple organs and tissues. Somites and somite boundaries were formed in Pez morphants, but the boundaries were irregular (Fig. 2, D and E). In the ventricular/subventricular zone of the brain, a region with strong Pez expression in wild-type embryos at 24 hpf (Fig. 1 A), there was an increase in cellular density and expansion of the ventricular zone at the expense of the cortex (Fig. 2, F and G) in the Pez morphants compared with uninjected or control MO- injected embryos. Similarly, structural defects were observed in the formation of the pharyngeal arches, which included a higher cellular density, shortening, and poor formation of the cartilage (Fig. 2, F and G).
The severe pericardial edema observed in Pez morphants (Fig. 2, B and C; Fig. S2) is symptomatic of defects in cardiac function. To further evaluate cardiac function in Pez morphants, we used fli1-GFP transgenic zebrafish, which express GFP in the endothelial cells comprising the endocardium (Lawson and Weinstein, 2002), to visualize structure and function of the atrium, ventricle, and the atrio-ventricular (A-V) valve in live embryos. By 53 hpf, the developing heart of control-injected embryos had begun to loop and the atrium and ventricle were partially superimposed (Fig. 2 H). The A-V valves between the two chambers were well developed and opened and shut in synchrony with rhythmic contractions (Fig. 2 H; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200705035/DC1). In contrast, the heart of Pez morphants remained tubular, suggesting a defect in looping, and they lacked patent A-V valves (Fig. 2 I; Video 2), which normally function to prevent regurgitation of blood back into the atrium during rhythmic contractions. We also observed a loss of unidirectional blood flow in the atrium of Pez morphants when the movements of blood cells in the atrium were tracked (Fig. 2 J), supporting a lack of functional A-V valve.
Pez-overexpressing MDCK epithelial cells undergo EMT
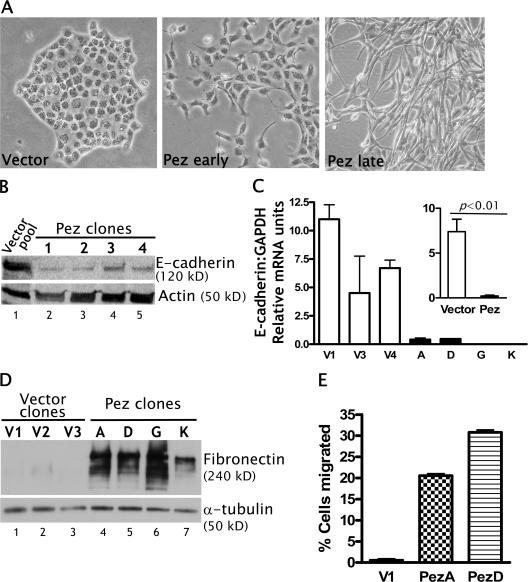
The expression profile of Pez during zebrafish development indicates that its expression is low or absent until required, whereupon it is highly up-regulated in specific tissues and contributes to organogenesis. To elucidate the molecular events induced by such a marked increase in Pez expression, we overexpressed Pez in MDCK epithelial cells and selected for clonal lines (referred to as Pez-MDCK cells). Pez overexpression transformed the normal epithelial morphology of MDCK cells into a fibroblast-like morphology (Fig. 3 A), whereas cells transfected with the empty expression vector (vector-MDCK) remained indistinguishable from parental MDCK cells. The transformation to fibroblast-like morphology appeared to occur in two steps: an initial scattering phenotype (Fig. 3 A, middle) that was evident ∼2–3 wks after transfection and clonal selection was followed by a more spindly and elongated morphology (Fig. 3 A, right) typical of epithelial cells that have undergone EMT. The Pez-MDCK cells appeared to have lost cell–cell contact, consistent with loss of junctional E-cadherin visualized by indirect immunofluorescence (Khew-Goodall and Wadham, 2005). Total E-cadherin protein expression was examined in four representative Pez-MDCK clones and found to be highly down-regulated compared with a pool of vector-MDCK cells (Fig. 3 B), suggesting that the loss of junctional E-cadherin was not merely due to a translocation of the protein away from the cell junctions. Similar results were obtained in a repeat of the experiment in which three individual vector-MDCK clones and another four representative Pez-MDCK clones were analyzed (unpublished data). Analysis of E-cadherin mRNA expression in these clones by qRT-PCR indicated that E-cadherin mRNA was also markedly down-regulated (Fig. 3 C). Averaging the data from four independent Pez-MDCK clones and three vector-MDCK clones, a >30-fold reduction in E-cadherin mRNA was observed in Pez-MDCK clones. The Pez-MDCK clones also had markedly up-regulated fibronectin (an extracellular matrix protein secreted by mesenchymal cells) compared with the vector-MDCK clones (Fig. 3 D). In addition, using a Boyden chamber assay to measure motility, two representative Pez-MDCK clones were found to be highly motile compared with vector-MDCK cells (Fig. 3 E). The alteration of cellular morphology accompanying Pez overexpression in MDCK cells, together with the loss of the epithelial marker E-cadherin and gain of mesenchymal marker (fibronectin) and function (motility), suggests that Pez had induced an EMT.
Figure 3.
Pez overexpression induces an EMT in MDCK cells. (A) A representative clone of vector-MDCK cells (left) or Pez-MDCK cells at 2 wks (middle) or several weeks (right) post-transfection. (B) Western blot analysis of E-cadherin expression in a pool of vector-MDCK cells (lane 1) and four representative Pez-MDCK clones (lanes 2–5). Actin is shown as a loading control. (B) E-cadherin mRNAs in three representative vector-MDCK (white bars) and four Pez-MDCK (black bars) clones were measured by quantitative real-time PCR. Relative mRNA units represent mRNA of interest normalized to a GAPDH internal control. Error bars represent the SEM of samples assayed in duplicate. Insets show pooled values for the three vector- and four Pez-MDCK clones. P-values were derived using the t test, n = 6 for vector-MDCK, n = 8 for Pez-MDCK. (D) Western blot analysis of fibronectin expression in three representative vector-MDCK clones (lanes 1–3) and four representative Pez-MDCK clones (lanes 4–7). α-Tubulin is shown as a loading control. (E) Migratory capacity of one representative vector-MDCK clone and two representative Pez-MDCK clones. “% cells migrated” represents the number of cells migrated in a modified Boyden chamber assay, expressed as a percentage of the initial number of plated cells. Error bars represent the SEM of samples assayed in quadruplicate, and data are representative of three individual experiments.
Pez–MDCK cells express EMT-promoting transcription factors
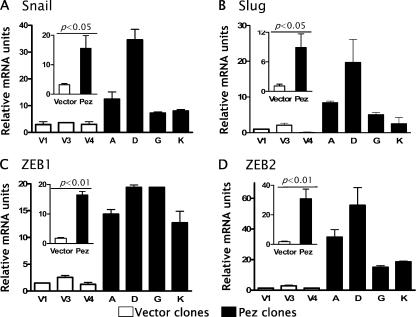
Many studies have shown that the loss of E-cadherin expression associated with EMT is a consequence of transcriptional repression of the E-cadherin gene, particularly by the zinc finger transcriptional repressors such as Snail and Slug (for review see Peinado et al., 2004). We compared the mRNA levels of the E-cadherin repressors Snail, Slug, ZEB1, and ZEB2 between Pez-MDCK and vector-MDCK cells. Snail and Slug expression were elevated in all four Pez-MDCK clones analyzed, averaging ∼5- and ∼8-fold higher, respectively, than expression in the vector-MDCK clones (Fig. 4, A and B), while ZEB1 and ZEB2 were induced ∼9- and ∼30-fold, respectively (Fig. 4, C and D). The marked loss of E-cadherin mRNA along with the induction of expression of the E-cadherin repressors Snail, Slug, ZEB1, and ZEB2 suggest that the Pez-induced reduction in E-cadherin is due to transcriptional repression.
Figure 4.
Effect of Pez overexpression on Snail, Slug, ZEB1, and ZEB2 expression. mRNAs from three representative vector-MDCK clones (white bars) and four representative Pez-MDCK clones (black bars) were measured by qRT-PCR. Relative mRNA units represent mRNA of interest normalized to a GAPDH internal control. Normalizing to other internal controls (β-actin, HPRT, and ribosomal RNAs) gave essentially identical results to GAPDH. Error bars represent the SEM of samples assayed in duplicate. Insets show pooled values for the three vector- and four Pez-MDCK clones. P-values were derived using the t test, n = 6 for vector-MDCK, n = 8 for Pez-MDCK.
We noted that ZEB1 levels were relatively constant between the Pez-MDCK clones whereas Snail, Slug, and ZEB2 mRNA levels were highest in clone D, intermediate in clone A, and lower in clones G and K (Fig. 4, A, B, and D). Upon further passaging, Snail expression in both clones A and D declined to the lower levels expressed by clones G and K (unpublished data), whereas ZEB1 expression remained relatively unchanged. These observations are consistent with the transient nature of Snail and ZEB2 expression noted previously (Peinado et al., 2004), and suggest that at the time of the analysis shown in Fig. 4, down-regulation of Snail and ZEB2 expression in individual Pez-MDCK clones had proceeded to different extents.
Pez induces TGFβ production
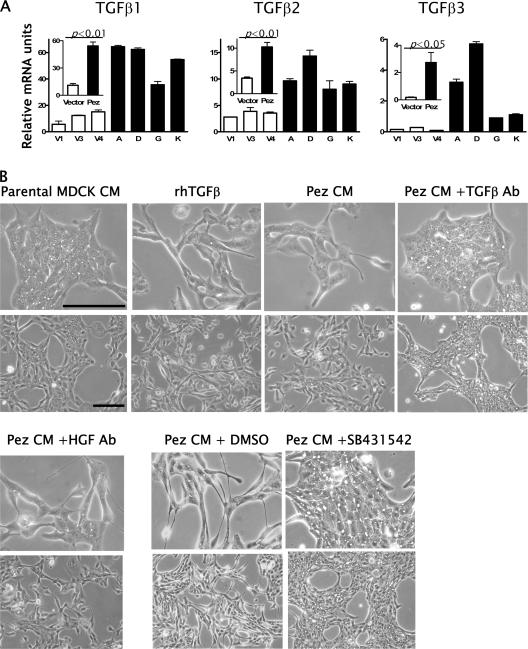
TGFβ has previously been shown to induce an EMT in MDCK and other epithelial cells through induction of all or a subset of the transcription repressors Snail, Slug, ZEB1, and ZEB2 (Zavadil and Bottinger, 2005), all of which were induced in Pez-MDCK cells. We therefore investigated the possibility that Pez may be mediating its effects through the action of TGFβ. By qRT-PCR, we found that the mRNAs for TGFβ-1, -2, and -3 were 5, 3, and 15× higher, respectively, in the Pez-MDCK cells than in the vector-MDCK cells (Fig. 5 A). To determine whether increased TGFβ mRNA expression in the Pez-MDCK cells was accompanied by increased secretion of active TGFβ capable of inducing an EMT, conditioned medium from the Pez-MDCK cells cultured for 72 h was collected and used to culture parental MDCK cells. After 1–2 d, parental MDCK cells cultured in Pez-MDCK–conditioned medium began to lose cell–cell contact and scatter, whereas cells cultured in control parental MDCK cell conditioned medium remained unaltered (unpublished data). After 4–6 d in culture in Pez-MDCK conditioned medium (Fig. 5 B, Pez CM), the parental MDCK cells underwent a transformation to a spindly fibroblast-like morphology resembling that of Pez-MDCK cells (Fig. 3 A) and cells treated with rhTGFβ (Fig. 5 B, rhTGFβ) that have undergone an EMT. In contrast, parental MDCK cells cultured in conditioned medium from parental (Fig. 5 B, parental MDCK CM) or vector-MDCK cells (unpublished data) remained epithelial and indistinguishable from cells cultured in normal medium (unpublished data). These observations suggest that Pez-MDCK conditioned medium contained a soluble factor that was able to promote EMT in a similar fashion to rhTGFβ, and that this factor was not present at levels sufficient to induce EMT in conditioned medium obtained from parental MDCK or vector-MDCK cells. The transition to a mesenchymal morphology induced by Pez-MDCK conditioned medium was inhibited by the addition of the inhibitor of TGFβ receptor kinase activity, SB-431542, and by a blocking pan-TGFβ Ab to the Pez-MDCK–conditioned medium, but not by an anti-HGF Ab (Fig. 5 B), suggesting that the EMT-promoting factor secreted by Pez-MDCK cells was active TGFβ.
Figure 5.
Pez overexpression induces active TGFβ expression. (A) TGFβ1, -2, and -3 mRNAs were analyzed and presented as described in the legend to Fig. 4 B. Parental MDCK cells cultured in Pez-MDCK conditioned medium undergo a TGFβ-dependent transformation to a fibroblast-like morphology. Parental MDCK cells were grown for 6 d in conditioned medium from either normal parental MDCK cells (denoted parental MDCK CM) or Pez-MDCK cells (Pez CM). Conditioned media from parental MDCK or Pez-MDCK cells were harvested after 3 d in culture. Where indicated, anti-TGFβ Ab (50 μg/ml), anti-HGF Ab (50 μg/ml), or the TGFβ receptor kinase inhibitor SB-431542 (1.0 μM) were added before culture in Pez-MDCK conditioned medium. DMSO was used as a vehicle control for SB-431542. Where indicated, rhTGFβ (10 ng/ml) was added to parental cells in normal culture medium. Top panels in each set show high magnification and bottom panels show low magnification images. Bars = 0.05 mm.
Transient Pez overexpression activates canonical TGFβ signaling
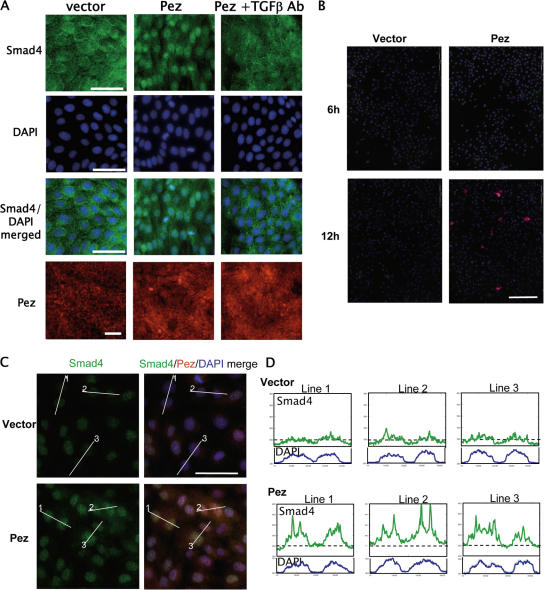
In Pez-MDCK cell lines, Pez expression has persisted over a long period of time, raising the possibility that TGFβ secretion and subsequent signaling may be an indirect consequence of Pez overexpression. In mammalian cells, ligand binding to the TGFβ receptor triggers downstream signaling by phosphorylation of the R-Smads, which then complex with the coSmad, Smad4, and translocate into the nucleus (for review see Massague and Chen, 2000). To address the kinetics of Pez-induced TGFβ signaling, we initially examined the effect of Pez expression on Smad4 nuclear translocation in transient transfection assays at 24 h post-transfection, when at least 50% of the Pez-transfected cells showed elevated Pez expression compared with empty vector-transfected cells (Fig. 6 A). In control empty vector-transfected MDCK cells, Smad4 remained predominantly cytoplasmic (Fig. 6 A). In contrast, in MDCK cells transfected with Pez, Smad4 was translocated into the nucleus (Fig. 6 A), suggesting that Pez overexpression induces Smad-dependent TGFβ signaling. Addition of a neutralising TGFβ Ab following Pez transfection blocked nuclear translocation of Smad4 (Fig. 6 A), confirming that the nuclear localization of Smad4 was TGFβ specific. The ablation of Smad nuclear translocation by the exogenous addition of TGFβ Ab also indicates that the activation of TGFβ signaling by Pez involves the secretion of active TGFβ.
Figure 6.
Transient Pez overexpression induces Smad4 nuclear translocation. (A) Immunofluorescence images of MDCK cells transiently transfected with either an empty vector, pCDNA3 (denoted vector), or pCDNA-Pez (denoted Pez), showing Smad4 subcellular localization detected by indirect immunofluorescence using a Smad4 mAb (green, top row). The cells were counterstained with DAPI (blue, second row) and a merged image (third row) showing nuclear Smad4 in Pez-transfected cells but not cells transfected with empty vector or where the anti-TGFβ Ab was added to the Pez-transfected cells. Vector- and Pez-transfected cells were also stained with a Pez Ab to show endogenous Pez expression in vector-transfected and elevated levels of Pez expression in the Pez-transfected cells (red, bottom row). Cells were processed for immunofluorescence 24 h post-transfection. The figure shows one representative experiment of three separate transfections. Bars = 0.05 mm. (B) Immunofluorescence images of MDCK cells at 6 and 12 h post-transfection with an empty vector or pCDNA-Pez showing Pez expression detected with an anti-Pez Ab (red). Cells were counterstained with DAPI (blue). Bars = 0.2 mm. (C) Immunofluorescence images of vector- or Pez-transfected MDCK cells at 12 h post-transfection showing Smad4 (green) localization and Pez (red) expression. Cells were counterstained with DAPI. Bars = 0.05 mm. (D) Fluorescence intensity profiles corresponding to the indicated lines in C showing Smad4 fluorescence intensity (green line) and the corresponding DAPI staining intensity (blue line) to indicate the position of the nuclei. The horizontal broken line marks 250 units of fluorescence intensity in each panel and scale markings on the Y-axis are at 50 fluorescence intensity unit intervals. Fluorescence intensity quantitated using Analysis software (Soft-Imaging Systems, Olympus).
Examination of Pez expression and TGFβ signaling at earlier time points after Pez or vector transfection showed no detectable increase in Pez expression in the Pez-transfected cells compared with vector-transfected cells at 6 h post-transfection, but by 12 h post-transfection a small proportion of cells with elevated Pez expression was evident (Fig. 6 B), indicating that elevated Pez expression occurs between 6 to 12 h after transfection. Examination of Smad4 subcellular localization at 12 h post-transfection showed an increase in nuclear Smad in cells with elevated Pez expression, suggesting that canonical TGFβ signaling is induced shortly after Pez expression (Fig. 6, C and D). The expression of TGFβ mRNAs was also assayed by qRT-PCR up to 24 h post-transfection, but no induction was detected (unpublished data), suggesting that TGFβ production induced by Pez initially occurs by post-transcriptional mechanisms. No analyses were performed beyond 24 h post-transfection, as Pez expression began to decline after this point (unpublished data), making the interpretation of data obtained beyond 24 h post-transfection equivocal.
Pez regulates TGFβ3 expression in zebrafish embryos
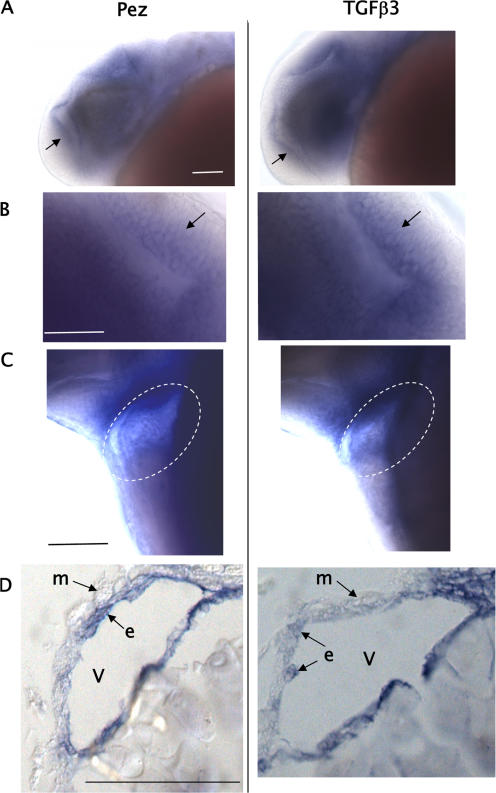
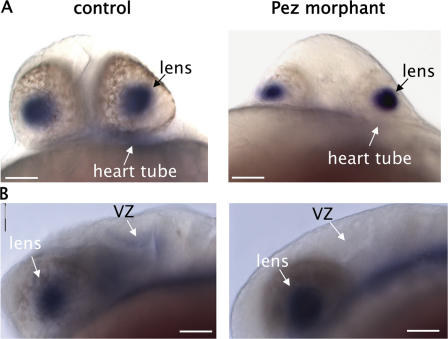
The induction of TGFβ signaling and mRNA synthesis by Pez in vitro prompted us to investigate if this is paralleled in vivo. Published data on the pattern of TGFβ3 expression (Cheah et al., 2005) revealed that its expression closely resembled that seen for Pez in zebrafish embryos. We therefore compared the profiles of Pez and TGFβ3 expression during zebrafish development and found that they coexpressed in the same subset of cells in the ventricular zone of the brain at 24 hpf (Fig. 7, A and B) and in the endocardial cells of the heart at 44 hpf (Fig. 7, C and D). We also noted that TGFβ3 was expressed in some tissues where Pez was not expressed, such as in the lens of the developing eye at 24 hpf (Fig. 8 and Fig. 1 A). In addition to the co-temporal expression of Pez and TGFβ3, the high degree of spatial correlation, particularly in the ventricular zone of the brain (Fig. 7 B) and in the heart (Fig. 7 D), is highly consistent with the notion that in these developing tissues Pez may up-regulate TGFβ3 expression. To explore this further, we examined the expression of TGFβ3 in Pez morphants at 24 hpf. Unlike embryos at later stages of development, the morphology and architecture of the developing brain and heart tube of the Pez morphants at 24 hpf are not overtly different from control MO-injected or wild-type embryos. Although morphologically similar at 24 hpf, expression of TGFβ3 was undetectable in the ventricular zone of the brain and the heart tube in the Pez morphants, in contrast to control embryos where TGFβ3 was clearly expressed in these tissues (Fig. 8). It is also worth noting that in the lens of the developing eye where Pez is not expressed, TGFβ3 expression was unaffected by knocking down Pez expression (Fig. 8).
Figure 7.
TGFβ3 expression correlates with Pez expression in zebrafish embryos. (A–C) Whole-mount in situ hybridisations using Pez (left) or TGFβ3 (right) probes. A, low magnification; B, higher magnification. Lateral views of 24-hpf embryos showing Pez and TGFβ3 colocalization in the ventricular zone (arrow) of the brain. (C) Lateral view of 44-hpf embryo showing coexpression of Pez and TGFβ3 in the heart. (D) Longitudinal sections of the embryos shown in C through the ventricles (V) (represented by the region marked by the ellipse in C) showing Pez and TGFβ3 staining in the endocardial cells (e) lining the cardiac chamber, but not myocardial (m) cells. Anterior to the left. Embryos were BBA cleared post-staining. Bars = 0.05 mm.
Figure 8.
Loss of TGFβ3 expression in the brain and heart of Pez morphants. Whole-mount in situ hybridizations of 24-hpf control (left) or Pez morphants (right) showing TGFβ3 expression. A, ventral view; B, lateral view. Anterior to the left. Embryos were BBA cleared post-staining. VZ, ventricular zone. Bars = 0.05 mm.
Discussion
EMT is a process shown to be essential for cell movement to facilitate the formation of new tissues and organs during development. Collectively, several lines of evidence generated from this study indicate that Pez is a novel regulator of TGFβ-mediated EMT in vitro and of organogenesis during embryonal development. Epithelial MDCK cells overexpressing Pez underwent morphological and functional changes, including the acquisition of cell motility, characteristic of an EMT. The morphological and functional changes were accompanied by and are likely to be a consequence of changes in gene expression typical of EMT, including loss of E-cadherin and up-regulation of the EMT-inducing transcription factors, Snail, Slug, ZEB1, and ZEB2. The transient nature of Pez expression during zebrafish development which correlated with specific stages of organ development suggests that Pez plays a role in regulating organogenesis. This is further supported by data obtained from zebrafish hypomorphic for Pez expression where we observed developmental defects in all organs that expressed Pez during development. Given that Pez can promote EMT in vitro, some or all of the defects observed in these morphant embryos may be due to an inability to induce EMT during organ development.
The role of TGFβ as a driver of developmental EMT is well recognized although little is known of the mechanisms that initiate TGFβ activation. In these studies we made several observations suggesting that Pez is a novel inducer of TGFβ signaling. In Pez-MDCK cell lines, TGFβ secretion is up-regulated to a level capable of promoting EMT. In addition, transient Pez overexpression induces TGFβ secretion and canonical Smad-dependent TGFβ signaling within 6 h of its expression, suggesting that TGFβ signaling is activated as an immediate-early response to Pez expression. The early activation of TGFβ by Pez was, however, not accompanied by increases in TGFβ mRNAs, suggesting that this initial activation occurs by post-transcriptional mechanisms which may be due to activation of latent TGFβ (for review see Massague and Chen, 2000) or de-repression of translation (Fraser et al., 2002). At later stages following Pez expression, TGFβ mRNAs were up-regulated as seen in Pez-MDCK cell lines that had undergone an EMT. It is well documented that TGFβ induces its own transcription (Van Obberghen-Schilling et al., 1988; Bascom et al., 1989) and we suggest that the initial activation of canonical TGFβ signaling by Pez acts to induce subsequent de novo synthesis of TGFβ mRNAs by an autoinductive mechanism. This is supported by studies in our laboratory showing that exogenous addition of rhTGFβ1 to MDCK cells induced TGFβ2 and TGFβ3 mRNAs after several days of stimulation (unpublished data). Furthermore, in other studies we have also found that the exogenous addition of rhTGFβ1 to MDCK cells induced the downstream effectors of EMT, Snail1 (Fig. S3), ZEB1, and ZEB2 (unpublished data), albeit this also took 3 d or more of continuous stimulation to achieve substantial levels of induction. Together, these observations suggest that Pez initially induces TGFβ activation by post-transcriptional mechanisms and the resultant activation of canonical TGFβ signaling then acts in an autocrine feed-forward mechanism to activate subsequent transcription of TGFβs and the downstream effectors of EMT. Finally, the high degree of correlation between Pez and TGFβ3 expression in the developing brain and heart of zebrafish embryos and the loss of TGFβ3 expression in these tissues when Pez was knocked down strongly supports our in vitro data that Pez regulates TGFβ production.
In the developing heart, rhythmic contractions signifying the beginning of cardiac development are evident at ∼22 hpf and development continues for several days (Beis et al., 2005). Although Pez expression was evident at 24 hpf, strong expression was not observed until 36 hpf (unpublished data), suggesting that it is not essential for the early stages of cardiac development, a notion supported by the development of atrium, ventricle, and rhythmic contractions in the Pez morphants. Instead, strong Pez expression in the heart coincides with looping of the heart tube and specification of the AV canal, which begin around 36 and 37 hpf, respectively (Yelon and Stainier, 1999; Walsh and Stainier, 2001). In the early steps of A-V valve development, the designated endocardial cells of the A-V canal undergo a TGFβ-dependent EMT to form the endocardial cushions, which precede formation of the A-V valves (Romano and Runyan, 2000; Armstrong and Bischoff, 2004). In zebrafish development, this begins at ∼45–48 hpf when Notch1b expression, which is required in conjunction with TGFβ signaling to induce EMT (Timmerman et al., 2004), is restricted to the A-V canal (Westin and Lardelli, 1997; Walsh and Stainier, 2001). Our studies show that Pez expression persists until ∼48 hpf and in 44-hpf embryos both Pez and TGFβ3 colocalize to the endocardium, coinciding precisely with the timing of restricted Notch1b expression at the A-V canal and the onset of EMT. The defects in cardiac valve development and loss of TGFβ3 expression when Pez is knocked down strongly suggest that Pez plays a role in initiating TGFβ3- induced EMT in cardiac valve development. However, TGFβ isoforms have also been found to be expressed in newly formed mesenchyme as a consequence of EMT (Barnett et al., 1994; Camenisch et al., 2002) and that they play subsequent roles in further valve development and remodelling. If Pez expression results in activation of latent TGFβ, Pez may have a more far-reaching role in cardiac valve formation beyond the initial induction of EMT.
Zebrafish embryos hypomorphic for Pez expression showed a loss of TGFβ3 expression and increased cellularity in the ventricular zone of the developing brain at the expense of the cortex, indicative of increased proliferation of the progenitors and/or lack of migration into the cortical region. It is therefore of interest to note that TGFβ plays a dual role in neurogenesis of the developing cerebral cortex. It encourages the progenitor cells of the ventricular zone to exit cell cycle, thereby maintaining an appropriate number of cycling cells (Siegenthaler and Miller, 2005), and it promotes migration of the post-mitotic cells into the emerging cortex (Siegenthaler and Miller, 2004), where they can undergo differentiation into mature neurons. The increase in cellular density and expansion of the ventricular zone observed in the Pez morphants is therefore consistent with a loss of TGFβ3 signaling, leading to an inability of the neuronal precursors to exit cell cycle and migrate out of the proliferation zone.
Although our in vitro and in vivo data suggest that Pez is a crucial regulator of TGFβ signaling, the lack of correlation between Pez and TGFβ3 expression in some tissues such as the somites suggest that Pez may also mediate its effects by TGFβ-independent pathways or through TGFβ1- or TGFβ2-mediated pathways. Pez expression in the developing somites is restricted to the central mesenchymal cell compartment surrounded by the epithelium (see Fig. S1) and occurs in a very narrow window after all the somites have formed and when EMT and re-epithelialization by mesenchymal–epithelial transition is complete. Consistent with the expression profile of Pez, somite boundaries were formed and maintained in Pez morphants, albeit ragged and uneven in appearance. At this stage, it is unclear what the role of Pez is in somite development and further work is underway to elucidate this.
Finally, loss of E-cadherin accompanied by induction of its repressors, Snail, Slug, ZEB1, and ZEB2 in various combinations have been observed in a number of epithelial cancer cell lines that have undergone an EMT (Batlle et al., 2000; Peinado et al., 2004). Furthermore, TGFβ signaling is emerging as a dual regulator of oncogenesis and metastasis (Roberts and Wakefield, 2003). The data reported here suggests that Pez has the potential to play a role in oncogenic EMT. It is therefore of interest to note that (Wang et al., 2004) reported a number of somatic mutations in Pez associated with colorectal cancers, although how these mutations affect Pez expression or function is unknown. Based on our findings here, we would suggest that the mutations may cause changes either in Pez expression or activity that could lead to dysregulation of TGFβ signaling.
Materials and methods
Antibodies and inhibitors
Anti–E-cadherin and anti-fibronectin mAbs were from Transduction Laboratories, anti-tubulin mAb was from Abcam, goat anti-actin pAb was from Santa Cruz Biotechnology, Inc. Anti-TGF-β1,-β2, -β3, and anti-HGF mAbs were from R&D systems. The TGFβ receptor kinase inhibitor, SB-431542, was obtained from Tocris Bioscience, UK.
Cell culture and transfection
MDCK cells were maintained in DMEM (JRH Biosciences) supplemented with 10% fetal bovine serum (JRH Biosciences), or 5% serum for conditioned medium experiments. MDCK cells were transfected with pcDNA3 eukaryotic expression vector (Invitrogen) or N-terminal Flag-tagged human Pez cDNA cloned into pcDNA3 (Wadham et al., 2003) and stable transfectants selected by resistance to 500 μg/ml Geneticin (G418; Promega). Single clones of Pez-MDCK cells and either single clones or a pool of vector- MDCK clones were isolated for analysis.
Western blotting
Cells were lysed in Triton X-100 lysis buffer (50 mM Hepes, pH 7.5, 150 mM sodium chloride, 10 mM sodium pyrophosphate, 5 mM EDTA, 50 mM sodium fluoride, 1 mM sodium orthovanadate, and 1% Triton X-100 with protease inhibitor cocktails) and whole-cell lysates analyzed by Western blotting with the indicated primary antibodies and appropriate HRP-conjugated secondary antibodies detected using enhanced chemiluminescence (ECL Plus; GE Healthcare).
Quantitative real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 1–2 μg total RNA was primed with 10 μM pd(N)6 random hexamer (GE Healthcare) and reverse transcribed using Omniscript reverse transcriptase (QIAGEN) according to the manufacturer's instructions. 2 μl of each 20-μl reverse transcription reaction was used as cDNA template for qRT-PCR, using SYBR Green 2x RT-PCR mix (QIAGEN) and 1 μM of each primer. Primer sequences and annealing temperatures for primers are shown in Table I. Reactions were performed on a Rotor-gene RG-3000 (Corbett Research), with the following cycle parameters: 95°C, 15 min; 94°C, 30 s; 50–65°C, 20 s, 72°C, 20 s × 30–35 cycles. Melt curves were analyzed from 72–99°C and products run on 2% agarose gels to verify homogeneity and correct size of all products.
Table I.
qRT-PCR primer sequences and annealing temperatures
| Gene | Primer sequence | Annealing temperature |
|---|---|---|
| GAPDH | + 5′-CATCACTGCCACCCAGAAG-3′; −5′-CAGTGAGCTTCCCGTTCAG-3′ | 50–65°C |
| E-cadherin | + 5′-AAGCGGCCTCTACAACTTCA-3′; −5′-AACTGGGAAATGTGAGCACC-3′ | 50°C |
| Snail | + 5′-CCCAAGCCCAGCCGATGAG-3′; −5′-CTTGGCCACGGAGAGCCC-3′ | 65°C |
| Slug | + 5′-CGTTTTCCAGACCCTGGTTA-3′; −5′-TGACCTGTCTGCAAATGCTC-3′ | 55°C |
| ZEB1 | + 5′-CAAGGTGGCCATTCTGTTAT-3′; −5′-CTAGGCTGCTCAAGACTGTAG-3′ | 55°C |
| ZEB2 | + 5′-CGGTCCAGAAGAAATGAAGG-3′; −5′-TCCTCAAAGTCTGATGTGCAA-3′ | 55°C |
| TGFβ1 | + 5′-TGGACACGCAGTACAGCAA-3′; −5′-TAGTACACGATGGGCAGTGG-3′ | 55°C |
| TGFβ2 | + 5′-CGTTTACAGAACCCGAAAGC-3′; −5′-TCGTCTTGACGACTTTGCTG-3′ | 55°C |
| TGFβ3 | + 5′-CTTGCACCACCTTGGACTTT-3′; −5′-CTGTTGTAAAGGGCCAGGAC-3′ | 55°C |
Migration assay
Migration assays were performed using a modified Boyden chamber assay (Transwells, 6.5-mm diameter, 8-μm pore size; CoStar). 5 × 104 cells were plated in the upper chamber in serum-free medium (DMEM; JRH Biosciences), and migration toward 10% FBS quantified 24 h post-plating using an MTS/PMS-based assay (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay; Promega) according to the manufacturer's instructions. Percentage of migrated cells was derived by comparison to a standard curve generated using known cell numbers. Standards were assayed in duplicate, and MDCK clones in quadruplicate.
Whole-mount in situ hybridizations
Whole-mount in situ hybridization probe templates for zebrafish Pez (zfPez) and zfTGF-β (zfTGF-β3) were generated by PCR amplification from adult (1 yr) zebrafish cDNA, and cloned into pBluescript SK (+) vector (Stratagene). For zfPez, a 600-bp region of the zebrafish Pez 3′ UTR was amplified using forward primer: 5′-GTGTCCGCGGTCCAGCACCTGTCAGGATTT-3′ and reverse primer: 5′-GTGTGAATTCGATCAGTCTGGAGTTTTTCAGGA-3′, based on EST sequence BQ285767.1. For zfTGF-β3, the 1.2-kb coding region was amplified using forward primer: 5′-GTGTGAATTCATGCATTTGGGCAAAGGACTG-3′ and reverse primer: 5′-GTGTGAATTCCCGCGGTCAGCTGCACTTGCAGGATTTG-3′, Based on GenBank sequence AY614705.1. DIG-11-UTP–labeled (Roche) sense and antisense probes were generated by in vitro transcription (MAXIscript in vitro transcription kit; Ambion) according to the manufacturer's instructions.
Wild-type zebrafish embryos were collected 0–48 hpf, chorions removed, and embryos fixed in 4% formaldehyde/PBS for a minimum of 24 h. In situ hybridization was performed as described previously (Oxtoby and Jowett, 1993) but using 0.2–0.5ng/μl sense or antisense RNA probe for hybridization, 1:4,000 dilution of 0.75 U/μl alkaline phosphatase–conjugated anti-DIG antibody (Roche) at 4°C for 16–18 h for antibody binding, and 340 μg/ml 4-nitroblue tetrazolium chloride (NBT)/175μg/ml 5-bromo-4-chloro-3-indolyl-phosphate toluidine salt (BCIP) stain solution (Roche) for signal detection. Where indicated, yolk proteins were cleared with benzylbenzoate/benzyl alcohol (2:1) post-staining, as described in Schier et al. (1996) before image capture.
Antisense morpholino oligonucleotide injection of zebrafish embryos
The human Pez mRNA sequence (GenBank accession no. X82676.1) was used to blast the zebrafish EST database for homologous sequences. ESTs spanning the 5′ (GenBank accession no. AL914296) and 3′ (GenBank accession no. BQ285767.1) ends of the zebrafish Pez coding sequence were identified. Forward (5′-TCATGTGTTTGTCTTGTGGAG-3′) and reverse (5′-TGCTGGACACCGTGTATCTC-3′) PCR primers were designed to amplify a 172-bp region spanning the ATG start site. Based on sequence amplified from adult (1 yr) zebrafish cDNA, three independent antisense MOs (GeneTools) (PezMO1-3) were designed to block translation by binding upstream of or spanning the start site. Wild-type or fli1-GFP transgenic zebrafish embryos were collected and injected at one-cell stage with Pez-specific MO or an irrelevant control using standard procedures. 96-hpf embryos were fixed in 4% formaldehyde/phosphate-buffered saline (PBS) for 16–24 h and either resuspended in 80% glycerol/PBS or embedded in paraffin, and 3-μm sections were cut and stained with hemotoxylin and eosin (H&E). For live imaging, embryos were anaesthetised by the addition of 0.003% tricaine methanesulfonate to embryo medium and time-lapse fluorescent images were collected using a microscope (MVX 10; Olympus) illuminated with a mercury lamp. MO sequences were as follows: PezMO1 (nt −40 to −16): 5′-AGCTTTCCTCTGCGCTTAATCCATG-3′; PezMO2 (nt −70 to −46): 5′-CGACCTTCCTCCAACAGAACGATTC-3′; PezMO3 (nt −19 to +5): 5′-GGCATGTTGAACCCGCGCCGCGAC-3′ where +1 is the A of the ATG start site. GeneTools standard control MO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′. All zebrafish manipulations were performed according to guidelines set out and approved by the University of Adelaide Animal Ethics Committee.
The MOs were injected at 1, 0.75, and 0.5 mM, with survival rates as follows; 1 mM <20%, 0.75 mM ∼20%, 0.5 mM 70–100%. Surviving morphants injected with 0.75-mM MOs had severe defects and were therefore not analyzed further. Approximately 80% of morphants derived from injecting 0.5 mM MOs had defects of varying degrees and were analyzed further. Approximately 100 of each control MO- and Pez MO–injected embryos were analyzed.
Image acquisition and analysis
Phase-contrast images were captured using a microscope (IX70; Olympus), UPlanFl 10× NA 0.3 Ph1 and LCPlanFl 20× NA 0.4 Ph1 objectives, and a camera (DP12; Olympus). Color and fluorescent images were captured using a microscope (IX81; Olympus) with 10× UPLSApo NA 0.4, 20× UPLSApo NA 0.75, and 40× UAPO/340 NA 1.15w objectives and CC12 (Soft Imaging System, color) and Hamamatsu Orca-ER (fluorescent) cameras, respectively. Videos were generated from a microscope (MVX10; Olympus) and F-view camera (Soft Imaging System). Fluorescence images were acquired using CellR software (Olympus Soft Imaging System), all other images were acquired and processed using the Analysis software (Olympus Soft Imaging System). For immunofluorescence images where comparisons of staining intensities were made, the images were acquired using the same attenuator and exposure settings.
Online supplemental material
Two videos are included displaying cardiac morphology and pumping in live control (Video 1) and Pez morphant (Video 2) fli-1-GFP transgenic zebrafish at 53 hpf. In addition, there are three supplemental figures showing a high magnification view of Pez mRNA localization in somite of zebrafish (Fig. S1), pericardial edema in Pez morphants (Fig. S2), and time course of Snail mRNA induction by TGFβ in MDCK cells (Fig. S3), and a table summarizing the defects in Pez morphants (Table S1). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705035/DC1.
Supplementary Material
Acknowledgments
The authors thank Svanhild Nornes, Simon Wells, and Christine Hepperle (Centre for the Molecular Genetics of Development) for advice and help with zebrafish manipulations. We also thank Graham Lieschke (Ludwig Institute, Melbourne, Australia) for providing seeding embryos of fli1-GFP transgenic zebrafish. Zebrafish sections were prepared by Adelaide Microscopy and Department of Anatomical Sciences, University of Adelaide.
This work was supported by a grant from the National Health and Medical Research Council, Australia to Y. Khew-Goodall. L. Wyatt is a recipient of an Australian Postgraduate Award and a Royal Adelaide Hospital Dawes Postgraduate Top-up Scholarship, and C. Wadham was a recipient of a Royal Adelaide Hospital Dawes Postgraduate Scholarship.
The authors have no commercial affiliations or conflicts associated with this publication.
Abbreviations used in this paper: AJ, adherens junction; A-V, atrio-ventricular; EMT, epithelial–mesenchymal transition; hpf, hours post-fertilization; MO, morpholino oligonucleotide; PTP, protein tyrosine phosphatase.
References
- Armstrong, E.J., and J. Bischoff. 2004. Heart valve development: endothelial cell signaling and differentiation. Circ. Res. 95:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett, J.V., A. Moustakas, W. Lin, X.F. Wang, H.Y. Lin, J.B. Galper, and R.L. Maas. 1994. Cloning and developmental expression of the chick type II and type III TGF beta receptors. Dev. Dyn. 199:12–27. [DOI] [PubMed] [Google Scholar]
- Bascom, C.C., J.R. Wolfshohl, R.J. Coffey Jr., L. Madisen, N.R. Webb, A.R. Purchio, R. Derynck, and H.L. Moses. 1989. Complex regulation of transforming growth factor beta 1, beta 2, and beta 3 mRNA expression in mouse fibroblasts and keratinocytes by transforming growth factors beta 1 and beta 2. Mol. Cell. Biol. 9:5508–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle, E., E. Sancho, C. Franci, D. Dominguez, M. Monfar, J. Baulida, and A. Garcia de Herreros. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2:84–89. [DOI] [PubMed] [Google Scholar]
- Beis, D., T. Bartman, S.W. Jin, I.C. Scott, L.A. D'Amico, E.A. Ober, H. Verkade, J. Frantsve, H.A. Field, A. Wehman, et al. 2005. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 132:4193–4204. [DOI] [PubMed] [Google Scholar]
- Bolos, V., H. Peinado, M.A. Perez-Moreno, M.F. Fraga, M. Esteller, and A. Cano. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116:499–511. [DOI] [PubMed] [Google Scholar]
- Camenisch, T.D., D.G. Molin, A. Person, R.B. Runyan, A.C. Gittenberger-de Groot, J.A. McDonald, and S.E. Klewer. 2002. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev. Biol. 248:170–181. [DOI] [PubMed] [Google Scholar]
- Carver, E.A., R. Jiang, Y. Lan, K.F. Oram, and T. Gridley. 2001. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 21:8184–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah, F.S., E.W. Jabs, and S.S. Chong. 2005. Genomic, cDNA, and embryonic expression analysis of zebrafish transforming growth factor beta 3 (tgfbeta3). Dev. Dyn. 232:1021–1030. [DOI] [PubMed] [Google Scholar]
- Comijn, J., G.Berx, P.Vermassen, K.Verschueren, L.van Grunsven, E.Bruyneel, M.Mareel, D.Huylebroeck, and van Roy F. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7:1267–1278. [DOI] [PubMed] [Google Scholar]
- Fleisch, M.C., C.A. Maxwell, and M.H. Barcellos-Hoff. 2006. The pleiotropic roles of transforming growth factor beta in homeostasis and carcinogenesis of endocrine organs. Endocr. Relat. Cancer. 13:379–400. [DOI] [PubMed] [Google Scholar]
- Fraser, D., L. Wakefield, and A. Phillips. 2002. Independent regulation of transforming growth factor-beta1 transcription and translation by glucose and platelet-derived growth factor. Am. J. Pathol. 161:1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooteclaes, M.L., and S.M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 19:3823–3828. [DOI] [PubMed] [Google Scholar]
- Kaartinen, V., J.W. Voncken, C. Shuler, D. Warburton, D. Bu, N. Heisterkamp, and J. Groffen. 1995. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 11:415–421. [DOI] [PubMed] [Google Scholar]
- Khew-Goodall, Y., and C. Wadham. 2005. A perspective on regulation of cell-cell adhesion and epithelial-mesenchymal transition: known and novel. Cells Tissues Organs. 179:81–86. [DOI] [PubMed] [Google Scholar]
- Lawson, N.D., and B.M. Weinstein. 2002. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248:307–318. [DOI] [PubMed] [Google Scholar]
- Massague, J., and Y.G. Chen. 2000. Controlling TGF-beta signaling. Genes Dev. 14:627–644. [PubMed] [Google Scholar]
- Oxtoby, E., and T. Jowett. 1993. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21:1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, H., F. Portillo, and A. Cano. 2004. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 48:365–375. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno, M.A., A. Locascio, I. Rodrigo, G. Dhondt, F. Portillo, M.A. Nieto, and A. Cano. 2001. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 276:27424–27431. [DOI] [PubMed] [Google Scholar]
- Roberts, A.B., and L.M. Wakefield. 2003. The two faces of transforming growth factor beta in carcinogenesis. Proc. Natl. Acad. Sci. USA. 100:8621–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, L.A., and R.B. Runyan. 2000. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev. Biol. 223:91–102. [DOI] [PubMed] [Google Scholar]
- Savagner, P. 2001. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 23:912–923. [DOI] [PubMed] [Google Scholar]
- Schier, A.F., S.C. Neuhauss, M. Harvey, J. Malicki, L. Solnica-Krezel, D.Y. Stainier, F. Zwartkruis, S. Abdelilah, D.L. Stemple, Z. Rangini, et al. 1996. Mutations affecting the development of the embryonic zebrafish brain. Development. 123:165–178. [DOI] [PubMed] [Google Scholar]
- Shook, D., and R. Keller. 2003. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech. Dev. 120:1351–1383. [DOI] [PubMed] [Google Scholar]
- Siegel, P.M., W. Shu, R.D. Cardiff, W.J. Muller, and J. Massague. 2003. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc. Natl. Acad. Sci. USA. 100:8430–8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler, J.A., and M.W. Miller. 2004. Transforming growth factor beta1 modulates cell migration in rat cortex: effects of ethanol. Cereb. Cortex. 14:791–802. [DOI] [PubMed] [Google Scholar]
- Siegenthaler, J.A., and M.W. Miller. 2005. Transforming growth factor beta 1 promotes cell cycle exit through the cyclin-dependent kinase inhibitor p21 in the developing cerebral cortex. J. Neurosci. 25:8627–8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G.H. 1996. TGF-beta and functional differentiation. J. Mammary Gland Biol. Neoplasia. 1:343–352. [DOI] [PubMed] [Google Scholar]
- Thiery, J.P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2:442–454. [DOI] [PubMed] [Google Scholar]
- Timmerman, L.A., J.Grego-Bessa, A.Raya, E.Bertran, J.M.Perez-Pomares, J.Diez, S.Aranda, S.Palomo, F.McCormick, J.C.Izpisua-Belmonte, and J.L.de la Pompa. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Obberghen-Schilling, E., N.S. Roche, K.C. Flanders, M.B. Sporn, and A.B. Roberts. 1988. Transforming growth factor beta 1 positively regulates its own expression in normal and transformed cells. J. Biol. Chem. 263:7741–7746. [PubMed] [Google Scholar]
- Wadham, C., J.R. Gamble, M.A. Vadas, and Y. Khew-Goodall. 2000. Translocation of protein tyrosine phosphatase Pez/PTPD2/PTP36 to the nucleus is associated with induction of cell proliferation. J. Cell Sci. 113:3117–3123. [DOI] [PubMed] [Google Scholar]
- Wadham, C., J.R. Gamble, M.A. Vadas, and Y. Khew-Goodall. 2003. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol. Biol. Cell. 14:2520–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, E.C., and D.Y. Stainier. 2001. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 293:1670–1673. [DOI] [PubMed] [Google Scholar]
- Wang, Z., D. Shen, D.W. Parsons, A. Bardelli, J. Sager, S. Szabo, J. Ptak, N. Silliman, B.A. Peters, M.S. van der Heijden, et al. 2004. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 304:1164–1166. [DOI] [PubMed] [Google Scholar]
- Westin, J., and M. Lardelli. 1997. Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev. Genes Evol. 207:51–63. [DOI] [PubMed] [Google Scholar]
- Yang, J., S.A. Mani, J.L. Donaher, S. Ramaswamy, R.A. Itzykson, C. Come, P. Savagner, I. Gitelman, A. Richardson, and R.A. Weinberg. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939. [DOI] [PubMed] [Google Scholar]
- Yelon, D., and D.Y. Stainier. 1999. Patterning during organogenesis: genetic analysis of cardiac chamber formation. Semin. Cell Dev. Biol. 10:93–98. [DOI] [PubMed] [Google Scholar]
- Zavadil, J., and E.P. Bottinger. 2005. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.