Abstract
The sorting of transmembrane cargo proteins into the lumenal vesicles of multivesicular bodies (MVBs) depends on the recruitment of endosomal sorting complexes required for transport (ESCRTs) to the cytosolic face of endosomal membranes. The subsequent dissociation of ESCRT complexes from endosomes requires Vps4, a member of the AAA family of adenosine triphosphatases. We show that Did2 directs Vps4 activity to the dissociation of ESCRT-III but has no role in the dissociation of ESCRT-I or -II. Surprisingly, vesicle budding into the endosome lumen occurs in the absence of Did2 function even though Did2 is required for the efficient sorting of MVB cargo proteins into lumenal vesicles. This uncoupling of MVB cargo sorting and lumenal vesicle formation suggests that the Vps4-mediated dissociation of ESCRT-III is an essential step in the sorting of cargo proteins into MVB vesicles but is not a prerequisite for the budding of vesicles into the endosome lumen.
Introduction
Transmembrane proteins monoubiquitinated on their cytosolic domains are sorted into the lumenal vesicles of late endosomal multivesicular bodies (MVBs; for review see Babst, 2005). MVB vesicles and their cargoes are exposed to the hydrolytic interior of the lysosome upon fusion of the limiting endosomal membrane with the lysosomal membrane. The mechanism of MVB cargo sorting is conserved and mediated by class E Vps proteins originally identified in Saccharomyces cerevisiae. Most class E VPS genes encode soluble cytosolic proteins recruited transiently to endosomes. Genetic and biochemical data suggest a sequence that begins with recruitment of the Vps27–Hse1 complex, which recognizes monoubiquitinated cargoes, followed by recruitment of three distinct endosomal sorting complexes required for transport (ESCRTs; for review see Hurley and Emr, 2006). Like the Vps27–Hse1 complex, ESCRT-I and -II bind monoubiquitinated cargoes, whereas ESCRT-III lacks ubiquitin-binding subunits and functions downstream of cargo recognition.
ESCRT-III is comprised of the Vps20–Snf7 and Vps2–Vps24 subcomplexes (Babst et al., 2002a). Although its molecular function is not fully understood, one role for ESCRT-III is the recruitment of late-acting components of the sorting machinery. Snf7 recruits Bro1 (Kim et al., 2005), and Bro1 recruits Doa4, which deubiquitinates cargoes before their enclosure within MVB vesicles (Luhtala and Odorizzi, 2004). Vps4 is an ATPase that catalyzes the dissociation of class E Vps proteins from endosomal membranes, and, in the absence of Vps4 activity, ESCRT complexes accumulate on endosomes (Katzmann et al., 2001; Babst et al., 2002a,b).
A central question of Vps4 function concerns how its activity is coordinated to dissociate multiple protein complexes. We report that Did2, a protein related to ESCRT-III subunits (Amerik et al., 2000), directs Vps4 activity to the dissociation of ESCRT-III. In the absence of Did2, ESCRT-I and -II dissociation occurs, whereas ESCRT-III and downstream components accumulate on endosomes. Surprisingly, MVB vesicle budding proceeds in the absence of Did2 despite the requirement for Did2 in sorting cargoes, demonstrating that vesicle formation and MVB cargo sorting can be uncoupled.
Results and discussion
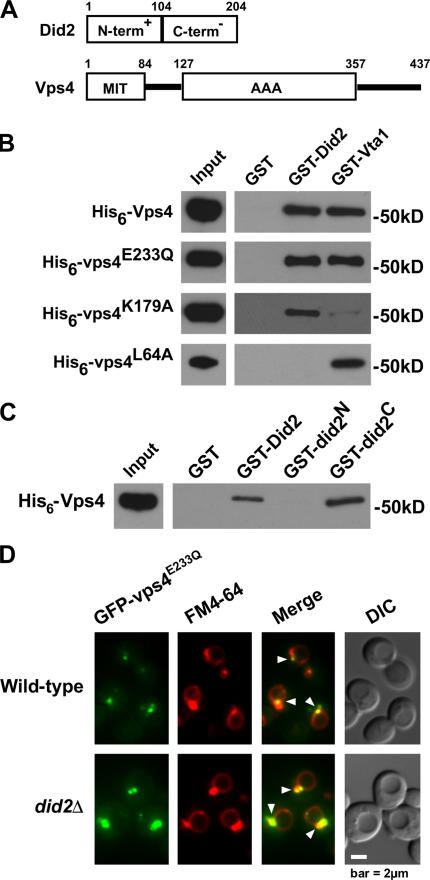
The C terminus of Did2 binds the MIT domain of Vps4
The N terminus of Did2 is predominantly comprised of basic amino acids, whereas its C terminus predominantly contains acidic residues (Fig. 1 A). As shown in Fig. 1 B, bacterially expressed His6-Vps4 copurified with GST-Did2 but not GST alone. This interaction occurred regardless of whether Vps4 was locked in its ATP-bound state (His6-Vps4E233Q) or was disabled from binding ATP (His6-Vps4K179A; Fig. 1 B). In contrast, GST-Vta1 showed a strong preference for binding His6-Vps4E233Q (Fig. 1 B), which is consistent with Vta1 interacting with ATP-bound Vps4 to stimulate its oligomerization (Azmi et al., 2006).
Figure 1.
The C terminus of Did2 binds Vps4. (A) Domain maps of Did2 and Vps4. (B and C) Western blots of in vitro glutathione-Sepharose pull downs of wild-type or mutant His6-Vps4 mixed with GST-Did2 (B) or wild-type His6-Vps4 mixed with wild-type or mutant GST-Did2 (C). (D) Fluorescence and DIC microscopy of FM 4-64–stained cells expressing GFP-Vps4E233Q. Arrowheads indicate endosomal membranes.
Studies of CHMP1b and Vps4a, the mammalian orthologues of Did2 and Vps4, respectively, demonstrated that the C terminus of CHMP1b binds Vps4a and that this interaction is disrupted by the mutation of leucine-64 in the microtubule interaction and trafficking (MIT) domain of Vps4a (Scott et al., 2005). This leucine is conserved in the MIT domain of yeast Vps4 (Fig. 1 A), suggesting that it is important for the interaction between Did2 and Vps4. Indeed, His6-Vps4L64A failed to bind GST-Did2 but still bound GST-Vta1 (Fig. 1 B), which is in agreement with Vta1 binding the AAA domain rather than the MIT domain of Vps4 (Yeo et al., 2003). We further observed that His6-Vps4 interacted with the C terminus of Did2 (GST-Did2104–204) but not its N terminus (GST-Did21–103; Fig. 1 C). Thus, the binding mechanism between Vps4 and Did2 appears conserved.
Because the MIT domain of Vps4 is essential for its localization to endosomes (Babst et al., 1998), we addressed whether its binding to Did2 mediates the endosomal recruitment of Vps4. Locked in the ATP-bound state, Vps4E233Q is unable to catalyze the dissociation of itself and its substrate proteins from endosomes (Babst et al., 1998). Thus, GFP fused to Vps4E233Q appeared concentrated at class E compartments stained with the lipophilic dye FM 4-64 (Fig. 1 D, arrowheads). GFP-Vps4E233Q also localized to endosomes in did2Δ cells (Fig. 1 D, arrowheads), indicating that the recruitment of Vps4 does not require Did2. Localization of Vps4E233Q to the endosomal membrane in the absence of Did2 was also observed by subcellular fractionation (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1), which was not surprising given that Vps4 binds multiple distinct ESCRT-III components. Indeed, the deletion of DID2 did not affect the ability of GST-Vps4 to pull down Snf7 from yeast lysates (Fig. S1 B), which is consistent with our observation that GST-Vps4 interacts directly with His6-Snf7 (Fig. S1 C) and with a previous study showing that Vps4 interacts directly with Vps20 (Yeo et al., 2003).
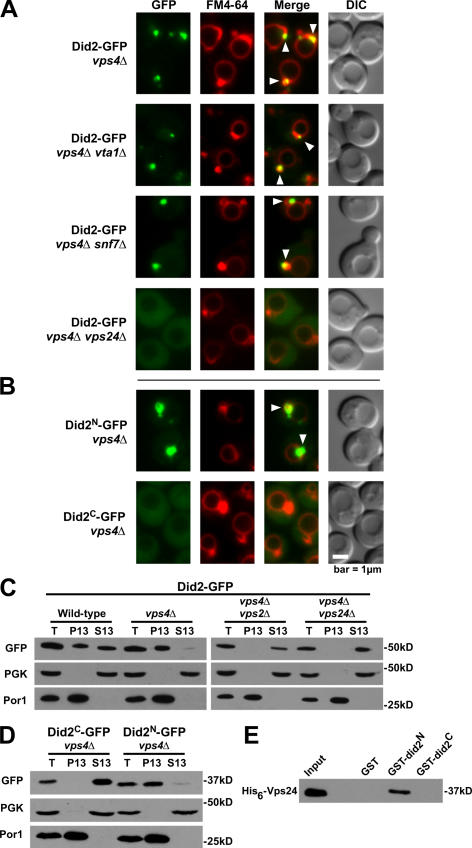
The Vps2–Vps24 subcomplex of ESCRT-III recruits Did2 to endosomes
Did2 binds Vta1 and is required for the interaction of Vta1 with the ESCRT-III component Snf7 (Lottridge et al., 2006). Therefore, we examined whether Vta1 or ESCRT-III proteins mediate the recruitment of Did2 to endosomes. Genetic and biochemical studies suggest that ESCRT-III consists of a Snf7–Vps20 subcomplex and a Vps2–Vps24 subcomplex (Babst et al., 2002a). Did2 accumulated at the class E compartment in vps4Δ cells regardless of whether VTA1, SNF7 (Fig. 2 A), or VPS20 (not depicted) had been deleted. In contrast, the deletion of either VPS24 (Fig. 2 A) or VPS2 (not depicted) caused Did2-GFP to remain cytosolic. Similarly, subcellular fractionation showed that Did2-GFP was concentrated in the membrane fraction in vps4Δ cell extracts but was soluble both in vps4Δ vps2Δ and vps4Δ vps24Δ extracts (Fig. 2 B).
Figure 2.
The N terminus of Did2 requires the Vps2–Vps24 subcomplex for recruitment to endosomes. (A and B) Fluorescence and DIC microscopy of FM 4-64–stained cells expressing GFP-tagged wild-type (A) or mutant (B) Did2 proteins. Arrowheads indicate endosomal membranes. (C and D) Subcellular fractionation and Western blot analysis of cells expressing GFP-tagged wild-type (C) or mutant (D) Did2 proteins. Cell lysates (T, total) were separated into membrane-associated pellet (P13) and soluble cytosolic (S13) fractions. PGK, 3-phosphoglycerate kinase. (E) In vitro glutathione-Sepharose pull downs of His6-Vps24 with GST-tagged wild-type or mutant Did2 proteins.
Fluorescence microscopy (Fig. 2 A) and subcellular fractionation (Fig. 2 B) indicated that the N terminus of Did2 is necessary and sufficient for endosomal localization. Therefore, the simplest explanation for the mislocalization of Did2 to the cytosol in vps4Δ cells lacking either Vps2 or Vps24 is that the N terminus of Did2 interacts directly with the Vps2–Vps24 subcomplex. Indeed, recombinant His6-Vps24 bound GST-Did21–103 but not GST-Did2104–204 or GST alone (Fig. 2 C). In contrast, recombinant His6-Snf7 failed to bind GST-Did2 (unpublished data).
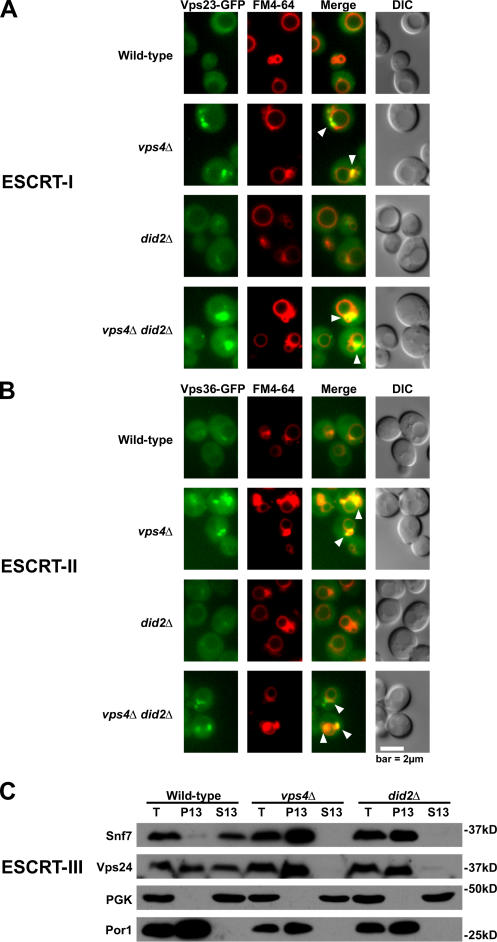
Endosomal dissociation of ESCRT-III requires Did2
To address the functional significance of the interaction between Did2 and Vps4, we examined the ability of Vps4 to mediate the dissociation of ESCRT complexes in the absence of Did2. As shown previously (Katzmann et al., 2001; Babst et al., 2002b), Vps23-GFP of ESCRT-I (Fig. 3 A) and Vps36-GFP of ESCRT-II (Fig. 3 B) in wild-type cells were predominantly cytosolic in addition to being localized weakly at punctate structures. As expected, Vps23- and Vps36-GFP in vps4Δ cells accumulated at class E compartments (Fig. 3, A and B; arrowheads). However, the distributions of both proteins in did2Δ cells appeared to be similar to their distributions in wild-type cells (Fig. 3, A and B). Thus, Did2 is not required for the dissociation of either ESCRT-I or -II. Moreover, Did2 has no role in the endosomal recruitment of ESCRT-I and -II because both Vps23- (Fig. 3 A) and Vps36-GFP (Fig. 3 B) accumulated at class E compartments in vps4Δ did2Δ cells.
Figure 3.
Did2 is required for the endosomal dissociation of ESCRT-III. (A and B) Fluorescence and DIC microscopy of FM 4-64–stained cells expressing Vps23- (A) or Vps36-GFP (B). Arrowheads indicate class E compartments. (C) Subcellular fractionation and Western blot analysis of endogenous Snf7 and Vps24. T, total; PGK, 3-phosphoglycerate kinase.
Fusion of GFP to ESCRT-III proteins disrupts their function in MVB sorting (unpublished data). Thus, we assessed the distributions of endogenous Snf7 and Vps24 by subcellular fractionation and Western blotting. As shown previously (Babst et al., 1998), Snf7 was predominantly soluble, and Vps24 was evenly distributed between membrane and soluble fractions in wild-type cells, whereas both proteins shifted entirely to the membrane pellet in vps4Δ cells (Fig. 3 C). Snf7 and Vps24 were similarly concentrated in the pellet fraction of did2Δ cells (Fig. 3 C), indicating that Did2 is essential for the membrane dissociation of both proteins. Likewise, Did2 was required for the endosomal dissociation of Bro1 and Doa4 (Table I), components that function downstream in the MVB pathway but depend on ESCRT-III for recruitment to endosomes (Luhtala and Odorizzi, 2004; Kim et al., 2005). The ability of Bro1 and Doa4 to dissociate from endosomes may require Did2 to coordinate the Vps4-mediated dissociation of ESCRT-III. Although Vta1 binds Did2 and requires Vps4 to dissociate from endosomes (Shiflett et al., 2004), Vta1 appeared predominantly cytosolic in the absence of Did2 (Table I), indicating that it does not need Did2 for dissociation. This quality makes Vta1 unique among ESCRT-III–associated Vps4 substrates acting late in the MVB pathway.
Table I.
Localization of Vps4 substrates in wild-type, vps4Δ, and did2Δ mutant cells
| Substrate | Complex | Wild type | vps4Δ | did2Δ |
|---|---|---|---|---|
| Vps23-GFP | ESCRT-I | Cytosolica | Punctateb | Cytosolica |
| Vps36-GFP | ESCRT-II | Cytosolica | Punctateb | Cytosolica |
| Snf7 | ESCRT-III | Solublec | Membraned | Membraned |
| Vps24 | ESCRT-III | Solublec | Membraned | Membraned |
| Bro1-GFP | NA | Cytosolica | Punctateb | Punctateb |
| Doa4-GFP | NA | Cytosolica | Punctateb | Punctateb |
| Vta1-GFP | With Vps60 | Cytosolica | Punctateb | Cytosolica |
NA, not applicable.
Predominantly cytosolic GFP signal.
Punctate GFP signal adjacent to the vacuole with a reduction in cytosolic signal.
Soluble as determined by subcellular fractionation.
Membrane bound as determined by subcellular fractionation.
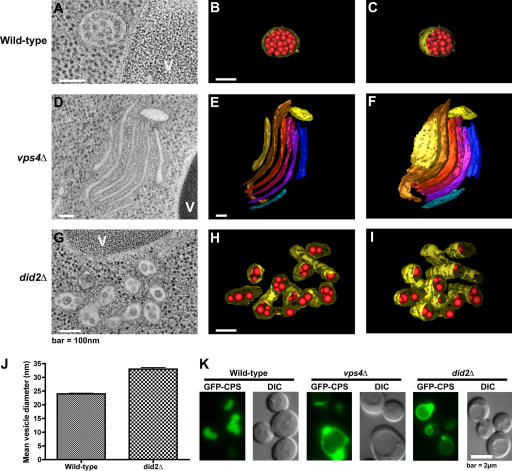
MVB vesicle budding in the absence of ESCRT-III dissociation
Class E compartments stained by FM 4-64 are a hallmark phenotype caused by mutations in VPS4 and other class E VPS genes. By EM, these abnormal late endosomes appear as flattened stacks of cisterna-like structures devoid of lumenal vesicles (Rieder et al., 1996; Odorizzi et al., 1998). We examined did2Δ cells versus wild-type and vps4Δ cells using high resolution EM and tomographic modeling. An example of a typical wild-type MVB is shown in the tomogram in Fig. 4 A and is modeled in Fig. 4 (B and C; and see Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1). The limiting membrane of this MVB is approximately spherical and surrounds numerous lumenal vesicles. As expected, no multivesicular endosomes were detected in vps4Δ cells, which instead contained class E compartments similar to the structures described previously in cells lacking Vps4 function (Odorizzi et al., 1998). An example of a class E compartment in vps4Δ cells is shown in the tomogram in Fig. 4 D and is modeled in Fig. 4 (E and F; and see Videos 3 and 4). Three-dimensional analysis indicated that its elongated cisternae-like elements did not connect with one another. Similar characteristics were observed in serial sections that included entire class E compartments (unpublished data), and lumenal vesicles were not observed in >300 class E compartments of vps4Δ cells examined by EM (including three structures modeled by tomography), which is consistent with an essential role for Vps4 function in the biogenesis of MVB vesicles.
Figure 4.
Tomographic analysis of endosome morphology. (A–I) Two-dimensional cross sections and three-dimensional models derived from 200-nm–thick section tomograms. V, vacuole. Models depict a wild-type MVB (B and C), a vps4Δ class E compartment (E and F), and a did2Δ VTE (H and I). Models in C, F, and I are rotated 25° relative to the models in B, E, and H. In MVB and VTE models, the endosomal limiting membrane is yellow, and lumenal vesicles are red. Cisternae in the vps4Δ class E compartment model are depicted in various colors to easily discriminate individual membrane structures. (J) Vesicle diameters in wild-type and did2Δ cells (n = 284 and 175, respectively). Mean values ± SEM (error bars; unpaired t test; P < 0.0001). (K) Fluorescence and DIC microscopy of cells expressing GFP-CPS.
Surprisingly, multivesicular endosomes were readily apparent in did2Δ cells, an example of which is shown in the tomogram in Fig. 4 G and modeled in Fig. 4 (H and I; and Videos 5 and 6, available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1). The limiting membrane, rather than being spherical as seen in wild-type cells, was typically elongated, which is similar to the cisternae-like elements of class E compartments. In >200 sections examined by EM, these vesicular tubular endosomes (VTEs) were most often observed crowded together, which is reminiscent of the compact organization displayed by class E compartments in vps4Δ cells (Fig. 4, D–F). Three-dimensional analysis of tomograms and serial sections that encompassed entire VTEs (unpublished data) indicated that the lumenal vesicles were not interconnected, nor were they connected to the limiting membrane. The interior of lumenal vesicles in VTEs of did2Δ cells appeared electron dense and were uniformly larger (by 38%) than lumenal vesicles of MVBs in wild-type cells (P < 0.0001; 23.98 ± 0.23 vs. 33.01 ± 0.56 nm, respectively; Fig. 4 J), raising the possibility that ESCRT-III, which is unable to dissociate from the membrane, is mistakenly packaged as cargo. However, ESCRT-III was only detected at the limiting membrane of VTEs in thin sections of did2Δ cells examined by immunogold labeling using antibodies against Vps24 (Fig. S2 D, available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1).
The similarity of the class E compartment and VTE when viewed by fluorescence microscopy (Fig. 1 D) underscores the need for EM when reaching any conclusion regarding endosome morphology. Moreover, the ultrastructural differences between class E compartments and VTEs suggest that the class E phenotype warrants subdivision based on endosome morphology. The absence of lumenal vesicles in class E compartments as a result of the loss of function of ESCRTs, Vps4, or Bro1 has been thought to signify that these components comprise the core class E Vps machinery required for vesicle budding (for review see Babst, 2005). However, the VTEs observed in did2Δ cells contradict the view that the dynamic cycling of ESCRT-III, a subset of this core machinery, is either a pre- or corequisite for MVB vesicle formation, although it remains likely that the assembly of ESCRT-III on endosomes is critical to the budding event.
Like Vps4, Did2 is required for efficient sorting of MVB cargoes, as indicated by the failure of GFP–carboxypeptidase S (CPS), a biosynthetic protein, to be sorted into the vacuole lumen in did2Δ cells (Fig. 4 K). Sna3-GFP (Fig. S2 A), another biosynthetic protein, as well as Ste3-GFP (Fig. S2 B), an endocytic protein, are also mislocalized, demonstrating that the loss of Did2 function causes a broad cargo-sorting defect. Intriguingly, the sorting of GFP-CPS in did2Δ cells was partially rescued upon in-frame fusion of ubiquitin to its cytosolic domain (Fig. S3 C, available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1). However, the mislocalization of Sna3- GFP suggests that the molecular basis for the cargo-sorting defect in did2Δ cells is not directly related to ubiquitination because Sna3 does not require ubiquitin to be sorted into the MVB pathway (Reggiori and Pelham, 2001). The ubiquitin-independent localization of Sna3-GFP to the vacuole lumen indicates, albeit indirectly, that cargo sorting can be uncoupled from lumenal vesicle formation in yeast. Indeed, MVB vesicles are observed in cells lacking functional Doa4 or Rsp5, the primary E3 ubiquitin ligase for MVB cargoes in yeast (unpublished data). Similarly, MVB vesicles are observed by EM despite deletion of the FAB1 gene (unpublished data), which blocks MVB sorting of CPS but not Ste2, an endocytic cargo protein (Odorizzi et al., 1998). In mammalian cells, the overexpression of a mutant form of Hrs that is defective in ubiquitin binding has no effect on MVB vesicle formation but reduces the efficiency of cargo sorting, perhaps because of a failure in the concentration of cargoes at the site of vesicle budding (Urbé et al., 2003). Although the nature of the sorting defect in did2Δ is not clear, it might be caused by the trapping of cargoes within an ESCRT-III network that is unable to release from endosomes, in which case the in-frame fusion of ubiquitin could promote sorting by enhancing cargo interactions with ESCRT-I and -II to the exclusion of ESCRT-III.
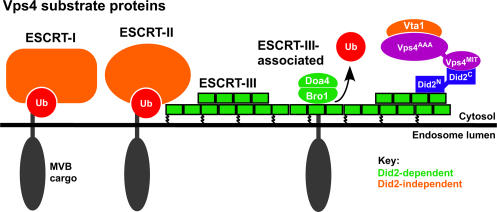
Our findings suggest that Did2 functions to coordinate Vps4 activity to ESCRT-III dissociation (Fig. 5). The C terminus of Did2 binds the MIT domain of Vps4, whereas the N terminus of Did2 binds Vps24 of ESCRT-III. Did2 has a position downstream of the Vps2–Vps24 subcomplex in order of assembly because Vps24 can be recruited to the membrane in the absence of Did2 but not vice versa. The significance of Did2 recruitment is that Vps4 requires Did2 to catalyze the endosomal dissociation of ESCRT-III as well as factors that function downstream. Therefore, this set of Vps4 substrates is Did2 dependent, which is in contrast with ESCRT-I and -II, which are Did2 independent (Fig. 5). The selective role Did2 plays in coordinating Vps4 with ESCRT-III dissociation implies that additional factors couple Vps4 function to the dissociation of ESCRT-I and -II.
Figure 5.
Model of Did2 function in MVB sorting. The Did2 C terminus interacts with Vps4, and its N terminus interacts with the ESCRT-III subunit Vps24. Did2 is required for the endosomal dissociation of ESCRT-III and downstream components, which are Did2-dependent Vps4 substrates (green). ESCRT-I and -II (orange) are Did2-independent Vps4 substrates.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1). Yeast manipulations were performed using standard protocols. Gene deletions and introduction of epitopes in yeast were constructed by homologous recombination of PCR products (Longtine et al., 1998). Genes PCR amplified from genomic DNA were TOPO cloned into pCR2.1 (Invitrogen) and subcloned using T4 DNA ligase into an expression vector.
Protein binding studies
BL21(DE3) cells transformed with pGEX4T1 or pET-His PL plasmids were grown at 37°C to logarithmic phase and were induced to express recombinant genes by the addition of 0.5 mM isopropyl-β-d-thiogalactoside. Cells were harvested 1–2 h later, lysed under native conditions, and cleared of cell debris. GST-tagged proteins were purified using GSTrap FF columns (GE Healthcare), and His6-tagged proteins were purified using Ni2+-agarose beads (QIAGEN). ∼1.2 μg of purified recombinant proteins were tested for stable interactions using glutathione–Sepharose beads (GE Healthcare) essentially as described previously (Yeo et al., 2003).
Subcellular fractionation
Fractionation of proteins into membrane-associated pellet and soluble cytosolic fractions was performed as described previously (Luhtala and Odorizzi, 2004). One-half of OD600 unit equivalent of each sample was resolved by SDS-PAGE and analyzed by Western blotting. Yeast 3-phosphoglycerate kinase and the mitochondrial porin (Por1) were examined as soluble and membrane-bound controls, respectively.
Fluorescence microscopy
Strains were grown to logarithmic phase at 30°C in synthetic medium before observation at room temperature in synthetic medium at 100× using a microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) equipped with an NA 1.40 oil immersion objective (Carl Zeiss MicroImaging, Inc.). Differential interference contrast (DIC) and fluorescence microscopy images were acquired with a digital camera (Cooke SensiCam; Applied Scientific Instruments) and processed using Slidebook (Intelligent Imaging Innovations) and Photoshop 7.0 software (Adobe). GFP-CPS was introduced by transforming cells with pGO45 (Odorizzi et al., 1998). Pulse-chase staining of cells with FM 4-64 has been described previously (Luhtala and Odorizzi, 2004).
Electron tomography and three-dimensional modeling
Cells were high-pressure frozen, freeze substituted with 0.1% uranyl acetate, 0.25% glutaraldehyde, and anhydrous acetone at −90°C, embedded in Lowicryl HM20, and polymerized under UV light at −50°C (Winey et al., 1995). 200-nm semithick sections were placed on Rhodium-plated Formvar-coated copper slot grids and mapped on an electron microscope (CM10 TEM; Phillips) at 80 kV. Dual tilt series images were collected from 60 to −60° with 1° increments at 200 kV using an electron microscope (Tecnai 20 FEG; FEI). Tomograms were imaged at 29,000× with a 0.77-nm pixel (binning 2). Sections were coated on both sides with 15-nm fiducial gold for the reconstruction of back projections using IMOD software (Kremer et al., 1996). 3dmod software was used for mapping structure surface areas. Mean z-scale values for wild-type and did2Δ sections were within 3%. Best fit sphere models were used to measure vesicle diameters to the outer leaflet of membrane bilayers. IMOD calculated limiting membrane surface areas using three-dimensional mesh structures derived from closed contours that were drawn each 3.85 nm using imodmesh software.
Online supplemental material
Table SI describes strains and plasmids used in this study. Fig. S1 shows that Did2 is not required for Vps4 to interact with ESCRT-III. Fig. S2 shows MVB cargo localization in did2Δ. Videos 1–6 depict the tomograms and three-dimensional models of wild-type, did2Δ, and vps4Δ endosomes shown in Fig. 4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200606113/DC1.
Supplementary Material
Acknowledgments
We thank Caitlin White-Root for constructing plasmids and the Boulder three-dimensional laboratory for tomography aid.
This work was funded by National Institutes of Health (NIH) grant GM065505. D.P. Nickerson is supported by NIH Training Grant GM07135, and G. Odorizzi is an Arnold and Mabel Beckman Foundation Young Investigator.
Abbreviations used in this paper: CPS, carboxypeptidase S; DIC, differential interference contrast; ESCRT, endosomal sorting complex required for transport; MIT, microtubule interaction and trafficking; MVB, multivesicular body; VTE, vesicular tubular endosome.
References
- Amerik, A.Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 11:3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi, I., B. Davies, C. Dimaano, J. Payne, D. Eckert, M. Babst, and D.J. Katzmann. 2006. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M. 2005. A protein's final ESCRT. Traffic. 6:2–9. [DOI] [PubMed] [Google Scholar]
- Babst, M., B. Wendland, E.J. Estepa, and S.D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, E.J. Estepa-Sabal, T. Meerloo, and S.D. Emr. 2002. a. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 3:271–282. [DOI] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, W.B. Snyder, B. Wendland, and S.D. Emr. 2002. b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., and S.D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., M. Babst, and S.D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. [DOI] [PubMed] [Google Scholar]
- Kim, J., S. Sitaraman, A. Hierro, B.M. Beach, G. Odorizzi, and J.H. Hurley. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 8:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J.R., D.N. Mastronarde, and J.R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Lottridge, J.M., A.R. Flannery, J.L. Vincelli, and T.H. Stevens. 2006. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA. 103:6202–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala, N., and G. Odorizzi. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., M. Babst, and S.D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., and H.R.B. Pelham. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, S.E., L.M. Banta, K. Köhrer, J.M. McCaffery, and S.D. Emr. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 7:985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A., J. Gaspar, M.D. Stuchell-Brereton, S.L. Alam, J.J. Skalicky, and W.I. Sundquist. 2005. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA. 102:13813–13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett, S.L., D.M. Ward, D. Huynh, M.B. Vaughn, J.C. Simmons, and J. Kaplan. 2004. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J. Biol. Chem. 279:10982–10990. [DOI] [PubMed] [Google Scholar]
- Urbé, S., M. Sachse, P.E. Row, C. Preisinger, F.A. Barr, G. Strous, J. Klumperman, and M.J. Clague. 2003. The UIM domain of Hrs couples receptor sorting to vesicle formation. J. Cell Sci. 116:4169–4179. [DOI] [PubMed] [Google Scholar]
- Winey, M., C.L. Mamay, E.T. O'Toole, D.M. Mastronarde, T.H. Giddings, K.L. McDonald, and J.R. McIntosh. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, S.C.L., L. Xu, J. Ren, V.J. Boulton, M.D. Wagle, C. Liu, G. Ren, P. Wong, R. Zahn, P. Sasajala, et al. 2003. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 116:3957–3970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.