Abstract
The endosomal sorting complex required for transport (ESCRT) complexes play a critical role in receptor down-regulation and retroviral budding. Although the crystal structures of two ESCRT complexes have been determined, the molecular mechanisms underlying the assembly and regulation of the ESCRT machinery are still poorly understood. We identify a new component of the ESCRT-I complex, multivesicular body sorting factor of 12 kD (Mvb12), and demonstrate that Mvb12 binds to the coiled-coil domain of the ESCRT-I subunit vacuolar protein sorting 23 (Vps23). We show that ESCRT-I adopts an oligomeric state in the cytosol, the formation of which requires the coiled-coil domain of Vps23, as well as Mvb12. Loss of Mvb12 results in the disassembly of the ESCRT-I oligomer and the formation of a stable complex of ESCRT-I and -II in the cytosol. We propose that Mvb12 stabilizes ESCRT-I in an oligomeric, inactive state in the cytosol to ensure that the ordered recruitment and assembly of ESCRT-I and -II is spatially and temporally restricted to the surface of the endosome after activation of the MVB sorting reaction.
Introduction
The multivesicular body (MVB) sorting pathway provides a mechanism for the lysosomal degradation of transmembrane proteins and plays a critical role in a diverse range of processes, including growth factor receptor down-regulation (Futter et al., 1996), antigen presentation (Kleijmeer et al., 2001), developmental signaling (Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001), and the budding of enveloped viruses (Garrus et al., 2001). The proteins that constitute the MVB sorting machinery were identified by a genetic screen in yeast for mutants that missort an MVB cargo (Odorizzi et al., 2003). Most of the mutants isolated were class E vacuolar protein sorting (vps) mutants, which accumulate enlarged endosomes and exhibit defects in the formation of MVB vesicles. Further characterization of the class E Vps proteins led to the identification of three high molecular weight cytoplasmic complexes that function in MVB sorting, the endosomal sorting complex required for transport (ESCRT) complexes I, II, and III (Katzmann et al., 2001; Babst et al., 2002a,b).
The ESCRT-I complex (Vps23, -28, and -37) is recruited to endosomes by Vps27, which interacts with ubiquitinated cargo and initiates the MVB sorting reaction (Katzmann et al., 2001). ESCRT-I also interacts with ubiquitinated cargo via the UEV domain of Vps23 (Katzmann et al., 2001). Genetic studies indicate that ESCRT-II (Vps36, -22, and -25) functions downstream of ESCRT-I (Babst et al., 2002b). ESCRT-II interacts with ubiquitinated cargo via the NZF domain of Vps36, and with phosphatidylinositol 3-phosphate (PtdIns3P) via the GRAM-like ubiquitin binding in EAP45 domain (Alam et al., 2004; Teo et al., 2006). ESCRT-II then recruits the ESCRT-III subunits (Snf7, Vps20, -2, and -24) to the endosome, where they oligomerize to form the ESCRT-III complex (Teo et al., 2004; Yorikawa et al., 2005). ESCRT-III, in turn, recruits accessory factors such as Bro1 (Kim et al., 2005), which, in turn, recruits Doa4 (Luhtala and Odorizzi, 2004), the deubiquitinating enzyme that removes ubiquitin from MVB cargo before their sorting into MVB vesicles. ESCRT-III also recruits the AAA-type ATPase Vps4, which catalyzes the disassembly of the ESCRT machinery and recycles the ESCRT complexes into the cytosol to allow further rounds of cargo sorting (Babst et al., 1998; Scott et al., 2005).
Recent studies on the architecture of the ESCRT machinery have enhanced our understanding of how the ESCRT complexes assemble and interact with ubiquitinated cargo and phosphoinositides (Hurley and Emr, 2006). The structures of the core complexes of yeast ESCRT-I and -II have been determined (Hierro et al., 2004; Teo et al., 2004; Kostelansky et al., 2006; Teo et al., 2006). The interaction between ESCRT-I and -II has been mapped to the C-terminal domain of Vps28 and the NZF-1 domain of Vps36 (Teo et al., 2006). Even though it has been shown in vitro that ESCRT-I and -II can form a stable complex in solution, no such complex has yet been detected in cytosolic extracts from yeast cells, suggesting that this interaction is tightly regulated. We report the identification of a new component of the ESCRT-I complex, multivesicular body sorting factor of 12 kD (Mvb12). We show that Mvb12 plays a role in assembling ESCRT-I into an oligomeric complex in the cytosol. In doing so, Mvb12 prevents premature assembly of ESCRT-I and -II to ensure their ordered and sequential recruitment onto the endosomal membrane during MVB sorting.
Results
Identification of Mvb12 as a new component of ESCRT-I
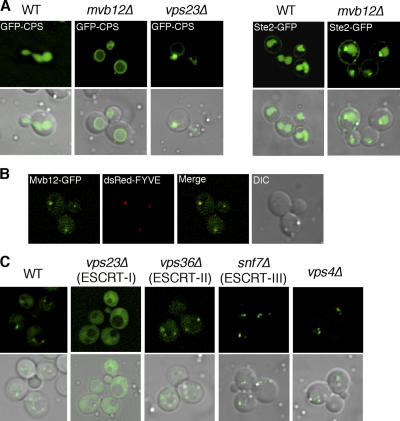
To identify novel regulators of the ESCRT machinery, we searched the Saccharomyces Genome Database for ORFs that show endosomal localization. We found eight uncharacterized ORFs and then tested to see if any of these ORFs are required for MVB sorting by examining the localization of a biosynthetic MVB cargo, the vacuolar hydrolase carboxypeptidase S (CPS), in deletion mutants lacking each of these ORFs. In wild-type cells, GFP-CPS is sorted into the MVB pathway and accumulates in the lumen of the vacuole (Fig. 1 A). In contrast, GFP-CPS accumulates on enlarged endosomes, as well as on the vacuolar-limiting membrane in ESCRT deletion mutants such as vps23Δ (Fig. 1 A). Interestingly, deletion of one of the uncharacterized ORFs, YGR206W, results in a defect in MVB sorting, and GFP-CPS is missorted to the limiting membrane of the vacuole (Fig. 1 A). We also examined the localization of the endocytic cargo Ste2, the yeast pheromone receptor, in the ygr206wΔ mutant. In wild-type cells, Ste2-GFP is endocytosed from the plasma membrane and sorted into the lumen of the vacuole. Sorting of Ste2-GFP is partially impaired in the ygr206wΔ mutant, as Ste2-GFP localized to both perivacuolar puncta and the vacuolar lumen (Fig. 1 A). The partial defect in MVB sorting, as well as the lack of observable enlarged endosomes, suggests that the Ygr206w protein is likely not a core component of the ESCRT machinery, but instead functions as a regulator of the ESCRT complexes. Sequence analysis indicates that YGR206W encodes a small protein with no well-characterized domains or motifs. We confirmed the predicted molecular weight and named the protein Mvb12. Mvb12 shares partial sequence homology with a set of genes in fungi and mammalian genomes, although the homology with mammalian genes is weak (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200608053/DC1).
Figure 1.
Mvb12 is a novel protein in the ESCRT pathway. (A) Sorting of GFP-CPS and Ste2-GFP in WT (SEY6210) and mvb12Δ (TCY246). (B) Yeast strain expressing Mvb12-GFP was transformed with dsRed-FYVE, and partial colocalization is observed. (C) Localization of Mvb12-GFP in WT, vps23Δ, vps36Δ, snf7Δ, and vps4Δ strains. Mvb12-GFP endosomal localization depends on Vps23 (ESCRT-I), but not on Vps36 (ESCRT-II), Snf7 (ESCRT-III), or Vps4.
To confirm the endosomal localization of Mvb12, we constructed a C-terminal GFP fusion of Mvb12 under the control of its endogenous promoter. Mvb12-GFP localized to the cytoplasm, as well as to perivacuolar puncta, many of which colocalized with dsRed-FYVE (EEA1), which is an endosomal marker (Fig. 1 B). The endosomal association of Mvb12 is likely transient and peripheral because the majority of Mvb12 was recovered from the cytosolic fraction in subcellular fractionation experiments, similar to the fractionation profile of ESCRT-I (unpublished data). We then tested the localization of Mvb12-GFP in different ESCRT deletion mutants. Interestingly, Mvb12-GFP failed to localize to endosomes and, instead, completely redistributed to the cytoplasm in the Vps23 (ESCRT-I) deletion mutant (Fig. 1 C). However, the endosomal localization of Mvb12-GFP is not dependent on downstream components of the MVB sorting machinery, including Vps36 (ESCRT-II), Snf7 (ESCRT-III), or Vps4 (Fig. 1 C). Collectively, these data indicate that the localization of Mvb12 to endosomes is dependent on ESCRT-I and suggest that Mvb12 functions either in a complex with or downstream from ESCRT-I in the MVB sorting pathway.
We next tested if Mvb12 is in a complex with ESCRT-I. By gel filtration chromatography, Mvb12 cofractionated with Vps23, suggesting that Mvb12 is likely a new component of ESCRT-I (Fig. 2 A). To confirm this, we performed a coimmunoprecipitation experiment in which we immunoprecipitated Flag-tagged Mvb12, and then probed for Vps23, which is a component of ESCRT-I (Fig. 2 B). Mvb12 coimmunoprecipitated with Vps23, indicating that Mvb12 is a new component of ESCRT-I. These data are consistent with the recent genome-wide analyses of protein–protein interactions in the yeast proteome that identified the Ygr206w protein as a new component of the ESCRT-I complex (Gavin et al., 2006; Krogan et al., 2006).
Figure 2.
Mvb12 is a new component of the ESCRT-I complex. (A) Gel filtration analysis of cell extracts from yeast strain expressing Mvb12-Flag (TCY257). Column fractions were analyzed by Western blotting using antibodies specific for Vps23 or Flag. Mvb12-Flag cofractionates with Vps23. (B) Cell extracts from WT (SEY6210) or strain expressing Mvb12-Flag (TCY257) were subjected to immunoprecipitation using antibody specific for Flag. Immunoprecipitates were analyzed by Western blotting using antibodies against Vps23 or Flag. Mvb12-Flag coimmunoprecipitates with Vps23. The asterisk highlights a nonspecific band.
Mvb12 negatively regulates the interaction between ESCRT-I and -II
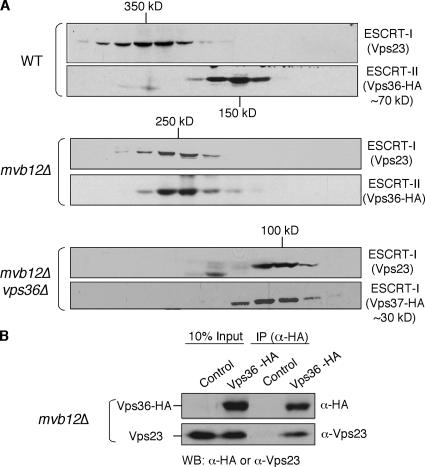
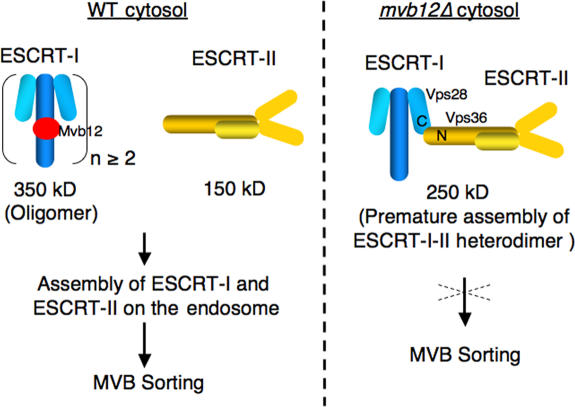
To determine the effect of deleting MVB12 on the size and stability of the ESCRT-I complex, we analyzed the elution profile of Vps23 by gel filtration. Deletion of MVB12 did not appear to affect the stability of Vps23, but did result in a change in the elution profile of Vps23, reducing its apparent molecular weight from ∼350 kD in wild-type cells to ∼250 kD in mvb12Δ cells (Fig. 3 A). Unexpectedly, the elution profile of Vps36, which is a subunit of ESCRT-II, shifted from 150 kD in wild-type cells to ∼250 kD in mvb12Δ cells (Fig. 3 A). The shift in the apparent molecular weight of both ESCRT-I and -II to ∼250 kD in mvb12Δ cells was quantitative, with most (if not all) of the cytoplasmic ESCRT-I and -II now eluting at this size (Fig. 3 A). Because ESCRT-I and -II are known to interact with each other in vitro (Teo et al., 2006), we suspected that the ∼250-kD complex might be a heterodimeric complex of ESCRT-I (∼100 kD) and -II (∼150 kD). This hypothesis is supported by the coimmunoprecipitation of Vps23 (ESCRT-I) with Vps36 (ESCRT-II) from mvb12Δ cell extracts, confirming the identity of the ∼250-kD species as a heterodimeric complex of ESCRT-I and -II (Fig. 3 B). To further characterize the ESCRT-I–II heterodimer, we generated a vps36Δ mvb12Δ double mutant to disrupt ESCRT-I–II interactions. The size of ESCRT-I, based on the elution profiles of Vps23 and Vps37, was determined to be ∼100 kD in the double mutant (Fig. 3 A). The ∼100-kD species of ESCRT-I observed in the vps36Δ mvb12Δ mutant most likely represents the monomeric form of ESCRT-I, as this size is consistent with the molecular weight of Escherichia coli–expressed monomeric ESCRT-I determined by equilibrium analytical ultracentrifugation (Kostelansky et al., 2006; Teo et al., 2006).
Figure 3.
Mvb12 negatively regulates the interaction between ESCRT-I and -II. (A) Gel filtration analysis of cell extracts from WT (SEY6210) strain expressing Vps36-HA, mvb12Δ strain expressing Vps36-HA, and mvb12Δ vps36Δ strain expressing Vps37-HA. Column fractions were analyzed by Western blotting using antibodies specific for Vps23 or HA. Vps36-HA does not cofractionate with Vps23 in WT, but cofractionates with Vps23 in mvb12Δ as a ∼250-kD complex in the cytoplasm. Deletion of VPS36 from the mvb12Δ strain disrupts the ESCRT-I–II heterodimer to form a ∼100-kD ESCRT-I monomer. (B) Cell extract from mvb12Δ strain expressing control plasmid or Vps36-HA plasmid were immunoprecipitated using antibody specific for HA. Immunoprecipitates were analyzed by Western blotting using antibodies against Vps23 or HA. Vps23 coimmunoprecipitates with Vps36-HA.
Yeast native ESCRT-I assembles into an oligomer in an Mvb12-dependent manner
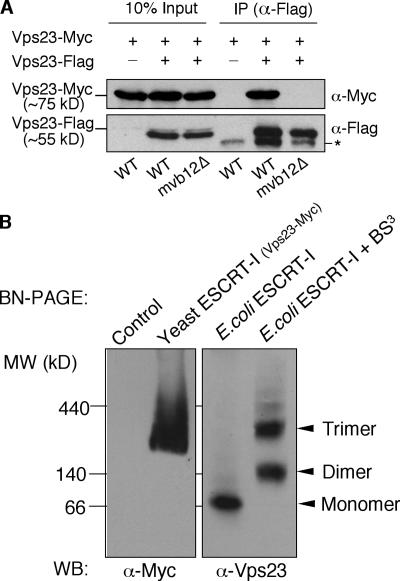
The presence of a ∼100-kD ESCRT-I complex in vps36Δ mvb12Δ mutant cells suggests that the cytosolic ∼350-kD species observed in wild-type cells may represent an oligomer of the ESCRT-I core complex (Vps23, -28, and -37). To test this hypothesis, we coexpressed Vps23-Flag and -Myc in the same cells and probed their ability to associate by performing coimmunoprecipitations. Cytosolic fractions were prepared from cell lysates and subjected to immunoprecipitation under native conditions using antibodies specific for the Flag tag. Vps23-Myc coimmunoprecipitated with Vps23-Flag in cytosolic fractions from wild-type cells, indicating that there are at least two copies of Vps23 in the ∼350-kD ESCRT-I complex (Fig. 4 A). In contrast, Vps23-Myc did not coimmunoprecipitate with Vps23-Flag in cytosolic fractions from mvb12Δ cells (Fig. 4 A). These results suggest that the ∼350-kD species is likely an oligomer of the ESCRT-I core complex. Deletion of MVB12 results in the dissociation of the oligomer into monomers of the ESCRT-I core complex, indicating that Mvb12 is required in assembling the ESCRT-I oligomer.
Figure 4.
Yeast native ESCRT-I assembles into an oligomer in an Mvb12-dependent manner. (A) Cytosolic ∼350-kD ESCRT-I complex is an oligomer. Cell extracts from Vps23-Myc strain (SJY027) expressing either empty vector or Vps23-Flag (pSJ106); mvb12Δ,Vps23-Myc strain (TCY274) expressing Vps23-Flag (pSJ106) were immunoprecipitated using antibody specific for Flag. Immunoprecipitates were analyzed by Western blotting using antibodies against Myc. Vps23-Flag coimmunoprecipitates with Vps23-Myc in wild-type cells, but not in mvb12Δ cells. The nonspecific band noted by the asterisk in the bottom gel likely corresponds to cross-reactivity of antibody heavy chains (last three lanes of the gel) that were loaded on the gel (anti-FLAG immunoprecipitations) with the goat anti–mouse antibodies used for this Western blot (in lane 5, a minor degradation product of the Vps23-FLAG also was detected at this position). (B; left) Cell extracts from WT cells (control) or cells expressing Myc-tagged Vps23 were prepared, analyzed by BN-PAGE, and immunoblotted using antibodies specific for the Myc tag. (right) Purified E. coli ESCRT-I was analyzed by BN-PAGE (left lane) or treated with 1 mM BS3 before BN-PAGE analysis. The monomeric form of E. coli ESCRT-I, as well as the dimeric and trimeric forms generated upon cross-linking, are labeled and indicated by the arrowheads.
In some cases, molecular weight estimates obtained by gel filtration chromatography is dependent on the shape of the protein–protein complex being analyzed; therefore, we used a second technique to analyze the molecular weight of yeast native ESCRT-I. Blue native gels (BN-PAGE) provide a reliable and reproducible means of estimating the molecular weight of native protein complexes. As shown in Fig. 4 B, BN-PAGE analysis of cytosolic ESCRT-I complex from yeast revealed the presence of a protein complex whose molecular weight is ∼300 kD. In contrast, the stable monomeric ESCRT-I purified from E. coli migrated at a significantly lower molecular weight of ∼80 kD, which is consistent with the molecular weight of ∼85 kD determined independently using equilibrium analytical ultracentrifugation (Kostelansky et al., 2006; Teo et al., 2006). The clear difference in the migration pattern of yeast ESCRT-I versus monomeric ESCRT-I from E. coli on BN-PAGE strongly suggests that the yeast complex is an oligomer. To get a rough estimate of whether the yeast ESCRT-I oligomer is a dimer, trimer, or a higher-order species, we induced oligomerization of the E. coli ESCRT-I monomer using the chemical cross-linker bis(sulfosuccinimidyl)suberate (BS3). As shown in Fig. 4 B, brief exposure to the chemical cross-linker generated clearly distinguishable dimeric and trimeric forms of the E. coli ESCRT-I, suggesting that the ESCRT-I core complex has an inherent propensity to oligomerize. Interestingly, the molecular weight of the yeast complex coincides best with that of the trimeric E. coli ESCRT-I. Collectively, the results of the coimmunoprecipitations and BN-PAGE strongly indicate that the ∼350-kD species observed in yeast represents an oligomeric complex consisting of at least two (but likely three) ESCRT-I core complexes (Vps23, -28, and -37).
To further test the possibility that ESCRT-I is a trimer, we performed a new coimmunoprecipitation experiment using a strain expressing two different tagged versions of Vps23, Myc-tagged chromosomal Vps23 and Flag-tagged Vps23 expressed from a low-copy plasmid. Using an antibody specific for Vps23, we were able to detect both tagged versions of Vps23, and we found that the plasmid-derived Vps23-Flag was expressed at lower levels (∼50–60%) than the chromosomal Vps23-Myc (unpublished data). As such, the differential expression of the two different (Myc- and Flag-tagged) pools of Vps23 provides an indirect way to examine the stoichiometry of the ESCRT-I complex by comparing the ratio of their incorporation into the oligomer. Consistent with our trimer prediction, immunoprecipitation using a Flag antibody coimmunoprecipitated Vps23-Myc. The ratio of Vps23-Myc to -Flag in the immunoprecipitate, as detected by an antibody specific for Vps23, was determined to be ∼2:1 by densitometric analysis (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200608053/DC1), suggesting that there are three copies of Vps23 in the ESCRT-I complex. Collectively, our data are consistent with the cytosolic ESCRT-I complex being a trimer.
The coiled-coil domain of Vps23 binds Mvb12 and mediates ESCRT-I oligomerization
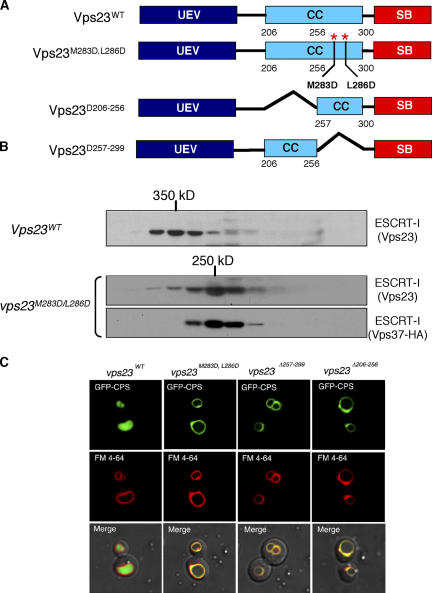
Vps23 is predicted to contain a coiled-coil domain between residues 212–304 (Babst et al., 2000). Coiled-coil domains are known to mediate protein oligomerization (Wolf et al., 1997). Bioinformatic analyses suggest that the coiled- coil domain of Vps23 has a high probability of forming a trimer (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200608053/DC1). Several signature heptad repeats were identified in the coiled-coil region of Vps23 and hydrophobic–hydrophilic mutations were introduced at positions a and d of the heptad repeat (abcdefg) to disrupt coiled-coil interactions. In addition to the point mutants, two coiled-coil domain deletion mutants were also generated to test the functional role of this region in Vps23/ESCRT-I oligomer formation (Fig. 5 A). Gel filtration analysis of the coiled-coil domain mutants Vps23(M283D/L286D; Fig. 5 B), Vps23(Δ206-256), and Vps23(Δ257-299; not depicted) displayed the formation of a ∼250-kD complex, which is consistent with the ESCRT-I–II heterodimer observed in mvb12Δ cells, demonstrating the importance of the coiled-coil region in maintaining the integrity of the ESCRT-I oligomer. Similar to the mvb12Δ strain, all of the Vps23 coiled-coil mutants displayed defects in MVB sorting with GFP-CPS missorted and colocalized with FM4-64 at the limiting membrane of the vacuole (Fig. 5 C). We also tested if the coiled-coil domain of Vps23 corresponds to the Mvb12 binding site by coimmunoprecipitation experiments. Both deletions and point mutations in the coiled-coil domain of Vps23 abolished interactions with Mvb12 (Fig. 6 A), suggesting that Mvb12 binds to the coiled-coil region. We further tested if this interaction between Mvb12 and Vps23 is direct by in vitro pull down assays. Mvb12 binds to His-tagged Vps23, but not to His-tagged Vps23(Δ206-299) missing the coiled-coil domain, indicating that Mvb12 binds to the coiled-coil domain of Vps23 (Fig. 6 B). These data suggest that the ESCRT-I oligomer is formed by the coiled-coil domains of Vps23 molecules and is stabilized by Mvb12.
Figure 5.
The coiled-coil domain of Vps23 mediates oligomerization of ESCRT-I. (A) Domain structure of Vps23 and coiled-coil domain mutants used in this study. (B) Mutations in the coiled-coil domain disrupt ESCRT-I complex assembly in vivo as determined by gel filtration analysis. Cell extracts were isolated from WT (SEY6210) and vps23Δ strain expressing Vps23M283D/L286D and Vps37-HA. Column fractions were analyzed by Western blotting using antibodies specific for Vps23 or HA. Both Vps23M283D,L286D and Vps37 were detected in a ∼250-kD subcomplex in the cytoplasm of vps23Δ strain similar to that of mvb12Δ strain. (C) Coiled-coil domain mutants displayed a MVB-sorting defect. MVB cargo is visualized using a GFP-CPS fusion, and the limiting membrane of the vacuole is visualized with FM4-64. Shown are Nomarski optics (bottom) and fluorescence localization of GFP-CPS (top) and FM4-64 (middle) in vps23Δ strain expressing WT Vps23, Vps23M283D,L286D, Vps23Δ257-299, or Vps23Δ206-256.
Figure 6.
Mvb12 interacts with the coiled-coil domain of Vps23. (A) Cell extract from strains coexpressing Mvb12-Flag and either wild-type Vps23 or Vps23 with coiled-coil mutations were immunoprecipitated using antibody specific for Flag. Immunoprecipitates were analyzed by Western blot using antibodies against Vps23 or Flag. Mvb12 fails to interact with Vps23 coiled-coil deletion and point mutants. (B) Direct Mvb12 and Vps23 interaction as determined by in vitro pull-down assay. Mvb12-Flag protein were incubated with equal amount of E. coli–expressed Vps23-His6 and Vps23(Δ206-299)-His6 proteins preconjugated with Ni2+ NTA beads. Bound proteins were eluted and analyzed by Western blot using antibodies against Flag. Mvb12 failed to interact with Vps23 coiled-coil deletion mutant.
Discussion
The ESCRT complexes appear to function in a sequential manner at the surface of the endosome during the formation of MVB vesicles and the sorting of cargo into these vesicles. Assembly of the ESCRT machinery on the endosomal membrane is regulated by both ubiquitin and lipid (PtdIns3P) signals and requires the coordinated recruitment of all ESCRT complexes and associated proteins (Hicke and Dunn, 2003; Katzmann et al., 2002; Raiborg et al., 2003). Interactions among the yeast ESCRT complexes and their mammalian homologues have been extensively investigated by yeast two-hybrid analysis (Martin- Serrano et al., 2003; von Schwedler et al., 2003; Bowers et al., 2004). ESCRT-I has been shown to interact with the Vps27 via interactions between the UEV domain of Vps23 and the PTVP motif in Vps27 (Bilodeau et al., 2003; Katzmann et al., 2003). Two-hybrid studies have indicated that ESCRT-I interacts with both ESCRT-II and -III components (Bowers et al., 2004). The interaction between ESCRT-I and -II has been characterized biochemically and mapped to Vps28 (ESCRT-I) and Vps36 (ESCRT-II; Kostelansky et al., 2006; Teo et al., 2006). In this study, we identify Mvb12 as a new component of ESCRT-I that interacts with the coiled-coil domain of Vps23 and stabilizes ESCRT-I as an inactive, soluble oligomer. In doing so, Mvb12 negatively regulates the interaction between ESCRT-I and -II. This ensures the ordered recruitment of ESCRT-I and -II and spatially restricts ESCRT-I–II assembly to the surface of the endosome. Premature assembly of the ESCRT-I–II complex in the cytosol of mvb12Δ cells likely interferes with the normal sequence of ESCRT-mediated cargo sorting events on the endosome, resulting in a defect in MVB sorting.
Both the yeast and mammalian ESCRT-I complexes have an apparent molecular weight of ∼350 kD by gel filtration (Babst et al., 2000; Katzmann et al., 2001). The structure of the ESCRT-I core complex has been determined, consisting of Vps23, Vps28, and Vps37 in a 1:1:1 stoichiometry (Kostelansky et al., 2006; Teo et al., 2006). In these studies, a truncated form of E. coli-expressed ESCRT-I was used and determined to be ∼85 kD by analytical ultracentrifugation, which is consistent with the monomeric form of ESCRT-I (Kostelansky et al., 2006; Teo et al., 2006). The difference between the size of E. coli–expressed ESCRT-I and yeast native ESCRT-I suggests that the native ESCRT-I complex is an oligomer of the ESCRT-I core complex. Indeed, the ability of E. coli ESCRT-I to assemble into clearly distinguishable dimeric and trimeric forms when treated with a chemical cross-linker (Fig. 4 B) strongly suggests that ESCRT-I monomers have a propensity to associate with each other and assemble into an oligomer. What, then, is the oligomeric status of the ESCRT-I complex in yeast? Is it a dimer, trimer, or a higher order oligomer? The results of a variety of approaches, including gel filtration, BN-PAGE, and coimmunoprecipitation experiments, presented in this study strongly argue that the ∼350-kD species observed in yeast is an oligomer that likely contains three copies of the ESCRT-I core complex.
The oligomer model for the yeast native ESCRT-I complex predicted by gel filtration chromatography and BN-PAGE is also strongly supported by the analysis of the coiled-coil domain of Vps23 using the MultiCoil prediction program (Wolf et al., 1997). Coiled-coil domains are known to mediate protein oligomerization in several cases including transcription factors and viral proteins. To predict the oligomeric state of Vps23/ ESCRT-I, we used the MultiCoil prediction program to analyze the Vps23 sequence. The predicted dimeric or trimeric state of this domain was calculated and the probability of each residue in a dimeric (blue) or trimeric (red) coiled-coil was plotted against residue number (Fig. S2 A). The program predicted a coiled-coil region from residue 212–304. This region has a higher probability of forming a trimeric coiled-coil than a dimeric coiled-coil. As a control, we used this program to analyze gp41p of HIV, the trimeric structure of which has been determined by NMR and x-ray crystallography. The MultiCoil program also predicts a higher probability of trimer formation for the coiled-coil region of gp41p (Fig. S2 B). Typically, trimers are not packed as tightly as dimers, rendering them less stable and better suited for regulation. Thus, an ESCRT-I trimer would be more suitable for assembly/disassembly regulation. Furthermore, mutations of key residues within the coiled-coil domain of Vps23, as predicted by the MultiCoil program, recapitulated the phenotype of the mvb12Δ strain (Fig. 5, B and C). The ESCRT-I oligomer is destabilized by either Mvb12 deletion or Vps23 coiled-coil domain mutations. Collectively, our analyses of the coiled-coil domain of Vps23 indicate that the coiled-coil domain of Vps23 is required for the oligomerization of the ESCRT-I complex.
Although the oligomeric status of the yeast ESCRT-I complex has been established, it is not clear what role Mvb12 plays in the oligomerization process. Although it is difficult to address this question, there are some interesting clues in the data presented here. Our data indicate that Mvb12 (Fig. 4 A) and the coiled-coil domain of Vps23 (Fig. 5, B and C) both appear to be essential in mediating the formation of the ∼350-kD oligomer, and that Mvb12 interacts with the Vps23 coiled-coil domain (Fig. 6, A and B). However, the fact that we cannot detect the oligomeric form of ESCRT-I in the coiled-coil mutants of Vps23 suggests that the coiled-coil domains are critical in bringing the monomers together (possibly via hydrophobic interactions), at which point Mvb12 probably functions as a molecular clamp or adaptor that tethers three ESCRT-I core complexes together in a stable oligomer. Therefore, Mvb12 appears to play a structural role in stabilizing the ESCRT-I oligomer, as well as a regulatory role in preventing ESCRT-I from interacting with ESCRT-II in the cytosol. Consistent with the role of Mvb12 as a regulator of ESCRT-I function, loss of Mvb12 does not result in an absolute block in MVB sorting, as is seen in mutants lacking the core components of ESCRT-I.
The results of previous genetic and biochemical studies have suggested a model for the sequential assembly of the ESCRT complexes on the endosomal membrane. In this model, membrane-bound ESCRT-I recruits -II, which recruits and activates downstream ESCRT machinery, resulting in cargo sorting into MVB vesicles. If Mvb12 is a negative regulator of the interaction between ESCRT-I and -II, one would expect that the membrane association of ESCRT-I would be accompanied by the dissociation of Mvb12, allowing ESCRT-I to recruit -II. Consistent with this, subcellular fractionation experiments show that Vps23 and Mvb12 exhibit different levels of membrane association (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200608053/DC1). Although a small pool of Mvb12 can be detected on the endosomal membrane, which is consistent with the partial Mvb12-GFP localization to endosomes, Vps23 has a larger membrane-associated pool (∼25% of total protein) than Mvb12 (∼8%), and the difference in membrane association is enhanced in vps4Δ cells (∼73% for Vps23 vs. ∼10% for Mvb12). The significant difference in the membrane association of Vps23 versus Mvb12 argues that the majority of ESCRT-I on the endosome is not bound to Mvb12. Thus, the function of Mvb12 as a clamp for the ESCRT-I oligomer appears to be restricted to the cytosol. It is generally difficult to predict the stoichiometry of individual constituents in a native, higher-ordered oligomer. However, in vitro analyses of the interaction between E. coli–expressed ESCRT-I and Mvb12 suggest that Mvb12 exists in a 1:1 stoichiometry with other ESCRT-I subunits (unpublished data; Williams, R., personal communication).
Based on our data, we propose the following working model for the function of Mvb12 (Fig. 7). In the cytosol, Mvb12 associates with and stabilizes ESCRT-I in an oligomeric and inactive state. ESCRT-I is recruited to the endosomal membrane by Vps27 via the interaction of the UEV domain of Vps23 with the PTVP motif of Vps27 and with ubiquitinated cargo. A conformational switch in ESCRT-I results in the dissociation of Mvb12, as well as the disassembly of the ESCRT-I oligomer into monomers. Consequently, the ESCRT-II binding site within ESCRT-I (Vps28 C-terminal domain) is exposed, allowing ESCRT-I to recruit ESCRT-II to the endosome. This leads to the activation of ESCRT-II and the recruitment of downstream ESCRT machinery (ESCRT-III and Vps4), culminating in the formation of MVB vesicles and the sorting of cargo into these vesicles. Ultimately, it will require detailed structural and biochemical analyses of the native ESCRT-I complex to add mechanistic details and further refine this model.
Figure 7.
Working model for the function of Mvb12. In the cytosol, Mvb12 associates with and stabilizes ESCRT-I in an oligomeric, inactive state so that ESCRT-I cannot interact with ESCRT-II. Once recruited to the endosome, ESCRT-I assembles with ESCRT-II, leading to the activation of the downstream ESCRT machinery and cargo sorting into the MVB pathway. In the absence of Mvb12, the assembly of ESCRT-I and -II occurs prematurely in the cytosol, resulting in a defect in MVB sorting.
Materials and methods
Plasmid construction and yeast strains
DNA encoding a HA, Myc, and Flag epitope were introduced just before the stop codon in the chromosomal copy of MVB12, VPS23, and VPS37, respectively. MVB12–Flag, MVB12-GFP, VPS23, VPS23–Flag, and VPS37-HA sequences were then amplified from genomic DNA. The SpeI–SacI- digested PCR products of MVB12-Flag and MVB12-GFP were ligated with SpeI–SacI-digested pRS414 and pRS415 to generate pTC74 and pTC75. The SmaI–XhoI-digested PCR products of MVB12-Flag were ligated with SmaI–XhoI-digested pGEX4T-1 to generate pTC76 E. coli expression vector. The NheI–XhoI-digested PCR products of Vps23 were ligated with NheI–XhoI-digested pET23b to generate pSJ033 E. coli expression vector. The SpeI–SacI-digested PCR products of VPS23, VPS23–Flag, and VPS37-HA were ligated with SpeI–SacI-digested pRS416 to generate pSJ102, pSJ106, and pSJ111, respectively. Vps23 coiled-coil point mutations were introduced into pSJ102 by QuikChange mutagenesis (Stratagene) to generate pSJ140(Vps23M283D,L286D). Vps23 coiled-coil deletion mutants were constructed by inverse PCR and blunt-end ligation using pSJ102 as the template to generate pSJ135(Vps23Δ257-299) and pSJ137(Vps23Δ206-256), using pSJ033 as the template to generate pSJ143(Vps23Δ206-299) E. coli expression plasmid. pGO45 GFP–CPS, dsRed-FYVEEEA1, and Ste2-GFP have been previously described (Odorizzi et al., 1998; Katzmann et al., 2003). The following yeast strains were constructed for this study: TCY246 (SEY6210.1; mvb12Δ∷HIS3); TCY257 (SEY6210.1; MVB12-Flag, HIS5); TCY274 (SEY6210.1; mvb12Δ∷HIS3, VPS23-Myc, HIS5); SJY027 (SEY6210.1; VPS23-Myc, HIS5); and SJY030 (SEY6210.1; mvb12Δ∷HIS3, vps36Δ∷HIS3). The following yeast strains were previously described (Babst et al., 2002a,b) SEY6210 (MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901lys2-801 suc2-Δ9); EEY6-2 (SEY6210; vps23∷HIS3); MBY30 (SEY6210; vps36Δ∷HIS3); MBY3 (SEY6210; vps4∷TRP1); and MBY24 (SEY6210.1; snf7∷HIS3).
Microscopy
Living cells expressing the GFP-CPS, Ste2-GFP, and Mvb21-GFP chimera were harvested at an A 600 of 0.4–0.6; some were labeled with FM4-64 for vacuolar membrane staining and resuspended in medium for visualization. Visualization of cells was performed on a fluorescence microscope (Axiovert S1002TV; Carl Zeiss MicroImaging, Inc.) equipped with FITC and rhodamine filters, captured with a digital camera (CH350 CCD; Photometrix), and deconvolved using Delta Vision software (Applied Precision, Inc.). Results presented were based on observations of >120 cells.
Gel filtration analysis
For gel filtration analysis, yeast cells were spheroplasted and lysed in PBS containing 0.1 mM AEBSF, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 mM benzamide, and protease inhibitor cocktail (Complete; Roche). The lysate was precleared for 5 min at 300 g, followed by a 100,000 g centrifugation. The resulting lysate was loaded onto a Sephacryl S300 column (16/60; GE Healthcare) and separated with PBS. Fractions were analyzed by Western blotting using anti-HA, anti-Flag monoclonal antibodies, and anti-Vps23 polyclonal antibody.
BN-PAGE
Soluble extracts were prepared from a yeast strain expressing Myc-tagged Vps23. Approximately 90 μl of the yeast extract or purified E. coli ESCRT-I was mixed with 10 μl of 10× sample buffer (5% Coomassie brilliant blue G-250, 100 mM Bis-Tris, pH 7.0, 500 mM [ɛ-aminohexanoic acid], and 10% glycerol). In the samples where cross-linking was performed, purified E. coli ESCRT-I was incubated with 1 mM BS3 at room temperature for 30 min. The samples were then analyzed using a 4–15% gradient gel using the same previously described technique (Frazier et al., 2003).
Immunoprecipitations
Immunoprecipitations under native conditions were performed essentially as previously described (Katzmann et al., 2001).
Pull-down assay
Equal amount of E. coli–expressed Vps23-His6 and Vps23(Δ206-299)-His6 proteins were incubated with Ni-NTA beads for 1 h in Ni-NTA Bind Buffer (Novagen), and then incubated with an equal amount of E. coli-expressed Mvb12-Flag proteins for an additional 4 h at room temperature. The beads were washed three times with 1 ml Ni-NTA Bind Buffer to remove unbound material. Bound proteins were eluted by incubating with 1 ml 0.5 M acetic acid for 30 min. Proteins conjugated to the beads were eluted with 1 ml elution buffer for 30 min. 10% TCA was added to all samples to precipitate proteins. Acetone-washed protein samples were boiled in sample buffer and analyzed by 10% PAGE and Western blotting using anti-Flag monoclonal antibodies and anti-Vps23 polyclonal antibody.
Online supplemental material
Fig. S1 shows a multiple sequence alignment of Mvb12 with its putative homologues. Fig. S2 displays the result of the MultiCoil analysis of the coiled-coil domain of Vps23. Fig. S3 displays data that supports a model in which ESCRT-I forms a trimer in the cytosol. Fig. S4 shows that Mvb12 likely dissociates from ESCRT-I upon membrane association. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200608053/DC1.
Supplementary Material
Acknowledgments
We thank Roger Williams for providing purified recombinant ESCRT-I, James H. Hurley for discussions on unpublished data, David Teis for discussions and comments on the manuscript, and Peter Rehling and Nathan Alder for their expertise on BN-PAGE.
This research was supported by fellowships from the Howard Hughes Medical Institute (J. Sun, T. Chu, and S. Saksena). S.D. Emr is supported as an investigator of the Howard Hughes Medical Institute.
T. Chu and J. Sun contributed equally to this paper.
Abbreviations used in this paper: BN, Blue native; BS3, bis(sulfosuccinimidyl) suberate; CPS, carboxypeptidase S; ESCRT, endosomal sorting complex required for transport; MVB, multivesicular body; PtdIns3P, phosphatidylinositol 3-phosphate; Vps, vacuolar protein sorting.
References
- Alam, S.L., J. Sun, M. Payne, B.D. Welch, B.K. Blake, D.R. Davis, H.H. Meyer, S.D. Emr, and W.I. Sundquist. 2004. Ubiquitin interactions of NZF zinc fingers. EMBO J. 23:1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., B. Wendland, E.J. Estepa, and S.D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., G. Odorizzi, E.J. Estepa, and S.D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 1:248–258. [DOI] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, E.J. Estepa-Sabal, T. Meerloo, and S.D. Emr. 2002. a. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282. [DOI] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, W.B. Snyder, B. Wendland, and S.D. Emr. 2002. b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., S.C. Winistorfer, W.R. Kearney, A.D. Robertson, and R.C. Piper. 2003. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K., J. Lottridge, S.B. Helliwell, L.M. Goldthwaite, J.P. Luzio, and T.H. Stevens. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 5:194–210. [DOI] [PubMed] [Google Scholar]
- Deblandre, G.A., E.C. Lai, and C. Kintner. 2001. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell. 1:795–806. [DOI] [PubMed] [Google Scholar]
- Frazier, A.E., A. Chacinska, K.N. Truscott, B. Guiard, N. Pfanner, and P. Rehling. 2003. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell. Biol. 23:7818–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter, C.E., A. Pearse, L.J. Hewlett, and C.R. Hopkins. 1996. Multivesicular endosomes containing internalized EGF–EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132:1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus, J.E., U.K. von Schwedler, O.W. Pornillos, S.G. Morham, K.H. Zavitz, H.E. Wang, D.A. Wettstein, K.M. Stray, M. Cote, R.L. Rich, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:55–65. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L.J. Jensen, S. Bastuck, B. Dumpelfeld, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature. 440:631–636. [DOI] [PubMed] [Google Scholar]
- Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141–172. [DOI] [PubMed] [Google Scholar]
- Hierro, A., J. Sun, A.S. Rusnak, J. Kim, G. Prag, S.D. Emr, and J.H. Hurley. 2004. Structure of the ESCRT-II endosomal trafficking complex. Nature. 431:221–225. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., and S.D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., M. Babst, and S.D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., G. Odorizzi, and S.D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., C.J. Stefan, M. Babst, and S.D. Emr. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., S. Sitaraman, A. Hierro, B.M. Beach, G. Odorizzi, and J.H. Hurley. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 8:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijmeer, M., G. Ramm, D. Schuurhuis, J. Griffith, M. Rescigno, P. Ricciardi-Castagnoli, A.Y. Rudensky, F. Ossendorp, C.J. Melief, W. Stoorvogel, and H.J. Geuze. 2001. Reorganization of multivesicular bodies regulates MHC class II antigen presentation by dendritic cells. J. Cell Biol. 155:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky, M.S., J. Sun, S. Lee, J. Kim, R. Ghirlando, A. Hierro, S.D. Emr, and J.H. Hurley. 2006. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 125:113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A.P. Tikuisis, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440:637–643. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., G.A. Deblandre, C. Kintner, and G.M. Rubin. 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell. 1:783–794. [DOI] [PubMed] [Google Scholar]
- Luhtala, N., and G. Odorizzi. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano, J., T. Zang, and P.D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., M. Babst, and S.D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., D.J. Katzmann, M. Babst, A. Audhya, and S.D. Emr. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos, E., C. Pitsouli, K.M. Klueg, M.A. Muskavitch, N.K. Moschonas, and C. Delidakis. 2001. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell. 1:807–816. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., T.E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446–455. [DOI] [PubMed] [Google Scholar]
- Scott, A., H.Y. Chung, M. Gonciarz-Swiatek, G.C. Hill, F.G. Whitby, J. Gaspar, J.M. Holton, R. Viswanathan, S. Ghaffarian, C.P. Hill, and W.I. Sundquist. 2005. Structural and mechanistic studies of VPS4 proteins. EMBO J. 24:3658–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, H., O. Perisic, B. Gonzalez, and R.L. Williams. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 7:559–569. [DOI] [PubMed] [Google Scholar]
- Teo, H., D.J. Gill, J. Sun, O. Perisic, D.B. Veprintsev, Y. Vallis, S.D. Emr, and R.L. Williams. 2006. ESCRT-I Core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 125:99–111. [DOI] [PubMed] [Google Scholar]
- von Schwedler, U.K., M. Stuchell, B. Muller, D.M. Ward, H.Y. Chung, E. Morita, H.E. Wang, T. Davis, G.P. He, D.M. Cimbora, et al. 2003. The protein network of HIV budding. Cell. 114:701–713. [DOI] [PubMed] [Google Scholar]
- Wolf, E., P.S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorikawa, C., H. Shibata, S. Waguri, K. Hatta, M. Horii, K. Katoh, T. Kobayashi, Y. Uchiyama, and M. Maki. 2005. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem. J. 387:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.