Abstract
Cellular senescence is a tumor-suppressing mechanism that is accompanied by characteristic chromatin condensation called senescence-associated heterochromatic foci (SAHFs). We found that individual SAHFs originate from individual chromosomes. SAHFs do not show alterations of posttranslational modifications of core histones that mark condensed chromatin in mitotic chromosomes, apoptotic chromatin, or transcriptionally inactive heterochromatin. Remarkably, SAHF-positive senescent cells lose linker histone H1 and exhibit increased levels of chromatin-bound high mobility group A2 (HMGA2). The expression of N-terminally enhanced green fluorescent protein (EGFP)–tagged histone H1 induces premature senescence phenotypes, including increased levels of phosphorylated p53, p21, and hypophosphorylated Rb, and a decrease in the chromatin-bound endogenous histone H1 level but not in p16 level accumulation or SAHF formation. However, the simultaneous ectopic expression of hemagglutinin-tagged HMGA2 and N-terminally EGFP-tagged histone H1 leads to significant SAHF formation (P < 0.001). It is known that histone H1 and HMG proteins compete for a common binding site, the linker DNA. These results suggest that SAHFs are a novel type of chromatin condensation involving alterations in linker DNA–binding proteins.
Introduction
Normal human cells gradually lose replicative capacity after a finite number of cell divisions as a result of telomere shortening and irreversibly enter the growth-arrested state called replicative senescence (Hayflick and Moorhead, 1961). This process is accompanied by a series of changes, including cell enlargement, increased acidic β-galactosida se activity (senescence-associated β-galactosidase [SA–β-gal]), and alterations of chromatin structures. Replicative senescence has been suggested to represent certain aspects of organismal aging (Campisi, 2005).
It is known that cells also enter a state that is indistinguishable from replicative senescence after exposure to sublethal doses of various types of stress, including reactive oxygen species, oncogenic activation, and inappropriate culture conditions. Such a stress-induced state is called premature senescence or stress-induced senescence (Serrano et al., 1997; Ramirez et al., 2001). Accordingly, cellular senescence refers to both replicative and premature senescence. Although the stimuli inducing the two types of senescence appear to be considerably diverse (telomere dependent and independent), the stress-induced MAPK p38 plays a key role in signaling such diverse stimuli to induce both types of senescence (Ishikawa, 2003).
Cellular senescence is not a condition that simply causes organismal aging to endanger the survival of individuals. Instead, it has been hypothesized that cellular senescence plays an adaptive role in preventing cells from immortalization and neoplastic transformation in vitro and plays a role in tumor suppression in vivo (Lowe et al., 2004; Campisi, 2005). Indeed, it was recently reported that senescent cells exist in several types of premalignant tumors in vivo and that cellular senescence is required to prevent tumorigenesis in vivo (Braig and Schmitt, 2006). Therefore, cellular senescence acts as a barrier to tumorigenesis as apoptosis does by stably suppressing the growth of stressed cells.
The contribution of cellular senescence to tumor suppression depends on the stable maintenance of growth arrest. Alterations of chromatin structure are believed to account for the irreversible nature of the senescent state (Narita et al., 2003). In the nucleus, DNA is wrapped around histone octamers to form nucleosomes, an array of which, in turn, is packed into higher ordered chromatin structures by the association of linker histone H1 and other chromatin proteins (Thoma et al., 1979). Senescent cells form characteristic heterochromatin structures called senescence-associated heterochromatic foci (SAHFs; Narita et al., 2003; Zhang et al., 2005). SAHFs contain methylated histone H3 at Lys9 (H3(K9)me) and heterochromatin protein 1, which are typically detected in heterochromatin. It was also reported that H3(K9)me is enriched at proliferation-promoting gene promoters specifically in senescent cells that are concomitant with the appearance of SAHFs but not in quiescent cells (Narita et al., 2003). Furthermore, Suv39h1, a histone methyltransferase for H3K9, is required for oncogene-induced senescence in mouse lymphocytes and for the suppression of lymphoma (Braig and Schmitt, 2006). Therefore, it is suggested that senescent cells maintain the growth-arrested state, at least in part, by stably forming heterochromatin at proliferation-promoting gene loci.
The molecular mechanism of SAHF formation remains poorly understood. Although histone chaperones Asf1a and histone regulation A have been reported to play a role in SAHF formation (Zhang et al., 2005), it is not known how these chaperones induce SAHFs. In this study, we have found an unexpected role of histone H1 in cellular senescence.
Results
Individual SAHFs originate from individual chromosomes
SAHFs are characterized by numerous, highly condensed granular DNAs spreading throughout the nucleus, as shown in WI-38 cells that were induced to senesce by the retroviral expression of oncogenic Ras (RasG12V; Fig. 1 A, bottom). SAHFs were observed in >80% of RasG12V-transfected cells as early as day 1 after drug selection. However, the degree of chromatin condensation appeared to increase during the following 7 d, during which the senescent state was stably maintained.
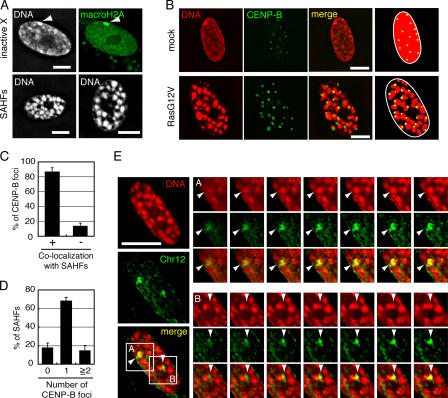
Figure 1.
A single SAHF originates from a single chromosome. (A) Similar sizes of SAHFs and inactive X chromosomes. The top two figures show a single mock-transfected WI-38 nucleus stained with DAPI (left) or anti-macroH2A antibody (right). The inactive X chromosomes are indicated by arrowheads. Two separate RasG12V-transfected WI-38 SAHF nuclei stained with DAPI are shown at the bottom. (B) Mock- or RasG12V-transfected WI-38 cells were stained with anti–CENP-B antibody and DAPI. Schematic drawings illustrating the localization of SAHFs and CENP-B foci are shown (right). (C) Percentages of CENP-B foci that colocalized with (+) or excluded (−) SAHF signals (n = 418 derived from 16 senescent cells). Error bars show the SD from the mean of three independent experiments. (D) Percentages of SAHFs that contained zero, one, or more than one CENP-B foci (n = 665 derived from 24 senescent cells). The mean number of SAHFs per optical section was 27.7 ± 5.8 (mean ± SD). (E) RasG12V-transfected senescent WI-38 cells were hybridized with a probe specific for chromosome 12 (Chr12) and stained with DAPI. The probe stained two equal-sized metaphase chromosomes in WI-38 cell metaphase spreads, showing its specificity (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200604005/DC1). Enlarged images of regions A and B are optical sections at 0.4-μm intervals (right). Arrowheads indicate the overlapping of a single SAHF and chromosome 12. The results are representative of observations of five independent cells. Bars, 10 μm.
Because SAHFs are similar in size to inactive X chromosomes in WI-38 female normal fibroblasts (Fig. 1 A, top), we hypothesized that each granular structure originates from individual chromosomes. It is known that individual chromosomes occupy nonoverlapping and discrete spaces called chromosome territories (Cremer and Cremer, 2001). Previously, it was reported that centromeres are localized at the periphery of chromosome territories in quiescent human lymphocytes and human fibroblasts (Weierich et al., 2003). We examined the number and position of centromeres in RasG12V-transfected senescent WI-38 cells using antibodies recognizing centromere protein B (CENP-B). Approximately 85% of CENP-B foci were colocalized with SAHF signals (Fig. 1, B and C). Conversely, ∼70% of SAHFs contained one CENP-B focus at their periphery (Fig. 1, B and D). These results suggest that individual SAHFs originate from individual chromosomes.
We also performed FISH analysis using a probe specific for chromosome 12. Optical sections of the nucleus of RasG12V-transfected senescent WI-38 cells at 0.4-μm intervals showed that two chromosome 12 signals completely overlapped with two SAHFs in all of the five senescent cells exained (Fig. 1 E). Together, we conclude that each SAHF represents a heterochromatinized portion of an individual chromosome.
SAHFs are surrounded by the ICD
It has been proposed that in interphase nuclei, individual chromosomes are surrounded by the interchromosome domain (ICD), where transcription and RNA splicing take place (Cremer and Cremer, 2001). To investigate which regions in the nucleus are responsible for transcription in SAHF-positive senescent cells, RNA polymerase II was subjected to immunofluorescence (IF) analysis using specific antibodies (Fig. 2 A). In mock-transfected WI-38 cells, anti-RNA polymerase II signals were diffusely distributed throughout the nucleus. In RasG12V-transfected senescent WI-38 cells, RNA polymerase II was specifically localized in the DAPI-negative regions in a mutually exclusive manner with SAHFs (Fig. 2 A, bottom). Quantitative analyses revealed that ∼90% of SAHFs excluded anti-RNA polymerase II signals (Fig. 2 B). This result is not caused by the inability of the antibody to penetrate SAHFs because anti-H3(K9)me antibody reacted with SAHFs in IF experiments (see Fig. 3 A).
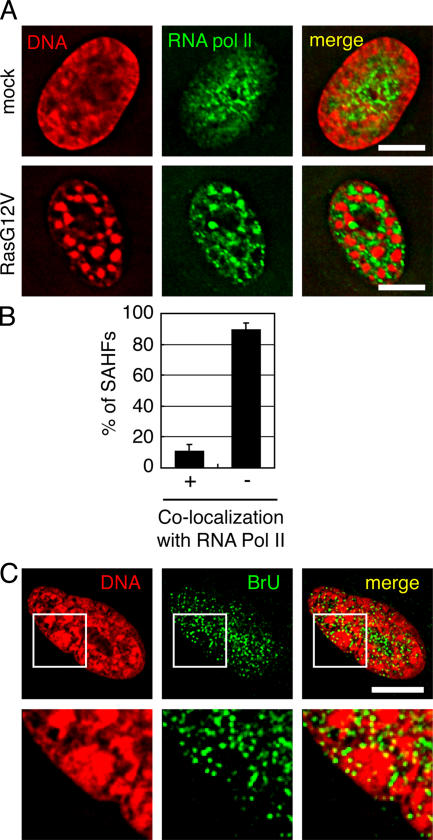
Figure 2.
SAHFs are surrounded by the ICD. (A) Mock- or RasG12V-transfected WI-38 cells were stained with anti-RNA polymerase II antibody and DAPI. (B) Percentages of SAHFs that overlapped with (+) or excluded (−) anti-RNA polymerase II signals (n = 341 derived from 13 senescent cells). Error bars represent SD. (C) Senescent cells were incubated with bromo-labeled UTP (BrU) and stained with anti-BrdU antibody. Enlarged images of the boxed areas are shown in bottom panels. No signals were observed when unlabeled UTP was used instead of BrU, when cells were treated with 1 μg/ml α-amanitin (the concentration that inhibits RNA polymerase II), or when they were treated with RNase A (Fig. S1 B, available at http://www.jcb.org/cgi/content/full/jcb.200604005/DC1). Bars, 10 μm.
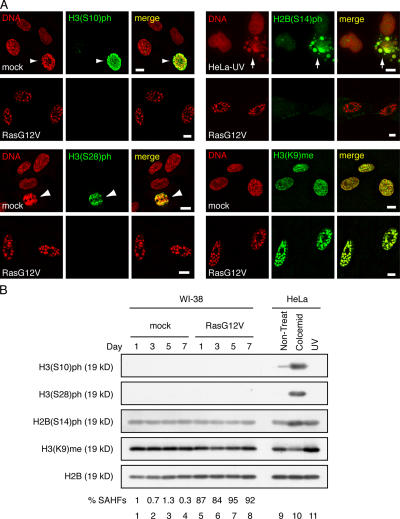
Figure 3.
SAHFs do not show histone modifications that characterize three types of chromatin condensation. (A) Mock- or RasG12V-transfected WI-38 cells as well as HeLa cells irradiated with 100 Jm−2 UV rays (HeLa-UV) were stained with antihistone antibodies and DAPI. Mitotic WI-38 nuclei and apoptotic HeLa nuclei are shown by arrowheads and arrows, respectively. (B) WI-38 cells were mock- or RasG12V-transfected and harvested at the indicated time points after drug selection. HeLa cells were untreated (non-treat), treated with 100 ng/ml colcemid, or irradiated with 100 Jm−2 UV rays (UV). Histones were prepared by acid extraction and analyzed by immunoblotting. Percentages of SAHF-positive cells are shown (bottom; n = 300). Bars, 10 μm.
Next, we performed in situ labeling of nascent RNAs to visualize the transcription sites (Fig. 2 C). RasG12V-transfected senescent WI-38 cells were permeabilized with 0.05% Triton X-100 and incubated with transcription buffer containing bromo-labeled UTP (BrU). The incorporation of BrU into nascent RNAs was visualized by IF analysis using anti-BrdU antibody. Although the shape of SAHFs was not well preserved after the transcription reaction, small dots of anti-BrU signals were exclusively localized in the DAPI-negative regions (Fig. 2 C). Interestingly, relatively large dots of anti-BrU signals were localized at the surface of SAHFs (Fig. 2 C, bottom enlarged images). Together, we conclude that in SAHF-positive senescent cells, active transcription occurs at the DAPI-negative inter-SAHF regions, particularly at the surface of SAHFs. This result is consistent with a previous report that SAHFs do not contain active sites for transcription (Narita et al., 2003) and with our hypothesis that the DAPI-negative regions correspond to ICDs (see Discussion).
SAHFs do not show histone modifications that characterize other types of chromatin condensation
Three types of condensed chromatin are characterized by specific posttranslational modifications of core histones: phosphorylations of histone H3 at Ser10 and Ser28 (H3(S10)ph and H3(S28)ph) in mitotic chromosomes, phosphorylation of histone H2B at Ser14 (H2B(S14)ph) in apoptotic chromatin, and methylation of histone H3 at Lys9 (H3(K9)me) in transcriptionally inactive heterochromatin (Peterson and Laniel, 2004). It has been argued that SAHFs represent transcriptionally inactive heterochromatin because SAHFs are colocalized with H3(K9)me and heterochromatin protein 1 (Narita et al., 2003; Zhang et al., 2005).
To investigate to which of these distinct chromatin condensations SAHFs are related, we examined the posttranslational modifications of histone H2B and histone H3 in RasG12V-transfected senescent WI-38 cells (Fig. 3). In IF experiments, neither anti-H3(S10)ph, anti-H3(S28)ph, nor anti-H2B(S14)ph antibodies stained SAHFs in senescent WI-38 cells, whereas they stained mitotic WI-38 chromosomes and apoptotic HeLa nuclei (Fig. 3 A). Similarly, in immunoblotting experiments, neither anti-H3(S10)ph nor anti-H3(S28)ph antibodies detected substantial protein bands in acid-extracted (AE) histone fractions derived from mock- or RasG12V-transfected WI-38 cells, whereas strong signals were observed in histone fractions from colcemid-treated HeLa cells (Fig. 3 B). Although anti-H2B(S14)ph signals were observed in histone fractions derived from mock- and RasG12V-transfected WI-38 cells, they were much weaker than those observed in colcemid- or UV-treated HeLa-derived histone fractions (Fig. 3 B). These results indicate that SAHFs are a type of chromatin condensation that is distinct from those found in mitotic chromosomes or apoptotic cells. Anti-H3(K9)me antibody reacted with SAHFs in senescent cells as well as the whole nuclear regions in mock-transfected cells (Fig. 3 A), as previously reported (Narita et al., 2003). Importantly, however, immunoblotting analysis of AE fractions using anti-H3(K9)me antibody revealed that the total amount of H3(K9)me per unit cell (calibrated by anti-H2B signals) was not substantially different between mock- and RasG12V-transfected WI-38 cells (Fig. 3 B), suggesting that the apparently strong anti-H3(K9)me signals at SAHFs reflect the condensation of DNAs.
Histone H1 does not exist in the senescent cell
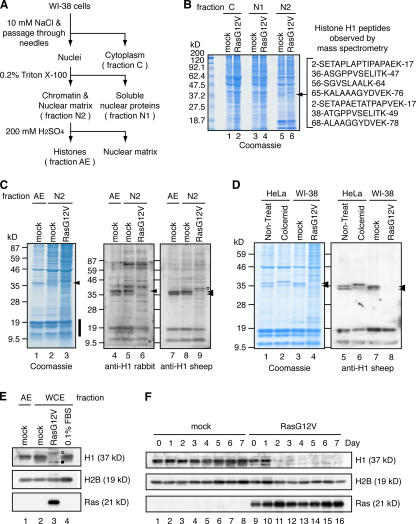
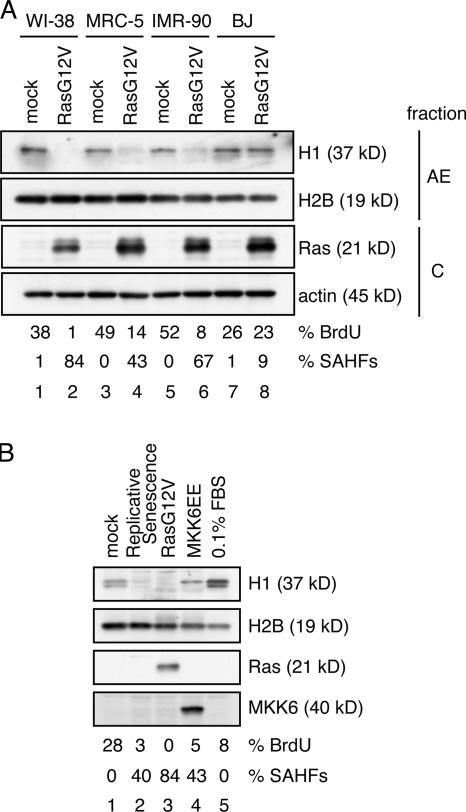
To analyze the molecular mechanisms involved in SAHF formation, we examined chromatin proteins in mock- and RasG12V-transfected WI-38 cells. The fractionation scheme used in this study is shown in Fig. 4 A. SDS-PAGE and Coomassie staining of the fractionated proteins revealed several protein bands showing distinct patterns in mock-transfected and senescent cells (Fig. 4 B). Notably, a protein band having an apparent molecular mass of 37 kD (hereafter referred to as a 37-kD protein) was found in chromatin-rich fraction N2 derived from mock-transfected cells but not from senescent cells (Fig. 4 B, arrow). Mass spectrometric analysis identified this protein as histone H1 (Fig. 4 B).
Figure 4.
Histone H1 is lost from the senescent cell. (A) Scheme showing the fractionation of WI-38 cells to isolate chromatin fractions. Hoechst 33342 staining of the chromatin-rich fraction (fraction N2) showed that SAHFs are retained in the nuclei present in this fraction (not depicted). (B) Coomassie staining of fractions C, N1, and N2 derived from mock- and RasG12V-transfected WI-38 cells. The 37-kD protein band from which peptide sequences of human histone H1 were obtained (right) is indicated with an arrow. (C) Chromatin-rich fractions (N2) were prepared as shown in A and analyzed by Coomassie staining and immunoblotting. AE fractions prepared from mock-transfected cells were used for comparison. The amount of loaded proteins was standardized with core histones (vertical bold line). Arrowheads indicate the positions of histone H1. The band of which signal intensity appeared to increase in RasG12V-transfected cells (lane 9, open circle) was detected in the absence of the first antibody, indicating that it is unrelated to histone H1 (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200604005/DC1). (D) Histones were prepared from mock- or RasG12V-transfected WI-38 cells by acid extraction and analyzed by Coomassie staining and immunoblotting. HeLa cells with or without colcemid treatment were used for comparison. (E) Quiescent cells were prepared by culturing mock-transfected cells in 0.1% FBS for 1 wk. WCEs were prepared from mock-transfected, RasG12V-transfected, or serum-starved quiescent cells and analyzed by immunoblotting. AE fractions obtained from mock-transfected cells were also examined. Two bands that showed slightly slower and faster migration than the 37-kD band were detected in RasG12V-transfected cells (lane 3; open and filled circles, respectively). However, these bands were also detected in the absence of the first antibody, indicating that they are unrelated to histone H1 (Fig. S2 B). (F) WCEs were prepared from mock- or RasG12V-transfected cells at the indicated time points after drug selection. Samples were analyzed by immunoblotting.
To confirm that the 37-kD protein is histone H1, we performed immunoblotting analyses using two different antihistone H1 antibodies prepared in rabbit or sheep (Fig. 4 C). Although the anti-H1 rabbit antibody detected a single band, the anti-H1 sheep antibody detected two bands showing apparent molecular masses of 37 kD (Fig. 4 C, lanes 4, 5, 7, and 8; arrowheads). This is probably the result of different specificities of these antibodies for histone H1 variants. In both cases, no anti-H1 signals were detected in fraction N2 derived from RasG12V-transfected senescent WI-38 cells (Fig. 4 C, lanes 6 and 9).
To further confirm that histone H1 is lost from senescent cell chromatin, histones were prepared from fraction N2 by acid extraction (Fig. 4 D). Histone H1 prepared from colcemid-treated HeLa cells (Fig. 4 D, lanes 2 and 6) showed lower mobility than that prepared from nontreated HeLa cells (Fig. 4 D, lanes 1 and 5) and mock-transfected WI-38 cells (Fig. 4 D, lanes 3 and 7), which is consistent with the hyperphosphorylation of histone H1 at metaphase. Histone H1 was not detected by Coomassie staining (Fig. 4 D, lane 4) or by anti-H1 sheep antibody (Fig. 4 D, lane 8). Collectively, we conclude that histone H1 is lost from the chromatin of RasG12V-transfected senescent WI-38 cells.
To determine whether histone H1 existed in any other fractions of RasG12V-transfected senescent WI-38 cells, we performed immunoblotting analyses of whole cell extracts (WCEs) using anti-H1 sheep antibody (Fig. 4 E). WCEs were prepared from mock-transfected, RasG12V-transfected, and serum-starved quiescent (0.1% FBS) WI-38 cells by directly suspending cell pellets in SDS sample buffer. No 37-kD histone H1 band was detected in the WCE from senescent cells (Fig. 4 E, lane 3). Time-course experiments showed that the majority of histone H1 was lost between days 2 and 3 after the completion of drug selection in the RasG12V-transfected cells (Fig. 4 F). These results indicate that histone H1 is lost from the entire cell in the Ras-induced senescence.
Histone H1 is lost from various types of senescent cells
We extended the experiments to include other types of normal human fibroblasts (MRC-5, IMR-90, and BJ; Fig. 5 A). The amount of histone H1 was moderately decreased in RasG12V-transfected MRC-5 and IMR-90 cells (Fig. 5 A, lanes 4 and 6) but not in BJ cells (Fig. 5 A, lane 8). Importantly, as shown in Fig. 5 A, the extent of histone H1 reduction is well correlated with the frequency of SAHF-positive cells after RasG12V expression. We also examined histone H1 in cellular senescence induced by other types of stimuli. Histone H1 was lost from or considerably decreased in senescent cells induced by extensive culturing (replicative senescence) or by the ectopic expression of a constitutive active form of MKK6 (MKK6EE) that activates the stress-induced MAPK p38 (Fig. 5 B; Wang et al., 2002; Iwasa et al., 2003). Moreover, the extent of histone H1 reduction was correlated with the frequency of SAHF-positive cells. Together, the loss of histone H1 is observed not only in RasG12V-transfected WI-38 cells but also in other types of RasG12V-transfected normal human fibroblasts and in senescent cells induced by other stimuli, extending its correlation with SAHF formation.
Figure 5.
Histone H1 is lost from various types of senescent cells. (A) Mock- or RasG12V-transfected cells (WI-38, MRC-5, IMR-90, and BJ) were fractionated as shown in Fig. 4 A at day 5 after drug selection. AE and cytoplasmic fractions (C) were analyzed by immunoblotting. The amounts of loaded proteins were calibrated by anti-H2B or antiactin signals, respectively. Transfected cells were scored for the percentages of BrdU- and SAHF-positive cells at harvest time (bottom; n = 200). (B) WI-38 cells were induced to senesce by the retroviral expression of RasG12V or a constitutive active form of MKK6 (MKK6EE). Replicative senescent cells were analyzed at 57 population doubling levels, at which cells could not reach confluence for 3 wk. Quiescent cells were prepared by culturing the mock-transfected cells in 0.1% FBS for 1 wk. WCEs were analyzed by immunoblotting. The percentages of BrdU- and SAHF-positive cells are indicated (bottom; n = 200).
Loss of histone H1 from senescent cell chromatin is posttranslationally regulated
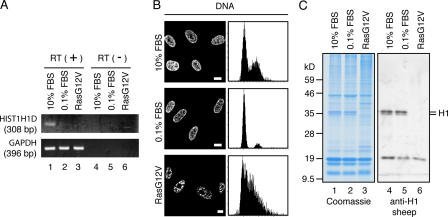
The expressions of most histone genes are coupled with DNA replication (Gunjan et al., 2005). As senescent cells do not undergo DNA replication (Serrano et al., 1997; Narita et al., 2003), it is expected that histone H1 is transcriptionally repressed. Indeed, RT-PCR analysis showed that the mRNA expression of HIST1H1D, one of the major histone H1 genes encoding histone H1.3, was repressed in RasG12V-transfected senescent WI-38 cells compared with young proliferating cells (10% FBS; Fig. 6 A, lanes 1 and 3). This raised the possibility that the loss of histone H1 from senescent cell chromatin is simply caused by the repression of de novo histone H1 synthesis. We found that the mRNA expression of HIST1H1D was not detected in quiescent WI-38 cells that had been serum starved for 1 wk (note that 1 wk is long enough for RasG12V expression to cause histone H1 loss; Figs. 4 F and 6 A, lane 2), as expected from the observation that the fraction of quiescent cells in S phase was very small (Fig. 6 B, right). Nevertheless, the serum-starved quiescent WI-38 cells contained the same amount of chromatin-bound histone H1 protein as proliferating WI-38 cells cultured with 10% FBS (Fig. 6 C), arguing that repression of the de novo histone H1 synthesis itself is not sufficient for the loss of histone H1 protein from chromatin. Therefore, the loss of histone H1 from senescent cells is most likely posttranslationally regulated.
Figure 6.
Loss of histone H1 from senescent cell chromatin is posttranslationally regulated. (A) Mock- and RasG12V-transfected WI-38 cells in 10% FBS and quiescent cells prepared by culturing noninfected WI-38 cells in 0.1% FBS for 1 wk were analyzed for mRNA expressions of HIST1H1D (encoding histone H1.3) and GAPDH by RT-PCR from total RNA in the presence (lanes 1–3) or absence (lanes 4–6) of reverse transcriptase (RT). (B) Cells were stained with DAPI (left) or propidium iodide and were analyzed for DNA contents with a laser-scanning cytometer (right). Although RasG12V-transfected cells completely stopped dividing, the laser-scanning cytometer analysis showed a large percentage of the cells in S phase. It is not clear whether this reflects cell cycle arrest in S phase or whether the laser-scanning cytometer did not accurately measure DNA contents in senescent cells, presumably as a result of the highly irregular patterns of DNA staining. (C) AE fractions were prepared from cells described in A and were analyzed by Coomassie staining (left) and immunoblotting (right). Bars, 10 μm.
Ectopic expression of histone H1 does not prevent RasG12V-induced senescence
To examine whether the loss of histone H1 has a causative role in senescence phenotype induction and SAHF formation, we prepared WI-38 cells that express short hairpin RNA to knock down the expression of histone H1. However, despite repeated trials, we did not succeed in knocking down the genes (unpublished data), probably as a result of the presence of multiple histone H1 variant genes.
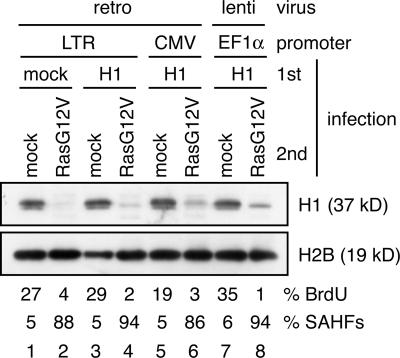
We next examined whether or not the ectopic expression of histone H1 prevented RasG12V-induced senescence. Young proliferating WI-38 cells were first infected with retrovirus (driven by the weak long terminal repeat promoter or the strong cytomegalovirus promoter) or lentivirus (driven by the strong EF1α promoter) expressing untagged histone H1 and were infected with RasG12V-expressing retrovirus (Fig. 7). The ectopic expression of histone H1 only slightly increased the amount of chromatin-bound histone H1 in RasG12V-transfected cells (Fig. 7, compare lane 2 with lanes 4, 6, and 8). However, it did not substantially prevent the RasG12V-induced loss of histone H1, growth arrest, or SAHF formation (Fig. 7) even when histone H1 was expressed by the strong cytomegalovirus and EF1α promoters. Together, these experiments do not conclude whether or not histone H1 loss has any causative role in cellular senescence because we could not manipulate the histone H1 level sufficiently to induce phenotypic changes.
Figure 7.
Ectopic expression of histone H1 does not prevent RasG12V-induced senescence. WI-38 cells were first infected with retrovirus (retro) or lentivirus (lenti) expressing histone H1 under the control of the indicated promoters and were infected with RasG12V-expressing retrovirus. Cell lysates were prepared from transfected cells at day 7 after drug selection and analyzed by immunoblotting. Transfected cells were scored for the percentages of BrdU- and SAHF-positive cells at day 3 after drug selection (bottom; n = 200). CMV, cytomegalovirus; LTR, long terminal repeat.
Ectopic expression of N-terminally EGFP-tagged histone H1 induces senescence phenotypes
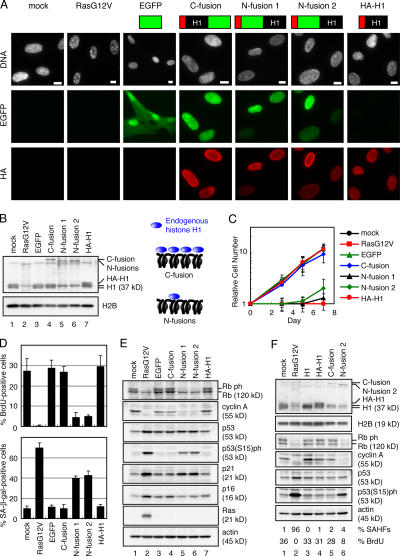
We next examined whether the ectopic expression of various types of recombinant histone H1 alone had any effects on WI-38 cells. Young proliferating WI-38 cells were transfected with one of four retroviruses expressing histone H1 fused to EGFP or HA (Fig. 8 A, top). Fluorescence microscopy revealed that all histone H1 fusion proteins were localized in nuclei and colocalized with mitotic chromosomes (Fig. 8 A and Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200604005/DC1), indicating their proficient chromatin binding in nuclei. Unexpectedly, cells expressing N-terminal EGFP-H1 fusion proteins (N fusions 1 and 2) showed decreased levels of chromatin-bound endogenous histone H1 compared with cells expressing C fusion or HA-H1 (Fig. 8 B). N fusions 1 and 2 have different linker amino acid sequences between EGFP and the histone H1 open reading frame (see Materials and methods) but showed the same phenotypes in WI-38 cells.
Figure 8.
Ectopic expression of N-terminally EGFP-tagged histone H1 induces premature senescence. (A) Nuclear localization of histone H1 fusion proteins. Schematic structures of histone H1 (histone H1.3) fusion proteins are indicated (top). Transfected WI-38 cells were examined for EGFP and stained with anti-HA antibody (HA) and DAPI (DNA) at day 2 after drug selection. Images are projections of whole nuclei. (B) Cell lysates were prepared from transfected cells at day 3 after drug selection and analyzed by immunoblotting. The amounts of loaded proteins were calibrated by anti-H2B signals. Endogenous histone H1 (H1) and H1 fusion proteins were analyzed using anti-H1 sheep antibody. (C) The number of transfected cells was scored at the indicated time points after drug selection. Error bars represent SD. (D) Transfected cells were scored for the percentages of BrdU- (top) and SA–β-gal–positive cells (bottom) at days 3 and 5 after drug selection, respectively (n = 200). (E) Cell lysates were prepared from transfected cells at day 3 after drug selection and analyzed by immunoblotting. The amounts of loaded proteins were calibrated by antiactin signals. (F) Cell lysates were analyzed as described in B and E. The percentages of BrdU- and SAHF-positive cells are indicated (bottom; n = 200). Bars, 10 μm.
Interestingly, concomitant with the decreases in histone H1 protein levels, the expression of N fusions caused severe growth defects in young WI-38 cells as revealed by the growth curves (Fig. 8 C) and the BrdU incorporation assay (Fig. 8 D, top). N fusions also induced a large, flat morphology and increased the percentage of SA–β-gal–positive cells (Fig. S3 B and Fig. 8 D, bottom), suggesting that the ectopic expression of N fusions induced premature senescence in WI-38 cells. Consistent with this idea, cells expressing N fusions showed decreased levels of phosphorylated Rb and cyclin A and increased levels of p21 and phosphorylated p53 (Fig. 8 E), which are typical molecular markers for senescent cells. In contrast, the ectopic expression of C fusion or HA-H1 did not induce senescence markers or decrease the levels of endogenous histone H1. The ectopic expression of untagged histone H1 slightly increased the levels of histone H1 protein but did not induce senescence phenotypes in WI-38 cells either (Fig. 8 F, lane 3). Collectively, these results suggest that the induction of senescence phenotypes is correlated with the decreased levels of chromatin-bound endogenous histone H1. Notably, N fusions did not induce the accumulation of p16 or SAHF formation (Fig. 8, E and A, DNA).
Coexpression of N-fusion histone H1 and HA-tagged HMGA2 induces SAHF formation
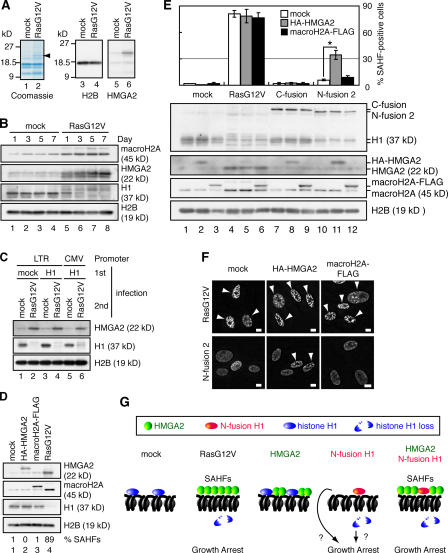
We found that a protein band showing an apparent molecular mass of 22 kD was accumulated in the AE fraction prepared from RasG12V-transfected senescent WI-38 cells (Fig. 9 A, arrowhead). Mass spectrometric analysis and immunoblotting analysis identified this protein as high mobility group A2 (HMGA2; Fig. 9 A). Similar observations have been reported recently (Narita et al., 2006). Time-course experiments indicated that the accumulation of chromatin-bound HMGA2 was inversely correlated with the loss of histone H1 from chromatin (Fig. 9 B, lanes 5–8). Interestingly, the amount of chromatin-bound HMGA2 was also increased in senescent WI-38 cells expressing both histone H1 and RasG12V (Fig. 9 C). We also showed that macroH2A was incorporated into chromatin in RasG12V-transfected cells as previously reported (Fig. 9 B, lanes 5–8; Zhang et al., 2005).
Figure 9.
Coexpression of HA-tagged HMGA2 and N-fusion histone H1 induces SAHF formation. (A) AE fractions were prepared from mock- and RasG12V-transfected WI-38 cells and analyzed by Coomassie staining and immunoblotting. A protein band having an apparent molecular mass of 22 kD from which peptide sequences of human HMGA2 were obtained is indicated with an arrowhead. (B) AE fractions were prepared from mock- and RasG12V-transfected WI-38 cells at the indicated time points after drug selection and were analyzed by immunoblotting. Histone H1 was analyzed using anti-H1 rabbit antibody. (C) Cell lysates were prepared from transfected WI-38 cells at day 7 after drug selection and were analyzed by immunoblotting as described in Fig. 8 B. CMV, cytomegalovirus; LTR, long terminal repeat. (D) Transfected WI-38 cells were analyzed as described in B. Histone H1 was analyzed using anti-H1 sheep antibody. The percentages of SAHF-positive cells are indicated (bottom; n = 200). (E) Transfected WI-38 cells were scored for the percentage of SAHF-positive cells at day 3 after drug selection (top; n = 200). *, P < 0.001 by t test. Error bars represent SD. Transfected WI-38 cells were analyzed as described in Fig. 8 B (bottom). (F) Representative images of SAHF-positive cells. Transfected WI-38 cells were stained with DAPI at day 3 after drug selection. Arrowheads indicate SAHF-positive nuclei. (G) A model of the role of histone H1 dissociation and HMGA2 accumulation in inducing growth arrest and SAHF formation. p53-p21-pRb–dependent growth arrest is mediated by either histone H1–free chromatin itself or signaling elicited by unbound histone H1. SAHFs are formed when histone H1 is replaced with HMGA2, which occurs only when RasG12V is expressed or N-fusion H1 and HMGA2 are coexpressed. Bars, 10 μm.
To investigate the role of macroH2A and HMGA2 in senescence phenotype induction and SAHF formation, young proliferating WI-38 cells were transfected with HA-tagged HMGA2 (HA-HMGA2) or FLAG-tagged macroH2A (macroH2A–FLAG; Fig. 9 D). The amounts of chromatin-bound HA-HMGA2 and macroH2A–FLAG were comparable with those of endogenous proteins observed in RasG12V-transfected cells (Fig. 9 D, lanes 2–4; and Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200604005/DC1). Furthermore, IF analysis revealed that both HA-HMGA2 and macroH2A–FLAG were localized in nuclei (Fig. S4 B and not depicted). The expression of HA-HMGA2 or macroH2A–FLAG did not induce senescence phenotypes, including growth defects, a large, flat morphology, or SAHF formation (Fig. 9 D, bottom; Fig. S4, C and D; and not depicted).
To investigate whether the overexpression of HMGA2 or macroH2A collaborates with the reduced endogenous histone H1 level found in N fusion–expressing cells, young WI-38 cells expressing HA-HMGA2 or macroH2A–FLAG were further transfected with the retrovirus expressing N fusion 2 and were scored for SAHF-positive cells (Fig. 9, E and F). The coexpression of HA-HMGA2 and N fusion 2 significantly increased the percentage of SAHF-positive cells (P < 0.001), whereas the coexpression of macroH2A–FLAG and N fusion 2 did not. The coexpression of HA-HMGA2 and N fusion 1 led to similar results, but the coexpression of HA-HMGA2 and HA-H1 did not (Fig. S5), suggesting that the removal of endogenous histone H1 from chromatin by N-fusion histone H1 rather than the production of recombinant histone H1 itself is important for SAHF formation. Together, these results indicate that the simultaneous overexpression of HA-HMGA2 and N-fusion histone H1 leads to SAHF formation. However, cells coexpressing HA-HMGA2 and N fusion 2 did not show the accumulation of p16, at least up to day 3 after drug selection (unpublished data).
Discussion
In this study, we found that the induction of senescence phenotypes is accompanied by the loss of linker histone H1 from and the accumulation of HMGA2 to chromatin. The loss of histone H1 is not simply through the repression of histone H1 mRNA expression but is most likely through a posttranslational mechanism. The ectopic expression of N-terminally EGFP-tagged histone H1 in WI-38 cells acutely induces senescence phenotypes except SAHF formation and p16 accumulation, which is concomitant with a decrease in the chromatin-bound endogenous histone H1 protein level. This result suggests that distinct pathways are responsible for p53-p21-pRb–dependent growth arrest and SAHF formation with or without p16 accumulation in cellular senescence. Notably, when HMGA2 was coexpressed in this condition, significant amounts of SAHFs were formed (P, 0.001), suggesting that the substitution of HMGA2 for histone H1 is required for SAHF formation. Previously, it was reported that the amount of histone H1 protein was decreased in replicative senescence (Mitsui et al., 1980). However, the biological significance of that observation has remained a mystery. We suggest that SAHFs are a novel type of chromatin condensation involving alterations in linker DNA–binding proteins.
SAHFs are a novel type of chromatin condensation
We propose that individual SAHFs originate from individual chromosomes. This proposition is based on several lines of evidence. The majority of SAHFs contained one centromere at their periphery and was surrounded by DAPI-negative regions where RNA polymerase II exists and de novo transcription occurs. Such features of the DAPI-negative regions have been proposed and described for ICD, a system of channels that run between chromosome territories and metabolize and transport RNA transcripts (Cremer and Cremer, 2001). Finally, the painting probe for chromosome 12 stained exactly one SAHF.
A recent study has demonstrated the presence of RNA transcripts and transcription factors inside the chromosome territory, challenging the conventional ICD model (Williams, 2003). A revised model has been developed in which a single chromosome territory is composed of condensed chromatin subdomains as well as channels canalizing the territory and ultimately draining into ICDs. Active genes are proposed to exist on the surface of the condensed chromatin subdomains inside the territory. We found that RNA polymerase II exists more or less diffusely throughout the nucleus in young WI-38 cells, supporting this revised model. However, we also found that RNA polymerase II and RNA transcripts are excluded from SAHFs in senescent cells. These observations suggest that SAHF is a heterochromatinized portion of an individual chromosome whose chromatin is so tightly condensed that active genes are virtually excluded.
SAHFs are a novel type of chromatin condensation not only from a cytological viewpoint but also from a biochemical viewpoint. We found that SAHFs do not show the phosphorylation of histone H3 at either Ser10 or Ser28 or the phosphorylation of histone H2B at Ser14, which are hallmarks of mitotic and apoptotic chromatins, respectively. It has been reported that SAHFs exhibit features of transcriptionally inactive heterochromatin (Narita et al., 2003; Zhang et al., 2005). Consistent with such earlier studies, we also observed that SAHFs were strongly stained with anti-H3(K9)me antibody. However, we found that the total amount of H3(K9)me remained unchanged at least for 1 wk after the induction of senescence, during which the majority of cells became SAHF positive. These results suggest that although de novo histone methylation may occur locally and repress gene expression at some particular gene loci, a genome-wide increase in the total amount of H3(K9)me does not occur in senescent cells. Although these results do not exclude the involvement of transcriptionally inactive heterochromatin in SAHFs, they suggest that SAHFs are generated by a mechanism that should be distinguished from those involved in conventional chromosome condensation and heterochromatinization.
Loss of histone H1 and senescence phenotypes
We found that linker histone H1 is lost from senescent cells. Because it is generally believed that histone H1 facilitates chromatin condensation and acts mainly as a transcriptional repressor (Hayes and Hansen, 2001), it was unexpected that histone H1 was lost instead of increased in SAHFs. Although we could not definitely conclude whether the loss of histone H1 is causally linked to the induction of senescence phenotypes and SAHF formation, the following observations suggest that the alterations in histone H1 metabolism are involved in a senescence-inducing mechanism. First, the amount of histone H1 protein was lost or considerably reduced in a variety of cellular senescence occurring in different cell types and through different stimuli. Second, the extent of histone H1 loss in such occasions was well correlated with the frequency of SAHF formation. Finally, senescence phenotypes except SAHF formation and p16 accumulation were induced in young WI-38 cells by the ectopic expression of N fusion but not of C fusion, HA-H1, or untagged recombinant H1 proteins. These proteins were produced at similar levels (Fig. 8, B and F) and were efficiently bound to chromatin (Fig. 8 A and Fig. S3 A), suggesting that senescence was not induced by nonspecific effects caused by the overproduction of nonfunctional histone H1 proteins.
Interestingly, the senescence induction and reduced level of endogenous histone H1 were well correlated: only N fusions showed both effects, whereas the other recombinant histone H1 proteins showed neither. We do not know why N fusions but not C fusion affect the level of endogenous histone H1 and induce SAHF formation. It has been reported that N-terminal GFP-tagged histone H1.1 binds to chromatin with higher affinity than C-terminal GFP-tagged histone H1.1 in SK-N-SH neuroblastoma cells (Hendzel et al., 2004). It is possible that such different affinities cause different stabilities of endogenous histone H1 protein. Alternatively, N-terminal GFP may perturb the incorporation of endogenous histone H1 into neighboring nucleosomes by steric hindrance, for example.
Embryonic stem cells homozygously deleted of three out of six somatic histone H1 variant genes were viable and showed apparently normal growth. Interestingly, however, the DNA methylation and expression patterns of some specific loci, including imprinted genes, were deregulated (Fan et al., 2005). These results raise the possibility that the absence of histone H1 in senescent cells may specifically affect the expression of genes involved in senescence phenotypes (Fig. 9 G).
The amounts of histone proteins, especially those of chromatin-unbound forms, are strictly regulated in cells. It has been reported that the chromatin-unbound free core histones are degraded in a Rad53-dependent manner in yeast. In rad53 cells, chromatin-unbound core histones persist at the completion of S phase, leading to impaired cell growth (Gunjan et al., 2005). Therefore, another interesting possibility is that the production of free histone H1, not the reduction of chromatin-bound histone H1, positively triggers a signal leading to senescence (Fig. 9 G). A hypothesis reminiscent of this case was reported previously: after exposure to x rays, histone H1.2, a histone H1 variant, moves to the cytoplasm, where it induces cytochrome c release from mitochondria to activate caspase-dependent destruction cascades (Konishi et al., 2003).
Altered balance between histone H1 and HMGA2 bound to chromatin in SAHF formation
Recent studies have emphasized the dynamic binding of histone H1 to chromatin. Histone H1 associates with and dissociates from chromatin in a rapid turnover (Lever et al., 2000; Bustin et al., 2005). Histone H1 dissociation from chromatin may be enhanced by activities that destabilize the histone H1–chromatin association. HMG proteins are the second most abundant chromatin proteins after histones and consist of three subfamilies: HMGA, HMGB, and HMGN. It was reported that HMG proteins and histone H1 compete with each other for the same binding site, the linker DNA. Specifically, microinjected excess amounts of HMG proteins leads to high turnover rates and reduces the residence time of endogenous chromatin-bound histone H1, presumably by being excluded from chromatin by the excess HMG proteins (Catez et al., 2004).
On the surface, the failure of the ectopic expression of histone H1 to prevent RasG12V-induced senescence appears to refute the causative role of histone H1 loss in senescence induction. However, the amounts of chromatin-bound HMGA2 and chromatin-bound histone H1 were similarly increased and reduced, respectively, in RasG12V-expressing cells irrespective of histone H1 coexpression (Figs. 7 and 9 C), suggesting that RasG12V expression shifts the balance of HMGA2 and histone H1 association with the linker DNA in favor of HMGA2 (Fig. 9 G). The ectopic expression of histone H1 may fail to restore the balance in favor of histone H1 to a degree that is sufficient to prevent senescence in the RasG12V-expressing condition. It is not clear why the ectopic expression of histone H1 even by the strong promoters failed to restore the chromatin-bound histone H1 protein level in RasG12V-transfected cells. It is possible that histone H1 is actively removed from chromatin in such cells. Alternatively, senescent cell chromatin may be incompetent to incorporate newly synthesized histone H1. It is also not clear how endogenous histone H1 disappears from senescent cells. Further studies are warranted to determine whether histone H1 loss plays a causative role in senescence induction.
When HMGA2 was ectopically expressed in young WI-38 cells to the level found in RasG12V-expressing cells, no senescence phenotypes were induced (Fig. 9 D and Fig. S4). Conversely, when the amount of chromatin-bound histone H1 was reduced by N-fusion expression, no SAHFs were formed either (Fig. 8 A). SAHFs were formed only when N fusions accompanying histone H1 loss and HMGA2 were coexpressed. We hypothesize that both the reduced association of histone H1 and the increased association of HMGA2 with chromatin, which reflect the substitution of HMGA2 for histone H1, are required for SAHF formation (Fig. 9 G). Therefore, SAHFs may be viewed as heterochromatin in which the balance of the two major linker DNA–binding proteins (histone H1 and HMGA2) is altered.
We also found that the coexpression of N fusion 2 and HA-HMGA2 did not induce p16 accumulation, at least up to day 3 after drug selection. Additional conditions may be required to trigger the full-blown senescence program, including the activation of p16.
Materials and methods
Cell culture and retroviral gene transfer
Normal human fibroblasts WI-38, MRC-5, and IMR-90 were purchased from Coriell Cell Repositories. BJ was purchased from American Type Culture Collection. The cells were cultured in DME supplemented with 10% FBS. Phoenix cells (provided by G. Nolan, Stanford University, Stanford, CA) were transfected with retroviral vectors using FuGene 6 (Roche). Culture supernatants were supplemented with 8 μg/ml polybrene (Sigma-Aldrich) and incubated with target cells. Infected cells were selected by incubating with 2.5 μg/ml puromycin (Sigma-Aldrich) or 600 μg/ml G418 (Nacalai Tesque) for 3 or 5 d, respectively.
Retroviral vectors used in this study are pMX-puro and pMX-neo (gift from T. Kitamura, University of Tokyo, Tokyo, Japan). MKK6EE cDNA was provided by E. Nishida (Kyoto University, Kyoto, Japan) and Y. Goto (University of Tokyo, Tokyo, Japan). MacroH2A cDNA was provided by K. Shibahara (National Institute of Genetics, Mishima, Japan). Amino acid sequences between EGFP and the histone H1 open reading frame in N and C fusions are as follows: EGFP-Leu-Glu-Ala-Ala-Ala-Ile-Gly-Ser-H1 for N fusion 1, EGFP-Pro-Glu-Phe-Thr-H1 for N fusion 2, and H1-Pro-Glu-Phe-Thr-EGFP for C fusion.
Antibodies
The following primary antibodies were used: anti-RNA polymerase II (8WG16), anti-HA (16B12), and anti-HMGA2 (Covance); anticyclin A, anti-p16, anti-p21, anti-p53 (Bp53-12), anti–H-Ras, and anti-MKK6 (Santa Cruz Biotechnology, Inc.); antiphospho-S10 histone H3, antiphospho-S14 histone H2B, antidimethyl-K9 histone H3, antihistone H2B, and antihistone macroH2A1 (Upstate Biotechnology); anti-BrdU (BMC9318; Roche); antiactin (Chemicon); antihistone H1 sheep (Abcam); anti-Rb (G3-245; BD Biosciences); antiphospho-S15 p53 (Cell Signaling); anti–CENP-B (provided by H. Masumoto, Nagoya University, Nagoya, Japan); antiphospho-S28 histone H3 (provided by T. Urano, Nagoya University); and antihistone H1 rabbit (provided by K. Ohsumi, Tokyo Institute of Technology, Kanagawa, Japan).
IF and DNA content analyses
Cells were fixed with 3% formaldehyde in PBS at 4°C for 20 min and permeabilized with Triton buffer (TB; 0.5% Triton X-100 in PBS) for 10 min. After incubation in TB containing 5% skim milk for 1 h, the cells were treated with primary antibody and incubated with AlexaFluor488- or Cy-3–conjugated secondary antibody (Invitrogen and Jackson ImmunoResearch Laboratories, respectively). DNA was stained with 1 μg/ml DAPI in PBS for 10 min. Images were acquired at room temperature with a deconvolution microscope (IX70; Olympus; and DeltaVision; Applied Precision) equipped with a 40× NA 1.35 oil immersion objective lens (UApo/340; Olympus) and a charge-coupled device camera (CH350; Photometrics). The images were deconvolved and processed by SoftWoRx (Applied Precision) and Photoshop (Adobe). Unless otherwise mentioned, images are single optical sections.
For DNA content analysis, cells were fixed with 3% formaldehyde and permeabilized with TB. Then, the cells were incubated in PBS containing 50 μg/ml propidium iodide and 200 μg/ml RNase A at 37°C for 30 min. DNA content was analyzed with a laser-scanning cytometer (LSC2; Olympus).
In situ labeling of nascent RNAs and BrdU incorporation assay
In situ labeling of nascent RNAs was performed as previously described (Wansink et al., 1993) with slight modifications. In brief, cells were rinsed with glycerol buffer (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol, and Complete protease inhibitors [Roche]) containing 0.05% Triton X-100 at 4°C for 1 min. Then, the cells were incubated in transcription buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.5 mM EGTA, 100 mM KCl, 25% glycerol, 25 μM S-adenosylmethionine [New England Biolabs, Inc.], 0.5 mM each of BrU [Sigma-Aldrich], ATP, CTP, and GTP, 8 U/ml RNase inhibitor [Takara], and Complete protease inhibitors) at 35°C for 10 min. Incorporated BrU was visualized by IF analysis using anti-BrdU antibody. For BrdU incorporation assay, cells were cultured in DME supplemented with 100 μM BrdU for 5 h, fixed with 2% formaldehyde at 4°C for 15 min, and permeabilized with TB at room temperature for 5 min. After treatment with 1.5 N HCl at room temperature for 10 min, incorporated BrdU was visualized by IF analysis using anti-BrdU antibody.
FISH
FISH was performed using a chromosome 12–specific probe conjugated with Cy-3 (StarFISH; Cambio). FISH on metaphase spreads was performed according to the manufacturer's protocol. FISH on senescent cells was performed as described previously (Croft et al., 1999).
Subcellular fractionation, mass spectrometry, and immunoblotting
Cells were harvested and incubated in buffer A (40 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.5 mM DTT, 1 mM sodium orthovanadate, 1 mM sodium fluoride, 100 nM okadaic acid, and Complete protease inhibitors) containing 10 mM NaCl at 4°C for 10 min. Then, the cells were passed through a 26- (mock-transfected cells) or 23-gauge (RasG12V-transfected cells) needle ∼20 times to disrupt the cell membrane and were centrifuged at 1,000 g for 10 min at 4°C. Supernatants were removed (fraction C), and pellets were incubated in buffer A containing 150 mM NaCl and 0.2% Triton X-100 at 4°C for 10 min. After centrifugation at 1,000 g for 10 min at 4°C, supernatants were removed (fraction N1). Pellets (fraction N2) were resuspended in the SDS sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, and 20% glycerol, colored with bromophenol blue) and sonicated. For acid extraction of histones, fraction N2 was incubated with 200 mM sulfuric acid at 4°C for 30 min and centrifuged at 20,000 g for 10 min at 4°C. Supernatants were supplemented with 1/4 vol of 100% trichloroacetic acid and incubated at 4°C for 10 min. After centrifugation at 20,000 g for 5 min at 4°C, pellets were rinsed with cold acetone, dried, and resuspended in the SDS sample buffer. For the preparation of cell lysates (Figs. 7–9 ), cells were suspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM DTT, 1 mM sodium orthovanadate, 1.25 mM sodium fluoride, 100 nM okadaic acid, 10 mM β-glycerophosphate, and Complete protease inhibitors) at 4°C for 30 min. After centrifugation at 20,000 g at 4°C for 10 min, the supernatants were used for immunoblotting analyses using anti-Rb, cyclin A, p16, p21, p53, P-p53, Ras, and actin antibodies. Pellets were suspended in the SDS sample buffer and used for immunoblotting analyses using anti-H1, HMGA2, and H2B antibodies.
For mass spectrometry, protein bands were cut out from SDS-PAGE gels and digested with trypsin (Promega). The eluted peptides were analyzed with a hybrid mass spectrometer (QSTAR XL; Applied Biosystems) equipped with a nanoelectrospray ion source (MDS Protana).
For immunoblotting, proteins were loaded onto SDS-PAGE gels, transferred onto polyvinylidene fluoride membranes (Millipore), and detected with ECL immunoblotting detection reagents (GE Healthcare).
RT-PCR and SA–β-gal staining
RT-PCR was performed with an RNA PCR kit (Takara) according to the manufacturer's protocol. PCR primers used in this study are HIST1H1D (5′-CAAGCCTAGGAAGCCTGCTG-3′ and 5′-AAAGGGGAACGTCCCGCCAG-3′) and GAPDH (5′-ATTTGGTCGTATTGGGCGCCTGGTC-3′ and 5′-TTGTCATACTTCTCATGGTTCACAC-3′).
For SA–β-gal staining, cells were fixed with 0.5% glutaraldehyde in PBS at room temperature for 10 min and incubated in staining solution (0.65 mg/ml X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactoside], 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 1 mM MgCl2 in PBS, pH 6.0) at 37°C overnight.
Online supplemental material
Fig. S1 shows the specificities of FISH signals and anti-BrdU IF signals. Fig. S2 shows the specificity of antihistone H1 sheep antibody. Fig. S3 shows the colocalization of N-fusion H1 and mitotic chromosomes and the enlargement of WI-38 cells expressing N-fusion H1. Fig. S4 shows the effect of HA-HMGA2 expression on cell growth and SAHF formation. Fig. S5 shows the effect of N-fusion 1 expression on SAHF formation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200604005/DC1.
Supplementary Material
Acknowledgments
We thank T. Kitamura for providing retroviral vectors, E. Nishida and Y. Goto for human MKK6EE cDNA, K. Shibahara for macroH2A cDNA, H. Masumoto for anti–CENP-B antibody, T. Urano for antiphospho-S28 histone H3 antibody, K. Ohsumi for antihistone H1 rabbit antibody, and G. Nolan for Phoenix cells. The excellent secretarial work of A. Katayama, M. Sasaki, and M. Sakamoto and the technical assistance of M. Tamura are also acknowledged.
This work was supported by a Center of Excellence grant and Grants-in-Aid for Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used in this paper: AE, acid extracted; CENP-B, centromere protein B; HMG, high mobility group; ICD, interchromosome domain; IF, immunofluorescence; SA–β-gal, senescence-associated β-galactosidase; SAHF, senescence-associated heterochromatic focus; TB, Triton buffer; WCE, whole cell extract.
References
- Braig, M., and C.A. Schmitt. 2006. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 66:2881–2884. [DOI] [PubMed] [Google Scholar]
- Bustin, M., F. Catez, and J.H. Lim. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell. 17:617–620. [DOI] [PubMed] [Google Scholar]
- Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 120:513–522. [DOI] [PubMed] [Google Scholar]
- Catez, F., H. Yang, K.J. Tracey, R. Reeves, T. Misteli, and M. Bustin. 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 24:4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., T. Nikitina, J. Zhao, T.J. Fleury, R. Bhattacharyya, E.E. Bouhassira, A. Stein, C.L. Woodcock, and A.I. Skoultchi. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 123:1199–1212. [DOI] [PubMed] [Google Scholar]
- Gunjan, A., J. Paik, and A. Verreault. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie. 87:625–635. [DOI] [PubMed] [Google Scholar]
- Hayes, J.J., and J.C. Hansen. 2001. Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev. 11:124–129. [DOI] [PubMed] [Google Scholar]
- Hayflick, L., and P.S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585–621. [DOI] [PubMed] [Google Scholar]
- Hendzel, M.J., M.A. Lever, E. Crawford, and J.P. Th'ng. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028–20034. [DOI] [PubMed] [Google Scholar]
- Ishikawa, F. 2003. Cellular senescence, an unpopular yet trustworthy tumor suppressor mechanism. Cancer Sci. 94:944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa, H., J. Han, and F. Ishikawa. 2003. Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells. 8:131–144. [DOI] [PubMed] [Google Scholar]
- Konishi, A., S. Shimizu, J. Hirota, T. Takao, Y. Fan, Y. Matsuoka, L. Zhang, Y. Yoneda, Y. Fujii, A.I. Skoultchi, and Y. Tsujimoto. 2003. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 114:673–688. [DOI] [PubMed] [Google Scholar]
- Lever, M.A., J.P. Th'ng, X. Sun, and M.J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 408:873–876. [DOI] [PubMed] [Google Scholar]
- Lowe, S.W., E. Cepero, and G. Evan. 2004. Intrinsic tumour suppression. Nature. 432:307–315. [DOI] [PubMed] [Google Scholar]
- Mitsui, Y., H. Sakagami, S. Murota, and M. Yamada. 1980. Age-related decline in histone H1 fraction in human diploid fibroblast cultures. Exp. Cell Res. 126:289–298. [DOI] [PubMed] [Google Scholar]
- Narita, M., S. Nunez, E. Heard, M. Narita, A.W. Lin, S.A. Hearn, D.L. Spector, G.J. Hannon, and S.W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 113:703–716. [DOI] [PubMed] [Google Scholar]
- Narita, M., M. Narita, V. Krizhanovsky, S. Nunez, A. Chicas, S.A. Hearn, M.P. Myers, and S.W. Lowe. 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 126:503–514. [DOI] [PubMed] [Google Scholar]
- Peterson, C.L., and M.A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546–R551. [DOI] [PubMed] [Google Scholar]
- Ramirez, R.D., C.P. Morales, B.S. Herbert, J.M. Rohde, C. Passons, J.W. Shay, and W.E. Wright. 2001. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 15:398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, M., A.W. Lin, M.E. McCurrach, D. Beach, and S.W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 88:593–602. [DOI] [PubMed] [Google Scholar]
- Thoma, F., T. Koller, and A. Klug. 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83:403–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., J.X. Chen, R. Liao, Q. Deng, J.J. Zhou, S. Huang, and P. Sun. 2002. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol. Cell. Biol. 22:3389–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink, D.G., W. Schul, I. van der Kraan, B. van Steensel, R. van Driel, and L. de Jong. 1993. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierich, C., A. Brero, S. Stein, J. von Hase, C. Cremer, T. Cremer, and I. Solovei. 2003. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 11:485–502. [DOI] [PubMed] [Google Scholar]
- Williams, R.R. 2003. Transcription and the territory: the ins and outs of gene positioning. Trends Genet. 19:298–302. [DOI] [PubMed] [Google Scholar]
- Zhang, R., M.V. Poustovoitov, X. Ye, H.A. Santos, W. Chen, S.M. Daganzo, J.P. Erzberger, I.G. Serebriiskii, A.A. Canutescu, R.L. Dunbrack, et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 8:19–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.