Abstract
Three components of the chloroplast protein translocon, Tic110, Hsp93 (ClpC), and Tic40, have been shown to be important for protein translocation across the inner envelope membrane into the stroma. We show the molecular interactions among these three components that facilitate processing and translocation of precursor proteins. Transit-peptide binding by Tic110 recruits Tic40 binding to Tic110, which in turn causes the release of transit peptides from Tic110, freeing the transit peptides for processing. The Tic40 C-terminal domain, which is homologous to the C terminus of cochaperones Sti1p/Hop and Hip but with no known function, stimulates adenosine triphosphate hydrolysis by Hsp93. Hsp93 dissociates from Tic40 in the presence of adenosine diphosphate, suggesting that Tic40 functions as an adenosine triphosphatase activation protein for Hsp93. Our data suggest that chloroplasts have evolved the Tic40 cochaperone to increase the efficiency of precursor processing and translocation.
Introduction
Most proteins in chloroplasts are encoded by the nuclear genome and synthesized in the cytosol as precursors with N-terminal targeting signals called transit peptides. Precursor import into chloroplasts is mediated by a protein translocon, which is composed of the Toc (translocon at the outer envelope membrane of chloroplasts) and the Tic (translocon at the inner envelope membrane of chloroplasts) proteins and stromal chaperones (Soll and Schleiff, 2004; Kessler and Schnell, 2006). During import, transit peptides of precursors first interact with the Toc and then with the Tic proteins (Schnell et al., 1994). When sufficient ATP is present in the stroma, precursors are translocated across the inner envelope membrane into the stroma (Theg et al., 1989) and the transit peptide is removed by the stromal processing peptidase during the translocation (Kessler and Blobel, 1996; Richter and Lamppa, 1998; Chou et al., 2003; Chen and Li, 2006). Although many Tic and Toc proteins have been identified, the interactions among the translocon components and the mechanistic steps of the import process are largely unknown.
Tic110 is the major Tic protein identified (Kessler and Blobel, 1996; Lübeck et al., 1996). Tic110 has an N-terminal membrane anchor and a large stroma-located hydrophilic domain (Jackson et al., 1998; Inaba et al., 2003). The N-terminal half of the Tic110 stromal domain binds transit peptides directly. Tic110 is therefore thought to be the stroma-side receptor for transit peptides and the first protein that binds precursors as they emerge from the inner membrane channel (Inaba et al., 2003, 2005). Hsp93 (ClpC) belongs to the Clp/Hsp100 subfamily of the AAA+ (ATPase associated with various cellular activities) family of proteins (Schirmer et al., 1996; Hanson and Whiteheart, 2005). It is present both in a soluble form in the stroma and in an inner membrane–tethered form (Nielsen et al., 1997; Kouranov et al., 1998). It is proposed to function as the motor for chloroplast protein translocation, as translocation requires ATP hydrolysis in the stroma (Theg et al., 1989) and Hsp93 is the only known ATPase stably associated with the entire translocon complex (Nielsen et al., 1997).
Tic40 has a similar topology to Tic110. The stroma-located hydrophilic domain of Tic40 is composed of a tetratricopeptide repeat (TPR) domain followed by a C-terminal domain homologous to the C terminus of cochaperones Sti1p/Hop (Hsp70/Hsp90-organizing protein) and Hip (Hsp70-interacting protein; Frydman and Höhfeld, 1997; Chou et al., 2003). TPR domains, which consist of highly degenerated 34-amino-acid repeats, are present in proteins of diverse functions and are known to mediate protein–protein interactions (Höhfeld et al., 1995; Lamb et al., 1995; Scheufler et al., 2000). In contrast, no molecular function had been assigned to the Hip and Hop C-terminal domain, even though this domain is highly conserved from yeast (Sti1p) to human (Hip and Hop) to plants (Tic40).
Tic40 has been shown to function at a similar stage of import to Tic110 and Hsp93 (Chou et al., 2003; Kovacheva et al., 2005). Arabidopsis thaliana tic40-null mutants are extremely pale green and retarded in development but still viable, suggesting that Tic40 plays a nonessential stimulatory/regulatory role during import. Chloroplasts isolated from tic40 mutants are normal in binding of precursors to the outer envelope membrane but are specifically defective in precursor translocation across the inner envelope membrane. Precursors tend to be released from tic40 mutant chloroplasts before the completion of translocation. Based on these results, we have proposed that Tic40 may function as a cochaperone to coordinate the action of Tic110 and Hsp93 (Chou et al., 2003). This hypothesis predicts that the Tic40 will physically interact with Tic110 and/or Hsp93 and the interaction will have regulatory or mechanical consequence to the import of precursor proteins into the stroma. In this work, we have analyzed the molecular interactions among Tic40, Tic110, and Hsp93. We provide evidence to show that Tic40 regulates transit peptide–Tic110 interaction and Hsp93 ATP hydrolysis.
Results
Tic40 TPR domain binds Tic110
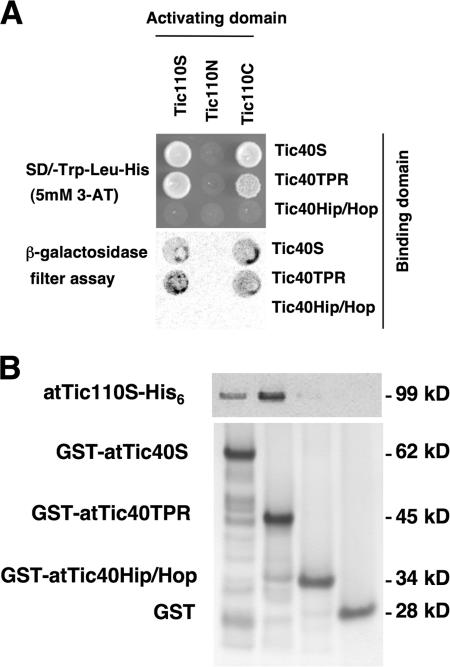
We first investigated the interaction between Tic40 and Tic110. We tested whether they could directly interact with each other by yeast two-hybrid assays. Pea Tic40 cDNAs encoding the entire stroma-located hydrophilic domain (Tic40S; Fig. 1), TPR subdomain (Tic40TPR), and C-terminal Sti1p/Hop/Hip-homologous subdomain (Tic40Hip/Hop, hereafter referred to as the Hip/Hop domain) were fused with the GAL4 DNA-binding domain. Pea Tic110 cDNAs encoding the entire stroma-located hydrophilic domain (Tic110S) and the N- and C-terminal parts of this stromal domain (Tic110N and Tic110C; Fig. 1) were fused with the GAL4 activation domain. As shown in Fig. 2 A, Tic40S interacted with Tic110S. This interaction is most likely due to the interaction between Tic40TPR and Tic110C (Fig. 2 A). We further confirmed this interaction by in vitro pull-down assays. GST fused with A. thaliana Tic40 stroma l hydrophilic domain, the TPR subdomain, or the Hip/Hop subdomain was produced and purified from Escherichia coli (GST-atTic40S, -atTic40TPR, and -atTic40Hip/Hop, respectively; Fig. 1). They were incubated with A. thaliana Tic110S fused to a C-terminal His6 tag (atTic110S-His6; identical to atTic11093-966 in Inaba et al. [2003]). As shown in Fig. 2 B, GST-atTic40S and -atTic40TPR, but not GST- atTic40Hip/Hop or GST, could pull down atTic110-His6.
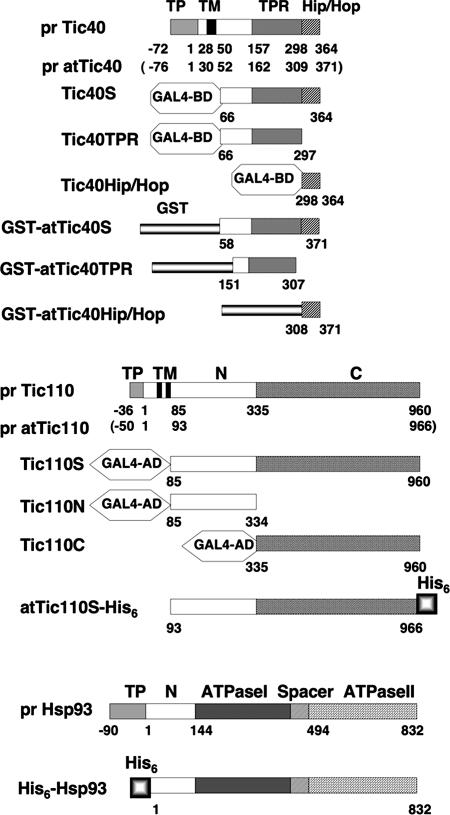
Figure 1.
Constructs of Tic40, Tic110, and Hsp93 used in this study. Numbers below each construct specify the amino acid residue numbers, with the first amino acid of the mature protein as 1 and the residues for transit peptides negative. Proteins from A. thaliana are specified with “at.” Others are from pea. Numbers in parentheses specify the corresponding residue numbers of the A. thaliana proteins. TP, transit peptide; TM, transmembrane region; pr, precursor form; GAL4-BD and -AD, GAL4 DNA binding and activation domains, respectively, for yeast two-hybrid assays.
Figure 2.
Tic40 TPR domain binds to Tic110. (A) Yeast two-hybrid analyses of interacting domains between Tic40 and Tic110. (top) Yeast cells containing each of the activation domain constructs were transformed with each of the binding domain constructs and plated on the synthetic dropout (SD) medium containing 5 mM 3-amino-1,2,4-triazole (3-AT) without tryptophan, leucine, and histidine. (bottom) Filter replicates from the SD plates were then assayed for β-galactosidase activity. (B) GST pull-down assay analyzing direct interaction between Tic40 and Tic110. GST fused to the entire Tic40 stromal soluble domain (GST-atTic40S), the TPR subdomain (GST-atTic40TPR), or the Hip/Hop subdomain (GST-atTic40Hip/Hop), or GST itself was incubated with atTic110S-His6 at 4°C for 1 h. GST fusion proteins were recovered by binding to glutathione resin and eluted with glutathione.
Tic40 does not bind precursors directly
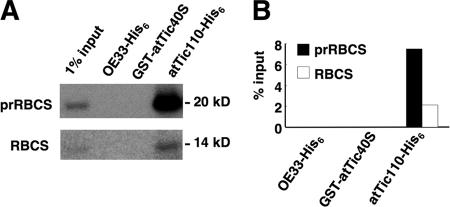
We next investigated whether Tic40 had a direct interaction with precursors. GST-atTic40S was incubated with in vitro–translated 35S-labeled precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase oxygenase (prRBCS), a representative chloroplast stromal protein imported from the cytosol. Recombinant atTic110S-His6 was used as a positive control, and the 33-kD subunit of the oxygen-evolving complex of the thylakoid membrane fused to a His6 tag (OE33-His6) was used as a negative control. As shown in Fig. 3, atTic110S-His6 indeed preferentially bound to prRBCS as previously reported (Inaba et al., 2003). GST-atTic40S did not bind to prRBCS or mature RBCS, suggesting that there is no direct contact between Tic40 and the importing substrates.
Figure 3.
Tic40 does not bind import substrates directly. 10 μl of [35S]prRBCS or an equal molar amount of [35S]RBCS was incubated with 48 pmol of OE33-His6, GST-atTic40S, or atTic110S-His6 at 4°C for 2 h. TALON or glutathione resin was then added, and incubation continued for another 30 min. Proteins eluted from resin were analyzed by SDS-PAGE (A) and quantified by a PhosphorImager (B).
Precursors increase the affinity of Tic110 to Tic40
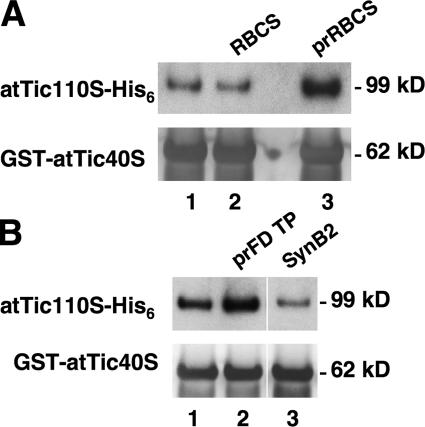
Because both transit peptides (Inaba et al., 2003) and Tic40 (Fig. 2) bind to Tic110, we investigated whether these two bindings affect each other. Preincubation of atTic110S-His6 with prRBCS, but not mature RBCS, increased the amount of atTic110S-His6 associated with GST-atTic40S (Fig. 4 A, lanes 2 and 3). This stimulatory effect could also be observed by incubating Tic110 with a synthetic peptide corresponding to the transit peptide of the precursor to ferrodoxin (prFD; Fig. 4 B, lane 2; Pilon et al., 1995). A control peptide, SynB2, had no effect (Fig. 4 B, lane 3). SynB2 has an artificially designed sequence that possesses a similar number of charges but a different conformation from a typical mitochondrial presequence, and it does not function as a mitochondrial presequence (Allison and Schatz, 1986). It is also discriminated by Toc75 from genuine chloroplast transit peptides (Hinnah et al., 2002). These results suggest that binding of Tic110 to transit peptides increases the affinity of Tic110 to Tic40.
Figure 4.
Precursors increase the affinity of Tic110 to Tic40. (A) 83 pmol of atTic110S-His6 was mixed with buffer (lane 1), 83 pmol of recombinant RBCS (lane 2), or prRBCS (lane 3). 83 pmol of GST-atTic40S was then added, and the mixture was incubated at 4°C for 2 h. Proteins associated with GST-atTic40S were recovered by glutathione resin, eluted by glutathione, and analyzed by immunoblotting using antibodies against Tic110 and Tic40. Approximately 0.5% of atTic110S-His6 added was coeluted with GST-atTic40S. (B) The experimental procedure was the same as in A, except RBCS and prRBCS were replaced with 83 pmol of synthetic prFD transit peptides (lane 2) or SynB2 control peptides (lane 3).
Tic40 causes the release of bound transit peptides from Tic110
We then investigated what happened to transit peptides bound by Tic110 after Tic40 binding. We tried to load Tic110 with as many transit peptides as possible by incubating atTic110S-His6 with 10-fold excess of 3H-labeled prFD transit peptides and then recovered atTic110S-His6 with bound transit peptides by metal-affinity resin. GST-atTic40S or GST control proteins were added to the resin suspension in increasing amounts. Increasing amounts of 3H were detected in the supernatant as increasing amounts of GST-atTic40S were added (Fig. 5 A), suggesting that GST-atTic40S caused the release of transit peptides from atTic110S-His6. These data suggest that binding of transit peptides by Tic110 recruits Tic40 to Tic110. Binding of Tic40 to Tic110 then causes transit peptides to be released from Tic110.
Figure 5.
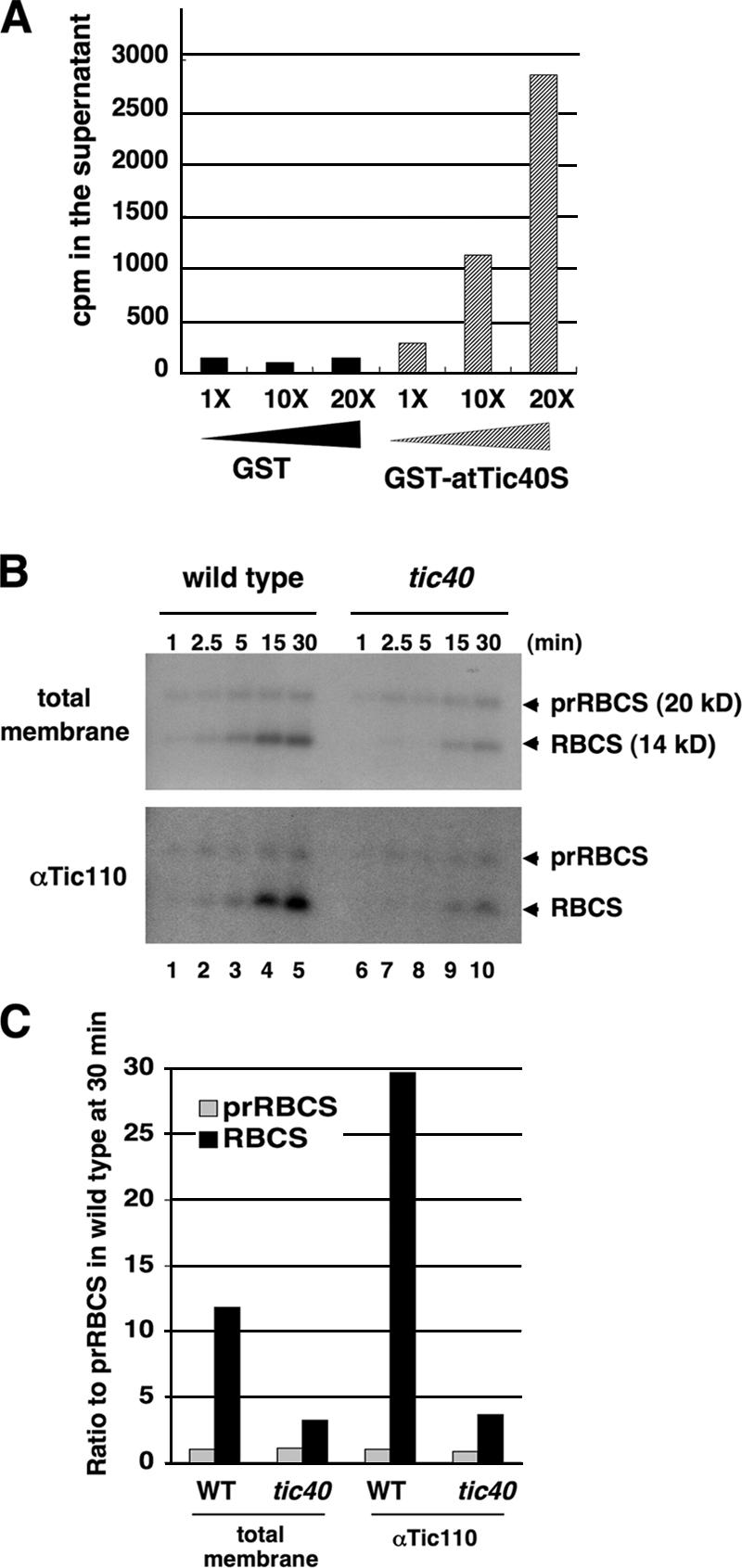
Tic40 causes the release of bound transit peptides from Tic110. (A) 48 pmol of atTic110S-His6 was incubated with 10-fold excess of 3H-labeled prFD transit peptides at 4°C for 2 h, TALON resin was added, and incubation continued for another 30 min. The resin was washed, and ∼3% of the added 3H-prFD transit peptides were bound to Tic110-His6 resin. Washed resin was then incubated with the same molar amount (48 pmol) or 10- or 20-fold excess of GST or GST-atTic40S at 4°C for 2 h. The resin was pelleted, and 3H counts in the supernatant were measured. Approximately 14% of bound 3H-prFD was released at 20-fold GST-atTic40S. (B) Membrane-bound processed mature proteins are delayed in appearance, and their association with Tic110 is reduced in the tic40 mutant chloroplasts. Isolated wild-type and tic40 mutant chloroplasts were used to perform an import time course experiment with [35S]prRBCS. Total membranes were isolated from chloroplasts of each time point and either directly analyzed by SDS-PAGE (top) or solubilized and immunoprecipitated by the anti-Tic110 antibody. (C) Quantification of the 30-min samples (B, lanes 5 and 10) of the gels shown in B. The amount of prRBCS in wild type at 30 min (prRBCS in lane 5) of each gel was taken as 1. The counts for RBCS have been corrected for the number of methionine residues compared with prRBCS.
Processing of transit peptides occurs when the importing precursors first emerge from the inner membrane while precursors are still bound to the inner membrane translocon (Kessler and Blobel, 1996; Chou et al., 2003; Chen and Li, 2006). If binding of Tic40 causes the release of transit peptides from Tic110 in vivo, then in a tic40 mutant, the release of transit peptides from Tic110, and therefore the processing of transit peptides, should be delayed and the amount of mature RBCS associated with Tic110 should be reduced. We thus compared the import of [35S]prRBCS into isolated A. thaliana wild-type chloroplasts and chloroplasts isolated from the tic40-1 mutant, which is a null mutant of Tic40 caused by transfer DNA insertion into the Tic40 gene (Chou et al., 2003). The import reaction was terminated at various time points by diluting with cold import buffer. Chloroplasts were reisolated and treated with 0.5 mM dithiobis(succinimidyl)propionate cross-linker to stabilize protein–complex interaction. Chloroplasts were lysed, and membrane fractions were isolated and analyzed (Chou et al., 2003). The amount of precursor proteins associated with the membranes was the same in the mutant and wild-type chloroplasts (Fig. 5 B, top). However, the appearance of processed mature proteins in the membranes was greatly delayed in the mutant chloroplasts. We have previously shown that bound precursors tend to be released from tic40 mutant chloroplasts before translocation is completed (Chou et al., 2003). Therefore, the fact that no accumulation of precursors was observed when production of processed mature proteins was delayed may be due to the release of the untranslocated precursors.
When the total membranes after import were further solubilized for immunoprecipitation, anti-Tic110 antibodies immunoprecipitated a high amount of mature RBCS in the wild-type chloroplasts (Fig. 5 B, bottom). Before immunoprecipitation, the ratio of RBCS to prRBCS was 12:1 at the 30-min import time point, and when precipitated by anti-Tic110 antibodies, the ratio increased to 29:1 (Fig. 5 C). This result indicated that there was a population of RBCS that was specifically associated with Tic110 and Tic110-associated translocon complexes. This population was likely absent in tic40 because in the mutant chloroplasts the ratio of RBCS to prRBCS remained similar in total membranes and in the population precipitated by anti-Tic110 antibodies (Fig. 5 C). It is possible that in the tic40 mutant chloroplasts, by the time transit peptides are spontaneously released by Tic110, the mature region of the precursor proteins has been translocated further into the stroma.
The Hip/Hop domain of Tic40 stimulates Hsp93 ATP hydrolysis
Because Hsp93 is the only chaperone molecule stably associated with the entire translocon complex, its ATPase activity should be important for precursor translocation. We therefore investigated which component that may have contact with Hsp93 could affect its ATPase activity. Recombinant pea Hsp93 with an N-terminal His6 tag, His6-Hsp93 (Fig. 1), was assayed for its ability to hydrolyze ATP. Addition of prFD transit peptides, Tic110, or Tic40 all had an approximately twofold stimulation on Hsp93 ATP hydrolysis compared with Hsp93 alone (Fig. 6 A). It is not clear whether such a small stimulation was specific or merely due to the presence of unfolded proteins. We further analyzed the effect of Tic40 subdomains. GST-atTic40TPR or -atTic40Hip/Hop was incubated with Hsp93. Interestingly, it was the Tic40 Hip/Hop domain, not the TPR domain, that had a clear stimulatory effect on Hsp93 ATP hydrolysis (Fig. 6 A). Various combinations of atTic110S-His6 plus domains of Tic40 or prFD transit peptides were also tested, and no additional stimulatory effect was observed (unpublished data). In support of the importance of the Tic40 Hip/Hop domain, A. thaliana tic40 mutant plants could be complemented by a full-length cDNA encoding the Tic40 precursor, but not by the same construct with the Hip/Hop domain deleted (unpublished data).
Figure 6.
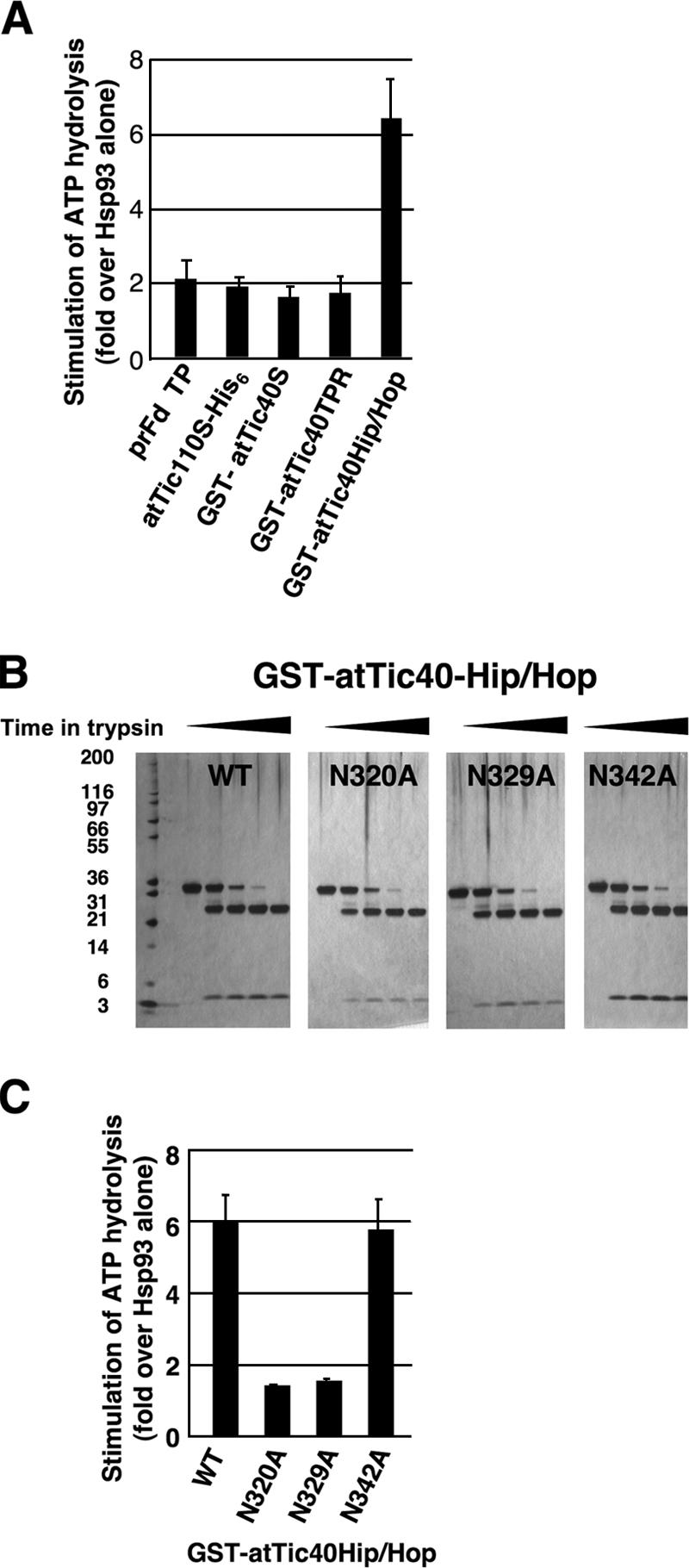
Tic40 Hip/Hop domain activates Hsp93 ATPase activity. (A) His6-Hsp93 was incubated with ATP or ATP plus various other proteins, as indicated at the bottom of the graph. The amount of Pi hydrolyzed by Hsp93 incubated with other proteins was divided by the amount of Pi hydrolyzed by Hsp93 alone. Values represent mean ± SEM; n = 3. (B) The three mutant forms of GST-atTic40Hip/Hop have the same protease sensitivity as the wild-type GST-atTic40Hip/Hop. GST-atTic40Hip/Hop and its three mutants, N320A, N329A, and N342A, were incubated with trypsin at a concentration of 4 U/g protein at 37°C for an increasing amount of time (0,10, 20, 30, and 40 min). The samples were analyzed by SDS-PAGE and silver staining. (C) His6-Hsp93 and ATP were incubated with the wild-type (WT) GST-atTic40Hip/Hop or one of the mutants, as indicated at the bottom of the graph. Stimulation of ATP hydrolysis was calculated as in A.
To further confirm the stimulatory function of the Tic40 Hip/Hop domain, three mutants of GST-atTic40Hip/Hop were generated. In the Hip/Hop domain of human Hip, the sequence DPEV occurs twice. Mutation of these two sequences to APAV inhibits progesterone receptor complex assembly with Hsp90 (Prapapanich et al., 1998). At the corresponding positions, atTic40 has N320PDV and N329PRV. (The subscript number stands for the residue number. It is equivalent to residue 396 and 405 if the first residue of atTic40 precursor, instead of mature protein, is counted as residue 1.) Another similar sequence of N342PMN is located further downstream. We mutated each of the asparagines to alanine. Limited proteolysis (Fig. 6 B) and secondary structure predictions (not depicted) indicated that the mutations did not affect the conformation of the three GST-atTic40Hip/Hop mutants. When assayed for stimulation of Hsp93 ATP hydrolysis, the N320A and N329A mutations abolished the stimulatory activity of GST-atTic40Hip/Hop, whereas the N342A mutation had no effect (Fig. 6 C).
The tic40 mutant is extremely pale and small but still viable (Chou et al., 2003), indicating that Tic40 plays a nonessential stimulatory/regulatory role during import. If Tic40 functions in stimulating Hsp93 ATP hydrolysis in vivo, either by promoting ATP/ADP exchange or by activating Hsp93 ATPase activity, it is likely that the protein import defect of tic40 mutants can be partially compensated by increasing the available amount of ATP in the stroma. We therefore preloaded isolated wild-type and tic40 mutant chloroplasts with 3 mM ATP and performed import experiments. Without ATP preloading, the amount of mature RBCS imported by the tic40 mutant chloroplasts was 26% of the wild-type chloroplasts. With ATP preloading, the tic40 mutant was increased to 38% of wild type (Fig. 7 A). We further confirmed the physiological significance of the results by testing whether Hsp93 ATPase activity was different in the two ATP concentrations before and after ATP preloading. Dark-adapted chloroplasts at the beginning of import have a stromal ATP concentration of ∼0.07 mM (Robinson and Portis, 1988; Theg et al., 1989; see Materials and methods). Preincubating chloroplasts in 3 mM ATP for 5 min at room temperature should increase the stromal ATP concentration to ∼2.1 mM (Leheny and Theg, 1994). The ATPase activity of Hsp93 was indeed increased significantly from an ATP concentration of 0.07 to 2.1 mM (Fig. 7 B). Stimulation of Hsp93 ATP hydrolysis by Tic40 provides another possible explanation for the release of untranslocated precursors from the tic40 mutant chloroplasts: a rapid ATP hydrolysis by Hsp93, stimulated by Tic40, may be important for a unidirectional translocation of precursors into the stroma.
Figure 7.
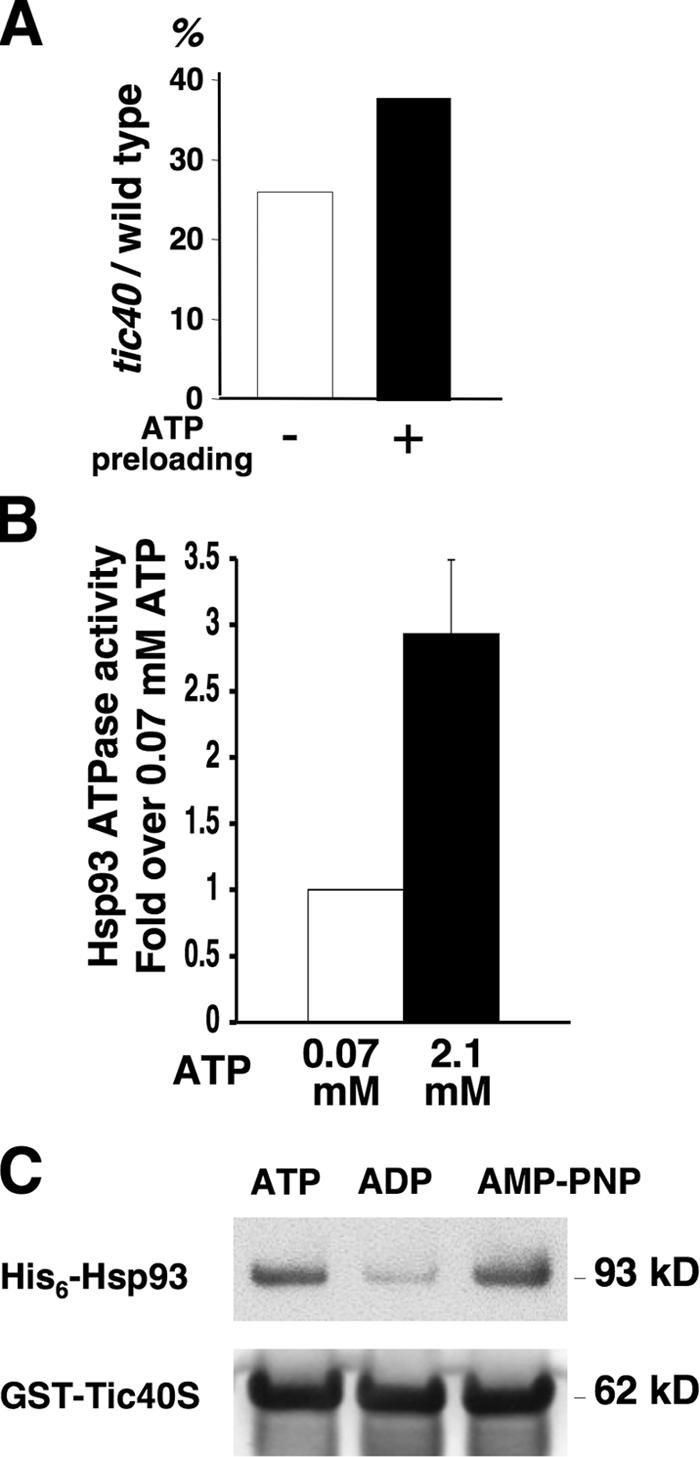
ATP preloading can partially rescue the tic40 mutant import defect. (A) Isolated chloroplasts from wild-type and tic40 mutant plants were preincubated with 3 mM ATP (closed bar) or import buffer (open bar) for 5 min at room temperature in the dark, reisolated, and used to perform import experiments with [35S]prRBCS. The amount of mature RBCS imported at 30 min was quantified, and the ratio of imported RBCS in tic40 chloroplasts to that in wild-type chloroplasts was plotted. (B) Hsp93 has a higher ATPase activity in the ATP concentration after chloroplasts have been preloaded with ATP. The ATP concentration within chloroplasts was calculated to be ∼0.07 mM before and 2.1 mM after preloading with 3 mM ATP (see Materials and methods). ATPase activity of Hsp93 was assayed under these two ATP concentrations. The ATPase activity at 0.07 mM ATP was taken as 1. (C) Association of Tic40 with Hsp93 decreases in the presence of ADP. 138 nM Hsp93-His6 was incubated with 690 nM ATP, ADP, or AMP-PNP at room temperature for 10 min. An equal amount of atTic110S-His6, prFD transit peptide, and GST-atTic40S was added with a 1-h incubation at 4°C after each addition. Tic40-containing complexes were recovered by glutathione resin, washed in PBS containing the nucleotides, eluted by glutathione, and analyzed by immunoblotting with antibodies against Hsp93 and Tic40.
Association of Hsp93 with Tic40 is reduced in the presence of ADP
We next investigated whether the nucleotide state of Hsp93 affected its association with Tic40. Hsp93 was preincubated with ATP, ADP, or the nonhydrolyzable ATP analogue adenylylimidodiphosphate (AMP-PNP). To mimic the in vivo situation, prFD transit peptides, atTic110S-His6, and GST-atTic40S were added to the reaction. Tic40-containing protein complexes were recovered using glutathione resin. The amount of Hsp93 recovered with Tic40 was higher in the presence of ATP or AMP-PNP and lower in the presence of ADP (Fig. 7 C), suggesting that Hsp93 may associate with Tic40 in its ATP state and dissociate from Tic40 after ATP is hydrolyzed to ADP. This result also suggests that Tic40 most likely functions in stimulating Hsp93 ATPase activity instead of facilitating ATP/ADP exchange.
Discussion
Stimulation of Hsp93 ATPase activity was only observed with the Tic40 Hip/Hop domain, not with the entire Tic40 stromal domain containing both the Hip/Hop and the TPR domains (GST-Tic40S; Fig. 6 A). It is possible that the Tic40 stromal domain is normally in a closed conformation in which the TPR domain shields the Hip/Hop domain. Binding of the Tic40 TPR domain to Tic110 causes a conformational change in Tic40, exposes the Hip/Hop domain, and allows the Hip/Hop domain to stimulate Hsp93. This mechanism would be similar to that found for protein phosphatase 5, in which its TPR domain normally engages with its phosphatase domain. Binding of the TPR domain to Hsp90 dissociates the TPR domain from the phosphatase domain and permits substrate access to the phosphatase domain (Yang et al., 2005).
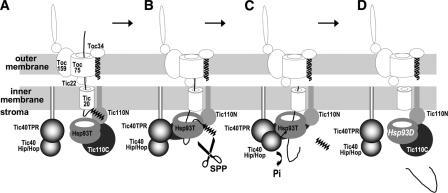
A working model for the sequential steps of protein translocation into the chloroplast stroma is proposed here (Fig. 8): as the transit peptide of a precursor protein emerges from the inner envelope membrane channel, it is bound by the N-terminal part of the Tic110 stromal domain (Fig. 8 A). This binding causes a conformational change in Tic110 and recruits Tic40TPR binding to Tic110 (Fig. 8 B). Binding of Tic40TPR to Tic110 causes release of the transit peptide from Tic110, freeing the transit peptide for cleavage by the stromal processing peptidase. Binding of Tic40TPR to Tic110 also unshields the Tic40 Hip/Hop domain, which then stimulates ATP hydrolysis by Hsp93 (Fig. 8 C). The energy of ATP hydrolysis by Hsp93 is most likely used to translocate the processed mature protein into the stroma (Fig. 8 D). Analyses of similar AAA+ proteins suggest that these proteins actively push substrates through its axial channel using conformational changes caused by ATP hydrolysis (Wang et al., 2001; Hanson and Whiteheart, 2005; Hinnerwisch et al., 2005). Hsp93-ADP may then dissociate from Tic40 (Fig. 8 D). Tic110 may also dissociate from Tic40 when there is no transit peptide bound. Under normal growth conditions in the light in which the stromal ATP concentration is high, Hsp93 may soon be reloaded with ATP and be ready for the next round of precursor translocation after Tic110 binds another incoming transit peptide. In our model, ATP is hydrolyzed only after the Tic40 TPR domain binds to Tic110, which only occurs after Tic110 binds to transit peptides. Therefore, Tic40 may function like a timing device to coordinate the sequential steps of translocation.
Figure 8.
A working model for sequential steps of protein translocation into the chloroplast stroma. The lines moving across the two membrane channels represent a precursor protein, with the zigzag line representing the transit peptide. SPP, stromal processing peptidase; Hsp93T and Hsp93D, Hsp93 bound with ATP or ADP, respectively. The names of other translocon components are labeled only in A, for simplicity.
The cochaperones Hip, Hop, and Tic40 all contain the TPR and Hip/Hop domains. The function of the TPR domains has been addressed extensively through structural and functional studies (Höhfeld et al., 1995; Scheufler et al., 2000). In contrast, no molecular function had been assigned to the Hip/Hop domain. Our data suggest that the Hip/Hop domain of Tic40 functions as an ATPase stimulation protein for Hsp93. Because the Hip/Hop domain is highly conserved, its identification as an ATPase stimulation protein may have implications in the functional mechanisms of Hip and Hop. It would be interesting to investigate whether the Hip/Hop domain in Hip and Hop can also stimulate the ATPase activity of Hsp70 and/or Hsp90. In addition, in vivo, whether the Tic40Hip/Hop domain has a stable association with Hsp93 like GroEL with GroES, or only interacts transiently with Hsp93 like DnaK with DnaJ, and which one of the two Hsp93 ATPase domains is activated by Tic40Hip/Hop, all require further investigations.
A possible similarity between protein translocation into the chloroplast stroma and retrotranslocation of misfolded proteins from the ER lumen is noted here. ER retrotranslocation requires the cytosolic ATPase p97/Cdc48, which is an AAA+ family protein, like Hsp93. VIMP (VCP interacting membrane protein), an ER-membrane protein with a sizeable cytosolic domain, acts as a receptor for p97, similar to Tic110 (Jackson et al., 1998). VIMP links p97 to Derlin-1, which is a proposed component of the ER retrotranslocation channel (Lilley and Ploegh, 2004; Ye et al., 2004). Derlin-1 is ∼25 kD with four predicted α-helical transmembrane domains, a size and structure very similar to Tic20, the proposed protein translocation channel across the chloroplast inner envelope membrane (Kouranov et al., 1998). Therefore, both systems contain an AAA+ type motor, a membrane protein with a large soluble domain as the receptor for the motor and a channel of a similar size and structure. It is possible that the substrates of both systems, the chloroplast precursors after being translocated across the outer membrane and the misfolded ER proteins after being recruited to Derlin-1, are in a similar folding state. Tic40 may have been evolved to meet some specific requirements of chloroplasts to increase the efficiency of precursor processing and translocation.
Materials and methods
DNA constructs, yeast two-hybrid analyses, and in vitro pull-down assays
All constructs used for the yeast two-hybrid analyses were made by PCR amplifying the respective DNA fragment using a forward primer that added an NcoI site and a reverse primer that added an EcoRI site to the amplified fragment. The fragment was digested and cloned into the NcoI–EcoRI site of plasmid pACT2 (for the activation-domain constructs) or pAS2-1 (for the binding-domain constructs). Other constructs were made similarly with the following specifications: GST fusion constructs of atTic40 were made in the BamHI–EcoRI site of pGEX-5X-1 (GE Healthcare), His6-Hsp93 was made in the NdeI–SalI site of pET28a (Invitrogen), and OE33-His6 (At5g66570, amino acids 86–332 with the first amino acid of the precursor as 1) was made in the NdeI–EcoRI site of pET22b (Invitrogen).
Two-hybrid assays were performed using the MatchMaker-II system (CLONTECH Laboratories, Inc.) according to the manufacturer's instructions. Except where specified in the figure legends, pull-down assays using His6-tagged proteins were performed using TALON (CLONTECH Laboratories, Inc.) resin by incubating the resin with the protein mixture at 4°C for 2 h. Pelleted resin was washed thrice with PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, and 2.7 mM KCl) containing 1 mM imidazole, and bound proteins were eluted with 50 mM imidazole in 50 mM Tris-HCl, pH 8.0. Pull-down assays using GST-tagged proteins were performed using Glutatione-4B resin (GE Healthcare) by incubating the resin with the protein mixture at 4°C for 30 min to 1 h. Pelleted resin was washed thrice with PBS, and bound proteins were eluted with 10 mM glutathione in 50 mM Tris-HCl, pH 8.0.
Recombinant prRBCS and RBCS were overexpressed and purified from E. coli (Tu et al., 2004). Antibodies against Tic110 were generated as described previously (Tu et al., 2004). Antibodies against Tic40 were produced by subcloning the coding region of atTic40 residues 227–447 into pET22b (Invitrogen), overexpressing the recombinant protein with a C-terminal His6 tag in E. coli, and injecting the purified recombinant proteins into mice. Anti-Hsp93 and anti-OE33 antibodies were generated by immunizing rabbits with the His6-Hsp93 and OE33-His6 recombinant proteins described in the previous paragraph.
Peptides and peptide labeling
The first 34 amino acids of the Silene prFD transit peptide without the initiation methionine (sequence ASTLSTLSVSASLLPKQQPMVASSLPTNMGQALF; Pilon et al., 1995) and the SynB2 peptide (sequence MLSRQQSQRQSRQQSQRQSRYLL; Allison and Schatz, 1986) were synthesized and purified by HPLC to >80% purity by SynPep. 1.5 mg of the prFD peptide was dissolved in 50 μl of water and mixed with 20 μl of boric acid (adjusted to pH 8.5 with NaOH), 20 μl of 0.5 M formaldehyde, and 15 μl of 3H-labeled 0.12 M NaBH4. The reaction was incubated on ice for 15 min and quenched by adding 50 μl of 1 M Tris-HCl, pH 7.5. 3H-labeled peptide and 3H-labeled NaBH4 were separated on a Sephadex G-10 column with 0.1 M NH4HCO3. The peptide fractions were collected and dried.
Protein import into isolated chloroplasts
[35S]prRBCS and [35S]RBCS were synthesized by in vitro transcription and translation (Perry et al., 1991). Growth of A. thaliana plants, isolation of chloroplasts from the WS2 wild type and the tic40-1 mutant, import assays and isolation, solubilization, and immunoprecipitation of membrane fraction from chloroplasts after import were performed as described previously (Chou et al., 2003). Stromal ATP concentrations used in Fig. 7 B is calculated as follows: internal ATP concentration of dark-adapted chloroplasts is ∼1.82 nmol/mg chlorophyll (Robinson and Portis, 1988; Theg et al., 1989). With a stroma volume of 25 μl/mg chlorophyll (Robinson, 1985), 1.82 nmol/mg chlorophyll is ∼0.07 mM. At room temperature for 5 min, ATP translocation across the envelope can achieve ∼70% (Leheny and Theg, 1994). Preincubating chloroplasts in 3 mM ATP for 5 min at room temperature should increase the stromal ATP concentration to ∼2.1 mM. Quantifications of gel images were performed on LAS-1000plus pictrography 3000 (Fuji).
Hsp93 ATPase assay
A 16-μl solution containing 0.5 μM of His6-Hsp93 in 75 mM Hepes-KOH, pH 7.0, and 4 mM MgCl2 was mixed with 4 μl of 1 mM ATP containing 0.078 μl of γ-[32P]ATP (5,000 Ci/nmol) and incubated at 37°C for 30 min. The reaction was stopped by adding 2 μl of 0.5 M EDTA. 0.35 μl of the reaction was spotted on a TLC plate. The plate was air-dried, and ATP and Pi were separated by a solution containing 0.5 M LiCl and 1 M formic acid. The TLC plate was dried. The amounts of ATP and Pi were quantified by a PhosphorImager (FLA-5000; Fuji). When other proteins were added to the reaction, they were present at the same concentration as His6-Hsp93, except for the prFD transit peptide, which was present at a 10× concentration of other proteins.
Acknowledgments
We thank Drs. Danny Schnell and Takehito Inaba for the construct of atTic110S-His6, Dr. Chih-wen Sun and Dr. Shih-Long Tu for advice in yeast two-hybrid analyses and GST pull down, respectively, Dr. Chung Wang for reagents and advice in peptide labeling and ATP hydrolysis analyses, Drs. Rey-Hui Chen and Chung Wang for critical reading of the manuscript, and Drs. Ken Deen and Harry Wilson for English editing.
H.-m. Li was supported by grants from National Science Council (NSC95-2321-B-001-004) and Academia Sinica of Taiwan. M. Akita acknowledges financial support during early parts of the work from U.S. Department of Energy and U.S. National Science Foundation grants to Dr. Ken Keegstra (Michigan State University).
M.-L. Chou and C.-C. Chu contributed equally to this paper.
M.-L. Chou's present address is Department and Institute of Life Science, Tzu-Chi University, Hualien 97004, Taiwan.
Abbreviations used in this paper: AMP-PNP, adenylylimidodiphosphate; Hip, Hsp70-interacting protein; Hop, Hsp70/Hsp90-organizing protein; prFD, precursor to ferrodoxin; prRBCS, precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase oxygenase; Tic, translocon at the inner envelope membrane of chloroplasts; Toc, translocon at the outer envelope membrane of chloroplasts; TPR, tetratricopeptide repeat.
References
- Allison, D., and G. Schatz. 1986. Artificial mitochondrial presequences. Proc. Natl. Acad. Sci. USA. 83:9011–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K.-Y., and H.-m. Li. 2006. Precursor binding to an 880-kD Toc complex as an early step during active protein import into chloroplasts. Plant J. In press. [DOI] [PMC free article] [PubMed]
- Chou, M.-L., L.M. Fitzpatrick, S.-L. Tu, G. Budziszewski, S. Potter-Lewis, M. Akita, J.Z. Levin, K. Keegstra, and H.-m. Li. 2003. Tic40, a membrane-anchored co-chaperone homologue in the chloroplast protein translocon. EMBO J. 22:2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman, J., and J. Höhfeld. 1997. Chaperones get in touch: the Hip-Hop connection. Trends Biochem. Sci. 22:87–92. [DOI] [PubMed] [Google Scholar]
- Hanson, P., and S. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529. [DOI] [PubMed] [Google Scholar]
- Hinnah, S.C., R. Wagner, N. Sveshnikova, R. Harrer, and J. Soll. 2002. The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys. J. 83:899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnerwisch, J., W. Fenton, K. Furtak, G. Farr, and A. Horwich. 2005. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 121:1029–1041. [DOI] [PubMed] [Google Scholar]
- Höhfeld, J., Y. Minami, and F.U. Hartl. 1995. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 83:589–598. [DOI] [PubMed] [Google Scholar]
- Inaba, T., M. Li, M. Alvarez-Huerta, F. Kessler, and D.J. Schnell. 2003. atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278:38617–38627. [DOI] [PubMed] [Google Scholar]
- Inaba, T., M. Alvarez-Huerta, M. Li, J. Bauer, C. Ewers, F. Kessler, and D.J. Schnell. 2005. atTic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 17:1482–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.T., J.E. Froehlich, and K. Keegstra. 1998. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273:16583–16588. [DOI] [PubMed] [Google Scholar]
- Kessler, F., and G. Blobel. 1996. Interaction of the protein import and folding machineries in the chloroplast. Proc. Natl. Acad. Sci. USA. 93:7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, F., and D.J. Schnell. 2006. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 7:248–257. [DOI] [PubMed] [Google Scholar]
- Kouranov, A., X. Chen, B. Fuks, and D.J. Schnell. 1998. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva, S., J. Bedard, R. Patel, P. Dudley, D. Twell, G.D. Rios, C. Koncz, and P. Jarvis. 2005. In vivo studies of the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41:412–428. [DOI] [PubMed] [Google Scholar]
- Lamb, J., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257–259. [DOI] [PubMed] [Google Scholar]
- Leheny, E.A., and S.M. Theg. 1994. Apparent inhibition of chloroplast protein import by cold temperatures is due to energetic considerations not membrane fluidity. Plant Cell. 6:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, B.N, and H.L. Ploegh. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 429:834–840. [DOI] [PubMed] [Google Scholar]
- Lübeck, J., J. Soll, M. Akita, E. Nielsen, and K. Keegstra. 1996. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., M. Akita, J. Davila-Aponte, and K. Keegstra. 1997. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16:935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, S.E., H.-m. Li, and K. Keegstra. 1991. In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol. 34:327–344. [DOI] [PubMed] [Google Scholar]
- Pilon, M., H. Wienk, W. Sips, M. de Swaaf, I. Talboom, R. van't Hof, G. de Korte-Kool, R. Demel, P. Weisbeek, and B. de Kruijff. 1995. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J. Biol. Chem. 270:3882–3893. [DOI] [PubMed] [Google Scholar]
- Prapapanich, V., S. Chen, and D.F. Smith. 1998. Mutation of Hip's carboxy-terminal region inhibits a transitional stage of progesterone receptor assembly. Mol. Cell. Biol. 18:944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, S., and G. Lamppa. 1998. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA. 95:7463–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, S. 1985. The involvement of stromal ATP in maintaining the pH gradient across the chloroplast envelope in the light. Biochim. Biophys. Acta. 806:187–194. [Google Scholar]
- Robinson, S., and A.J. Portis. 1988. Involvement of stromal ATP in the light activation of ribulose-1,5-bisphosphate carboxylase/oxygenase in intact isolated chloroplasts. Plant Physiol. 86:293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler, C., A. Brinker, G. Bourenkov, S. Pegpraro, L. Morder, H. Bartunik, F. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 101:199–210. [DOI] [PubMed] [Google Scholar]
- Schirmer, E., J. Glover, M. Singer, and S. Lindquist. 1996. Hsp100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289–296. [PubMed] [Google Scholar]
- Schnell, D.J., F. Kessler, and G. Blobel. 1994. Isolation of components of the chloroplast protein import machinery. Science. 266:1007–1012. [DOI] [PubMed] [Google Scholar]
- Soll, J., and E. Schleiff. 2004. Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5:198–208. [DOI] [PubMed] [Google Scholar]
- Theg, S.M., C. Bauerle, L.J. Olsen, B.R. Selman, and K. Keegstra. 1989. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264:6730–6736. [PubMed] [Google Scholar]
- Tu, S.-L., L.-J. Chen, M. Smith, Y.-s. Su, D. Schnell, and H.-m. Li. 2004. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell. 16:2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., J.J. Song, I.S. Seong, M.C. Franklin, S. Kamtekar, S.H. Eom, and C.H. Chung. 2001. Nucleotide-dependent conformational changes in a protease-associated ATPase HslU. Structure. 9:1107–1116. [DOI] [PubMed] [Google Scholar]
- Yang, J., S.M. Roe, M. Cliff, M. Williams, J. Ladbury, P. Cohen, and D. Barford. 2005. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 24:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., Y. Shibata, C. Yun, D. Ron, and T.A. Rapoport. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 429:841–847. [DOI] [PubMed] [Google Scholar]