Abstract
Retrograde flow of cortical actin networks and bundles is essential for cell motility and retrograde intracellular movement, and for the formation and maintenance of microvilli, stereocilia, and filopodia. Actin cables, which are F-actin bundles that serve as tracks for anterograde and retrograde cargo movement in budding yeast, undergo retrograde flow that is driven, in part, by actin polymerization and assembly. We find that the actin cable retrograde flow rate is reduced by deletion or delocalization of the type II myosin Myo1p, and by deletion or conditional mutation of the Myo1p motor domain. Deletion of the tropomyosin isoform Tpm2p, but not the Tpm1p isoform, increases the rate of actin cable retrograde flow. Pretreatment of F-actin with Tpm2p, but not Tpm1p, inhibits Myo1p binding to F-actin and Myo1p-dependent F-actin gliding. These data support novel, opposing roles of Myo1p and Tpm2 in regulating retrograde actin flow in budding yeast and an isoform-specific function of Tpm1p in promoting actin cable function in myosin-driven anterograde cargo transport.
Introduction
Retrograde, or centripetal, flow was originally identified as the net transport of surface receptors and the underlying actin cytoskeleton from the cell cortex toward the center of the cell (Bray, 1970; Yamada and Wessells, 1973). This process has been studied extensively in motile cells, including neuronal growth cones, cultured fibroblasts, and fish keratocytes (Heath, 1983; Forscher and Smith, 1988; Theriot and Mitchison, 1991), where it is required for the generation of traction forces on the substratum for cell movement and necessary for the maintenance of polarized, directional growth. Retrograde actin flow also occurs in nonmotile cells, such as sea urchin coelomocytes (Henson et al., 1999). In both motile and nonmotile cells, retrograde flow occurs as a result of two forces; the “push” of actin polymerization caused by the addition of G-actin at the barbed ends of elongating actin filaments at the cell cortex, and the “pull” of myosin molecules further back in the network. Numerous studies, including those using inhibitors for myosin or myosin light chain kinase, have implicated conventional type II myosin in retrograde actin flow (Lin et al., 1996; Gupton et al., 2002; Brown and Bridgman, 2003; Jurado et al., 2005; Medeiros et al., 2006). Other studies have also implicated tropomyosin in the regulation of actin flow, through the isoform-specific regulation of both the length of the actin network and the rate of retrograde flow (Gupton et al., 2005).
Retrograde actin flow similar to that observed in cortical actin networks in motile and nonmotile cells also occurs in polarized actin bundles within microvilli, filopodia, and stereocilia (for review see Lin et al., 2005). In each of these cortical protrusions, actin bundles elongate by the addition of new material to the tip of the bundle, which comprises F-actin barbed ends and a “tip complex” detected by ultrastructural analysis (Mallavarapu and Mitchison, 1999; Schneider et al., 2002; Tyska and Mooseker, 2002). Imaging of actin dynamics revealed that addition of material to the tip of actin bundles results in retrograde, or centripetal, flow of F-actin within the bundle from the tip toward the base of the bundle (Rzadzinska et al., 2004).
Several lines of evidence indicate that actin cables in budding yeast are conserved structures, which exhibit retrograde flow. Actin cables are parallel bundles of F-actin that align along the mother-bud axis and contain cross-linking and stabilizing proteins, including fimbrin (Sac6p), Abp140p, and two tropomyosin isoforms (Tpm1p and Tpm2p; Adams and Pringle, 1984; Drubin et al., 1988; Liu and Bretscher, 1989; Drees et al., 1995; Hermann et al., 1997; Amberg, 1998; Asakura et al., 1998; Pruyne et al., 1998; Singer and Shaw, 2003). Actin cables are required for the localization of vacuoles, secretory vesicles, mitochondria, late-Golgi components, spindle alignment elements, and ASH1 mRNA to the developing bud (Liu and Bretscher, 1989; Lazzarino et al., 1994; Hill et al., 1996; Simon et al., 1997; Takizawa et al., 1997; Pruyne et al., 1998; Rossanese et al., 2001; Schott et al., 2002). With the exception of mitochondria, which use Arp2/3 complex–mediated actin polymerization for anterograde movement (Boldogh et al., 2001), all of the other particles that rely on actin cables for distribution require type V myosin for localization in the bud. In the case of secretory vesicles and spindle-alignment elements, bud- directed movement is linear and occurs at a velocity that is directly proportional to the length of the type V myosin lever arm (Schott et al., 2002; Hwang et al., 2003). Thus, there is evidence that actin cables serve as tracks for myosin-driven, bud-directed particle movement. Moreover, because type V myosins are barbed-end–directed motors, it is likely that actin cables, like actin bundles in other eukaryotes, consist of actin filaments that are physically or functionally polarized with their barbed end oriented toward their assembly site in the bud.
Recent studies indicate that actin cables undergo retrograde flow. Previously, we showed that actin cables are labeled in living cells using an Abp140p-GFP fusion protein (Yang and Pon, 2002). Time-lapse imaging of Abp140p-GFP revealed that actin cables are dynamic structures that assemble in the bud and bud neck and exhibit assembly-dependent retrograde flow away from assembly sites along the mother-bud axis. Fluorescence loss in photobleaching experiments and analysis of the movement of fiduciary marks of Abp140p-GFP on motile actin cables revealed that insertion of material at the tip of the cable located within the bud results in retrograde movement of the entire cable toward the mother cell. Consistent with these findings, other laboratories showed that the yeast formins Bni1p and Bnr1p are required for actin cable stability, have the capacity to stimulate actin polymerization, and localize to the sites of actin cable assembly detected using Abp140p-GFP (Kohno et al., 1996; Imamura et al., 1997; Evangelista et al., 2002; Pruyne et al., 2002; Sagot et al., 2002a,b; Dong et al., 2003).
Our previous results also showed that actin cable movement could be mediated by assembly-independent mechanisms. Specifically, we found that fixed-length fragments of actin cables, which are produced by treatment with low levels of the actin monomer–sequestering factor latrunculin A, move along the cell cortex. This movement did not depend on actin cable assembly and occurred parallel to the long axis of the actin cable (Yang and Pon, 2002). These findings raised the possibility that myosin molecules that are anchored to the cortex could provide a pulling force for actin cable retrograde flow. Saccharomyces cerevisiae has five myosin genes: two type I myosin genes (MYO3 and MYO5), one type II myosin gene (MYO1), and two type V myosin genes (MYO2 and MYO4; Watts et al., 1987; Johnston et al., 1991; Haarer et al., 1994; Goodson and Spudich, 1995; Goodson et al., 1996). Type I myosins localize to actin patches (actin-coated endosomes), where they contribute to the control of actin organization, endocytosis, and activation of the actin nucleation activity of the Arp2/3 complex (Goodson et al., 1996; Evangelista et al., 2000; Lechler et al., 2000; Idrissi et al., 2002; Grosshans et al., 2006). The type II myosin of yeast localizes to the bud neck, and has been implicated in cytokinesis (Bi et al., 1998; Lippincott and Li, 1998; Vallen et al., 2000). Finally, type V myosins are motors for bud-directed cargo movement along actin cables, and accumulate in the bud tip (Lillie and Brown, 1994; Schott et al., 2002; Hwang et al., 2003). We studied the role of yeast myosins in actin cable dynamics.
In complementary studies, we evaluated the role of yeast tropomyosins in retrograde actin cable flow. The yeast tropomyosins share 64% homology, and show similar affinities for binding to F-actin. Deletion of both Tpm1p and Tpm2p results in cell death in budding yeast. These findings indicate that Tpm1p and Tpm2p together support an essential function in budding yeast. Because both proteins localize to actin cables, and actin cables are essential for cell viability in budding yeast, it is likely that the essential function supported by Tpm1p and Tpm2p is stabilization of actin cables. On the other hand, overexpression of TPM2 in yeast bearing a deletion in TPM1 does not suppress the defects in growth or chitin deposition associated with the tpm1Δ deletion. Moreover, actin cables are destabilized in tpm1Δ, but not in tpm2Δ mutants, (Drees et al., 1995). These findings indicate that the two tropomyosin isoforms of budding yeast carry out distinct functions, and that the primary function of Tpm1p is control of actin cable stability, along with its function in chitin deposition and bud growth.
We report novel roles for a type II myosin (Myo1p) and a specific tropomyosin isoform (Tpm2p) in retrograde actin cable flow in budding yeast. Our findings indicate that retrograde actin flow is conserved from yeast to humans, and raise the possibility that budding yeast may be useful as a model system for studying this fundamental process. Our studies also support an isoform-specific function for Tpm1p in promoting actin cable activity as a track for myosin-driven anterograde cargo movement.
Results
Role for type II myosin (Myo1p) in retrograde actin cable flow
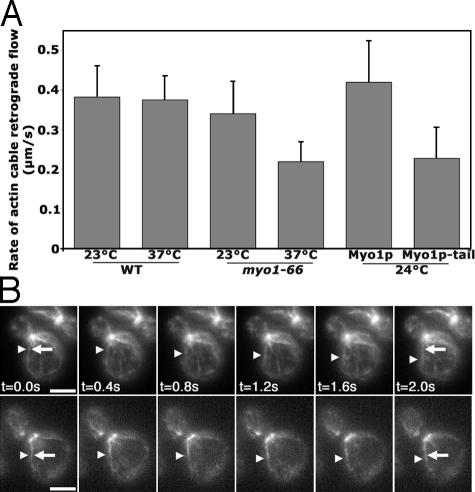
In wild-type cells, actin cables extend from the bud tip or bud neck along the mother-bud axis. In yeast bearing deletions in the type I myosins, actin cables do not align along the mother-bud axis and are less robust than those observed in wild-type cells; however, actin cables are present in these cells. To analyze the rate of retrograde actin cable flow, we monitored as a function of time the change in position of either the tips of Abp140p-GFP–labeled actin cables, or of fiduciary marks of Abp140p-GFP on actin cables (Fig. 1 A). The velocity of retrograde actin cable flow in various myosin mutants and their corresponding wild-type controls are shown (Table I). Because the velocity of retrograde actin cable flow was similar in all wild-type stains analyzed at 23 and 37°C (0.36–0.38 μm/s), the velocity of actin cable movement in various myosin mutants was compared with the mean velocity of all wild-type strains evaluated (Fig. 1 B).
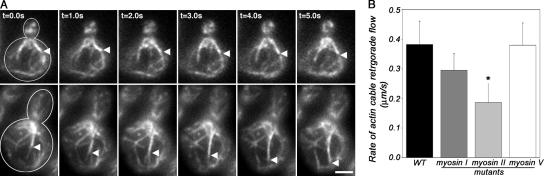
Figure 1.
Type II myosin is necessary for normal rates of actin cable retrograde flow. Wild-type and mutant strains were grown in selective media at RT to early log phase. For all mutants other than myo2-66 myo4Δ, time-lapse imaging was performed at RT. For temperature-sensitive myo2-66 myo4Δ mutants, strains were grown at RT and shifted to 37°C for 30 min before imaging. Imaging of Abp140p-GFP–labeled actin cables in a single cortical focal plane was performed as described in Materials and methods. (A) Still-frame images of a time-lapse series of Abp140p-GFP–labeled actin cables in myo1Δ (THY120; top) and the corresponding wild-type strain (YCY027; bottom). Arrowheads point to fiduciary marks, or regions of increased actin content in the actin cable, at t = 0 of image analysis. (B) Average rates of retrograde flow of Abp140p-GFP–labeled actin cables in wild-type cells (YCY027), a type I myosin mutant (myo3Δ myo5Δ; YCY009), and a type II myosin mutant (myo1Δ; THY120) at RT, as well as a type V myosin mutant (myo2-66 myo4Δ; YCY010) at 37°C. Rates of movement were determined as described in Materials and methods. Error bars represent the SD. n > 20 for each strain. Asterisk indicates statistically different rates, P < 0.01. Bar, 2 μm.
Table I.
Velocities of retrograde actin cable flow in myosin mutants and the corresponding wild-type strains
| Strain
|
Genotype
|
Retrograde actin cable flow rate
|
|
|---|---|---|---|
| 23°C | 37°C | ||
| μm/s | μm/s | ||
| S288C wild type for myo1Δ and myo2-66 myo4Δ (YCY027) |
MATα his3Δ leu2Δ met15Δ ura3Δ ABP140∷GFP∷KanMX6 | 0.38 ± 0.08 | 0.38 ± 0.08 |
| W303 wild type for myo3Δ myo5Δ
(YCY006) |
MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1
ABP140∷GFP∷KanMX6 |
0.36 ± 0.1 | nd |
| myo1Δ (THY120) | MATα leu2-3,112 ura3-52 ade1-101 myo1Δ∷URA3 ABP140∷GFP∷KanMX6 | 0.19 ± 0.07 | nd |
| myo2-66 myo4Δ (VSY21) | MATα leu2-3,112 his3-200 ura3-52 myo2-66∷HIS3 myo4Δ∷URA3 |
0.40 ± 0.06 | 0.38 ± 0.08 |
| myo3Δ myo5Δ (YCY009) | MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1
myo3Δ∷HIS3 myo5Δ∷TRP1 ABP140∷GFP∷KanMX6 |
0.29 ± 0.06 | nd |
nd, no data.
The retrograde flow rate of actin cables in strains bearing a deletion in the two type I myosins of yeast (myo3Δ myo5Δ) was not significantly different from rates observed in corresponding wild-type cells at 23°C. Because the type V myosin gene MYO2 is essential, we studied the effect of loss of type V myosin function using a yeast strain bearing a temperature-sensitive mutation in the MYO2 gene and a deletion in MYO4, which is the other, nonessential, type V myosin gene. We found that the rate of retrograde actin cable flow in this strain (myo2-66 myo4Δ) was similar to that observed in wild-type cells upon incubation at permissive (23°C) or restrictive (37°C) temperatures. In contrast, in the strain lacking the type II myosin (myo1Δ), the rate of retrograde actin cable flow of 0.19 μm/s was significantly lower than that observed in the wild-type control (P < 0.01). These findings indicate that type II myosin of budding yeast is required for normal rates of retrograde actin cable flow (Fig. 1 and Table I).
Type II myosin localizes to the bud neck from the time of commitment to a round of cell division (G1) to the end of the cell division cycle. We tested whether this localization of type II myosin is critical for retrograde actin cable flow by taking advantage of the finding that overexpression of the tail of type II myosin acts in a dominant-negative fashion and causes delocalization of the endogenous, full-length protein from the bud neck (Tolliday et al., 2003). We inserted the C-terminal 868 amino acids of the type II myosin into a multicopy plasmid under control of the galactose-inducible GAL1 promoter. The resulting plasmid, pGal-MYO1-tail, was used to evaluate the effect of regulated overexpression of the type II myosin tail on the localization of endogenous, GFP-tagged type II myosin and on the dynamics of actin cables.
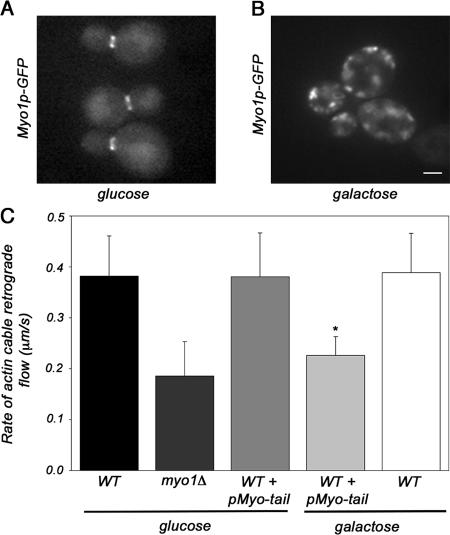
We confirmed that the addition of GFP to the C terminus of the endogenous type II myosin gene (MYO1) had no obvious effect on cell viability, polarity, or growth rates (unpublished data). As expected, we found that full-length, GFP-tagged type II myosin (Myo1p-GFP) forms a ring at the bud neck in untransformed cells, as well as in cells that were transformed with pGal-MYO1-tail and incubated under noninducing conditions (Fig. 2 A). Thus, endogenous type II myosin exhibits normal localization when the pGal-MYO1-tail expression is not induced. In contrast, galactose-induced overexpression of pGal-MYO1-tail caused mislocalization of endogenous, GFP-tagged type II myosin from the bud neck to the cytosol (Fig. 2 B).
Figure 2.
Normal localization of type II myosin is necessary for normal rates of actin cable retrograde flow. Wild-type cells expressing Myo1p-GFP from the genomic locus were transformed with pGal-MYO1-tail (THY159). Cells were grown in the glucose-based media to mid-log phase and shifted to fresh media containing either glucose or galactose for 4 h. Imaging was performed at a single central plane. (A and B) Fluorescence images from a single optical plane illustrating the localization of full-length, GFP-tagged type II myosin protein (Myo1p-GFP) in cells transformed with pGal-MYO1-tail after growth under noninducing conditions (glucose; A) and under inducing conditions (galactose; B). Bar, 1 μm. (C) The rates of retrograde flow of Abp140p-GFP–labeled actin cables were analyzed in wild-type cells (YCY027), myo1Δ cells (THY120), and cells bearing wild-type MYO1 that were transformed with pGal-MYO1-tail (THY160) upon growth in a carbon source that either induces (galactose) or does not induce (glucose) overexpression of the type II myosin tail construct. Analyses of the rates of retrograde flow were determined as in Fig. 1. In the absence of induction of pGal-MYO1-tail (glucose), normal rates of actin cable retrograde flow were observed. Growth for 4 h under inducing conditions (galactose) causes a significant reduction in the rate of retrograde flow of Abp140p-labeled actin cables, compared with that observed under noninducing conditions (P < 0.01). Error bars represent the SD. n > 20 for each strain.
Using Abp140p-GFP as a marker for actin cables, we found that wild-type cells bearing pGal-MYO1-tail showed normal rates of actin cable retrograde flow when grown under noninducing conditions. However, galactose-induced overexpression of pGal-MYO1-tail resulted in a significant decrease in the rate of retrograde actin cable flow (Fig. 2 C). Indeed, the retrograde flow rate in cells expressing the dominant-negative pGal-MYO1-tail construct was similar to that observed upon deletion of MYO1. This decrease in the rate of retrograde flow was not the result of incubation in galactose, as wild-type cells grown in galactose showed no change in actin cable retrograde flow rate. Instead, these findings indicate that localization of type II myosin at the bud neck is important for its function in retrograde actin cable flow.
The motor domain of type II myosin is required for its function in retrograde actin cable flow
Two approaches were taken in our study of the role of the myosin motor in retrograde actin cable flow. The first approach was based on recent findings that the Myo1p tail supports cytokinesis when expressed in lieu of full-length Myo1p and at a level similar to that of endogenous, wild-type Myo1p (Lord et al., 2005). The Myo1p tail and full-length Myo1p were expressed in myo1Δ cells using the plasmids constructed by Lord et al. (2005), and retrograde actin cable flow was examined in these cells using Abp140p-GFP. Full-length Myo1p and the Myo1p tail localize to the bud neck, as previously reported (Lord et al., 2005). The velocity of retrograde actin cable flow observed upon expression of full-length, plasmid-borne Myo1p (0.419 μm/s; n = 32) was similar to that observed in yeast expressing endogenous, wild-type MYO1. In contrast, expression of the Myo1p tail alone resulted in a reduced rate of retrograde actin cable flow (0.227 μm/s; n = 33) that was similar to that observed in untransformed myo1Δ cells and in myo1Δ cells that were transformed with the vector used for Myo1p and Myo1p tail expression. Thus, the motor domain of MYO1 is required for normal rates of retrograde actin cable flow.
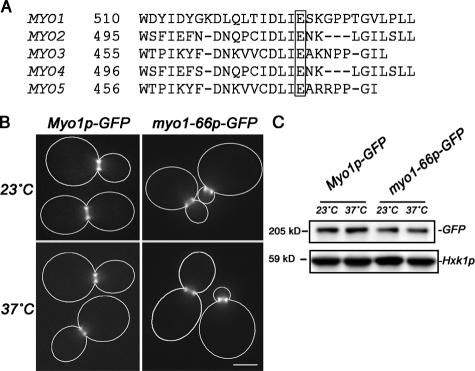
As a second approach to study the role for the type II myosin motor activity in retrograde actin cable flow, we constructed a mutant that carries a single amino acid substitution in the Myo1p motor domain, which inhibits the ATP-sensitive actin-binding activity of the protein. The site was chosen for mutagenesis based on the observation that the corresponding mutation in the essential type V myosin of budding yeast, the myo2-66 mutation, produces a temperature-dependent loss of cell viability (Johnston et al., 1991). The myo2-66 mutation is a substitution of lysine for glutamic acid 511 (Lillie and Brown, 1994), a residue conserved in all yeast myosin proteins (Fig. 3 A) that corresponds to Glu527 in chicken skeletal muscle myosin and Glu528 in yeast type II myosin. X-ray crystallography of chicken skeletal muscle myosin mapped this residue to the actin-binding face of the lower 50-kD component of the motor domain, and showed that this glutamic acid forms a salt bridge with Lys486, which is another highly conserved residue in the motor domain (Rayment et al., 1993).
Figure 3.
Construction and expression of the myo1-66 mutant. (A) Sequence alignment of the five S. cerevisiae myosin genes shows absolute conservation of a glutamate in the ATP-binding pocket of the motor domain. (B) myo1Δ cells expressing plasmid-borne, GFP-tagged wild-type (THY194) or myo1-66 mutant (THY195) type II myosin were grown at RT in selective media to mid-log phase and incubated at either RT or 37°C for 30 min. Optical z-series imaging was performed, and the resulting images were subjected to digital deconvolution. Images shown are 2D projections of 3D stacks and are representative of the larger cell population. Cell outlines are shown in white. GFP-tagged myo1-66p localizes to the bud neck, similar to wild-type Myo1p-GFP after incubation at RT or 37°C. Bar, 2 μm. (C) myo1Δ cells expressing plasmid-borne, GFP-tagged wild-type (THY194) or myo1-66 mutant (THY195) type II myosin were grown at RT in selective media to mid-log phase and incubated at either RT or 37°C for 30 min. Whole-cell lysates were evaluated by SDS-PAGE and Western blot analysis using antibodies specific for GFP to detect tagged myosin proteins and hexokinase (Hxk1p) as the loading control. myo1-66p-GFP is expressed at a slightly reduced level relative to wild-type Myo1p-GFP. Short-term incubation at 37°C had no obvious effect on the steady-state level of either Myo1p or myo1-66p.
To generate the mutant, the type II myosin gene (MYO1), which was tagged with GFP at its C terminus, was inserted into a low-copy (CEN) plasmid under control of the endogenous type II myosin promoter. Glu528 was than replaced with Lys in the plasmid-borne, MYO1 GFP-tagged gene. We refer to the mutated gene as myo1-66 to indicate that it shares the same mutation as myo2-66. The GFP-tagged myo1-66 mutant was expressed in lieu of wild-type myosin II, i.e., in a yeast strain bearing a deletion in the MYO1 gene (pmyo1-66 myo1Δ). Using Western blots probed with anti-GFP antibody, we found that the steady-state level of myo1-66p was 70% of the wild-type type II myosin level upon growth at permissive temperature (23°C) or after incubation at restrictive temperature (37°C) for 30 min (Fig. 3 C). Moreover, myo1-66 cells exhibited elevated levels of multibudded cells; the level of multibudded cells in these cultures increased upon incubation at 37°C (unpublished data). Nonetheless, the myo1-66 mutant protein localized normally to the bud neck throughout the cell cycle (Fig. 3 B). Moreover, the growth rate of these myo1-66 cells at 23°C was similar to that observed for the corresponding wild-type strain at 23°C. Finally, myo1-66 cells did not show temperature-dependent lethality, as expected, because type II myosin is not essential in the yeast genetic background used for these studies (Fig. 3).
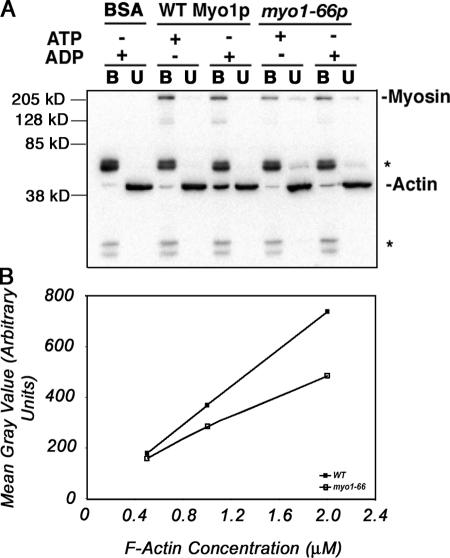
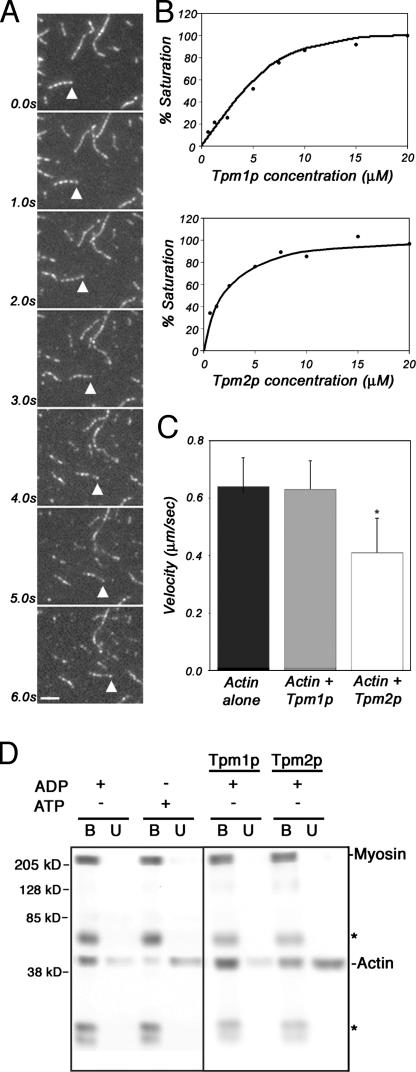
The effect of the myo2-66 mutation on the mechanochemical properties of type V myosin of yeast is not known. Therefore, we evaluated the rigor-binding activity of wild-type Myo1p and mutant myo1-66p to actin (Fig. 4 A). Wild-type myosin was immobilized on magnetic beads and incubated with yeast F-actin in the presence of ATP or ADP. In the presence of ATP, F-actin did not bind to wild-type type II myosin. However, in the presence of ADP, there was a significant increase in the amount of F-actin binding. Thus, we concluded that type II myosin exhibits ATP-sensitive actin-binding activity. Mutant myo1-66 protein also exhibited ATP-sensitive actin- binding activity under these experimental conditions. However, actin-binding activity of the mutant myo1-66 protein was lower than that observed in the wild-type protein (Fig. 4 A). This decrease in actin binding to mutant type II myosin was evident at all concentrations of actin tested (Fig. 4 B). Because wild-type protein lost activity after short-term incubation at elevated temperatures, we could not test whether the myo1-66 mutation confers temperature-dependent loss of actin-binding activity in vitro. Nonetheless, our results indicate that substitution of Glu528 with Lys in the actin-binding site of the type II myosin motor domain reduces the ATP-sensitive actin-binding activity of the protein.
Figure 4.
myo1-66p binds actin less effectively than wild-type Myo1p. Rigor-binding assays for GFP-tagged wild-type Myo1p and myo1-66p were performed by incubating myosin-bound magnetic beads with various concentrations of actin for 90 s in the presence of 1 mM ATP or ADP at 4°C, as described in Materials and methods. (A) Western blots showing bound and unbound fractions from a representative rigor-binding assay performed using 10 μM actin. Asterisks indicate IgG used to tether myosin to the beads. BSA-coated beads were used as a control for nonspecific actin binding. Magnetic bead fraction (B) and unbound fraction (U) were solubilized in SDS sample buffer. Equal volumes of solubilized samples were subjected to PAGE and Western blot analysis using antibodies specific for GFP and actin. (B) Quantitation by densitometric analysis of Western blots showing the amount of actin bound to immobilized wild-type or mutant type II myosin after incubation with 0.5–2 μM actin.
Given a clear understanding of the effect of the myo1-66 mutation on the type II myosin motor domain, we examined the retrograde flow rates of Abp140p-GFP–labeled actin cables in the myo1-66 mutant, relative to the wild-type (Fig. 5, A and B). At 23°C, the rate of retrograde actin cable flow in yeast expressing the myo1-66 mutant was similar to that observed in yeast expressing wild-type myosin II protein (Fig. 5 B). Thus, the mutant protein supports normal rates of retrograde flow at reduced temperatures, despite the fact that it is present at lower levels compared with wild-type cells. The rate of retrograde actin cable flow in wild-type cells was not affected by incubation at 37°C. In contrast, the rate of actin cable retrograde flow is significantly reduced in yeast expressing the myo1-66 mutant at 37°C. (Fig. 5, A and B). However, the rate of retrograde actin cable flow observed in the myo1-66 mutant at restrictive temperatures is similar to that observed in yeast bearing a deletion in MYO1 and in yeast expressing the Myo1p tail at wild-type levels in lieu of full-length Myo1p (Fig. 5 B). Thus, the myo1-66 mutant exhibits temperature- dependent reduction in the rate of retrograde actin cable flow. These data suggest that normal rates of actin cable retrograde flow require a functional type II myosin motor domain.
Figure 5.
Normal rates of actin cable retrograde flow require the actin-binding activity of type II myosin. The retrograde flow rate of Abp140p-labeled actin cables was examined in cells expressing wild-type MYO1 (YCY027) or myo1-66p (THY193) as the sole copy of type II myosin. Imaging was performed as described for Fig. 1. (A) Average rates of retrograde flow of Abp140p-GFP–labeled actin cables in wild-type cells, myo1-66 mutant at RT and 37°C, and myo1Δ cells that express full-length Myo1p (TLY002) or the Myo1p tail (TLY003) from low-copy, centromere-based plasmids (pTL2 and pTL3) under control of the MYO1 promoter. Rates of movement were determined as described in Materials and methods. At 37°C, there is a significant reduction in the rate of actin cable retrograde flow in myo1-66 cells compared with corresponding wild-type cells (P < 0.05). The rate of retrograde actin cable flow in the myo1-66 cell at restrictive temperatures is similar to that of the myo1Δ cell expressing Myo1p tail. Error bars represent the SD. n > 20 for each strain. (B) Still-frame images of a time-lapse series of Abp140p-GFP–labeled actin cables in the myo1-66 mutant at 23 (top) and 37°C (bottom). Arrowheads point to an Abp140p-GFP fiduciary mark on an actin cable undergoing retrograde flow. Arrows point to the position of the fiduciary mark at t = 0 of image analysis. Bar, 2 μm.
Isoform-specific role for Tpm2p in retrograde actin cable flow in vivo
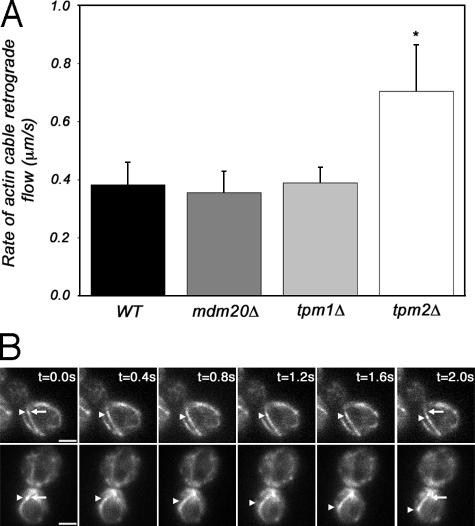
Earlier studies suggested that deletion of TPM1 results in loss of actin cables (Drees et al., 1995). However, with the advent of more sensitive optical imaging technology, it is now clear that deletion of TPM1 reduces the length and/or abundance of actin cables, but does not result in the complete loss of these structures. Time-lapse videos illustrating retrograde flow of actin cables in wild-type cells and tpm1Δ mutants are shown (Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200609155/DC1). Because actin cables are present in both tpm1Δ and tpm2Δ cells, we studied the effect of loss of the yeast tropomyosins on actin cable dynamics. We found that the rate of actin cable retrograde flow in tpm1Δ strains was not significantly different from that observed in wild-type cells (Fig. 6). In the tpm2Δ strain, however, the rate of actin cable retrograde flow was 0.78 μm/s, which is approximately twice that observed in wild-type cells (Fig. 6, A and B). This finding indicates that actin cable retrograde flow can occur at rates that are at least twice the mean rate of flow observed in wild-type cells. Equally important, this finding indicates that Tpm2p, but not Tpm1p, reduces the rate of retrograde actin cable flow.
Figure 6.
Loss of Tpm2p causes a significant increase in actin cable retrograde flow rate. The retrograde flow rate of Abp140p-labeled actin cables was examined in wild-type cells (YCY027) and in cells carrying a deletion of TPM1 (THY118), TPM2 (THY119), or MDM20 (THY114-2d). Imaging and analysis of retrograde actin flow rates were performed as described for Fig. 1. (A) Average rates of retrograde flow of Abp140p-GFP–labeled actin cables in wild-type cells, and mdm20Δ, tpm1Δ,or tpm2Δ mutants, at RT. Rates of movement were determined as described in Materials and methods. Deletion of TPM2 caused a significant increase in the rate of actin cable retrograde flow (P < 0.01), as indicated by the asterisk. Error bars represent the SD. n > 20 for each strain. (B) Still-frame images of a time-lapse series of Abp140p-GFP–labeled actin cables in wild-type cells (top) and the tpm2Δ mutant (bottom) at 23°C. Arrowheads point to an Abp140p-GFP fiduciary mark on an actin cable undergoing retrograde flow. Arrows point to the position of the fiduciary mark at t = 0 of image analysis. Bar, 2 μm.
To further explore the role of Tpm1p in retrograde actin cable flow, we studied Abp140p-GFP dynamics in yeast bearing a deletion in MDM20. Mdm20p was originally identified in a screen for mutations affecting mitochondrial inheritance (Hermann et al., 1997). Deletion of MDM20 results in reduced actin cable length, but has no effect on alignment of actin cables along the mother-bud axis. Recent evidence indicates that Mdm20p is associated with Nat3p, a protein that catalyzes acetylation of Tpm1p (Singer and Shaw, 2003). We find that deletion of MDM20 has no significant effect on the rate of retrograde actin cable flow (Fig. 6).
Isoform-specific role for Tpm2p in type II myosin–driven actin filament sliding in vitro
We found that the type II myosin Myo1p stimulates the rate of actin cable retrograde flow, whereas the tropomyosin Tpm2p has a negative effect on this process. Because both proteins bind to the lateral surface of F-actin, it is possible that Tpm2p regulates actin cable dynamics by competing with type II myosin for binding to F-actin within actin cables. One approach to test this model is to study actin cable dynamics in a cell bearing a deletion of MYO1 and TPM2. In the strains used for these studies, deletion of MYO1 and TPM2 produced synthetic lethality. Because Myo1p and Tpm2p have functions that are not related to their function in retrograde actin cable flow, and all strains used to generate the double mutant also carried a GFP tag on the ABP140 gene, the basis for the synthetic lethality is not readily apparent. Therefore, we could not evaluate retrograde actin cable flow in a myo1Δ tpm2Δ double mutant.
As an alternative approach to test this model, we performed in vitro assays to determine whether the tropomyosins exhibit isoform-specific effects on type II myosin motor–driven microfilament sliding and on the binding of type II myosin to F-actin. We found that type II myosin of budding yeast supports microfilament sliding in vitro. The assay used was a modification of a previously described assay (Reck-Peterson et al., 2001). GFP-tagged type II myosin from whole-cell extracts was immobilized in a microscope flow cell using anti-GFP antibody. The flow cell was treated with unlabeled F-actin and ATP to saturate ATP-insensitive, rigor-binding sites on immobilized type II myosin. Rhodamine-phalloidin–stained and stabilized yeast F-actin was then added in ATP- free buffer, and ATP was then added after washes to remove unbound material. Type II myosin–driven microfilament sliding was then monitored through time-lapse imaging.
Microfilament sliding driven by the yeast type II myosin was similar to that observed for other actin-based motors (Fig. 7 A). Microfilament sliding was unidirectional, continuous, and parallel to the long axis of the filament. The rate of movement of microfilament sliding was 0.63 ± 0.10 μm/s. This rate is similar to the rate of 0.5 μm/s observed for microfilament sliding by type II myosin isolated from the fission yeast Schizosaccharomyces pombe (Lord and Pollard, 2004). To our knowledge, this is the first direct evidence that the type II myosin of budding yeast has motor activity on actin.
Figure 7.
Tpm2p exerts isoform-specific effects on F-actin binding to myosin II and myosin II–mediated actin filament gliding. Myo1p-GFP was adsorbed from whole-cell lysate of yeast expressing GFP-tagged type II myosin protein (THY158) to a coverslip in an imaging flow cell using anti-GFP antibody. Chambers were preincubated with unlabeled actin and washed with ATP to block irreversible actin binding. Rhodamine-phalloidin–labeled actin filaments were added to the chamber. Upon addition of ATP, time-lapse imaging in a single focal plane was performed. (A) Still-frame images from a time-lapse series of rhodamine-phalloidin–labeled actin filaments gliding on a Myo1p-coated coverslip in the presence of ATP. Bar, 2 μm. (B) Tropomyosins purified from yeast bind to 10 μM actin with single micromolar affinity. Binding was determined by cosedimentation with F-actin in high-speed ultracentrifugation, followed by PAGE and quantitative densitometry of Coomassie-stained bands. Points shown are the means of three separate experiments. (C) Isoform-specific effects of Tpm2p on microfilament gliding on immobilized Myo1p. Average rates of actin filament gliding on a Myo1p-coated coverslip of actin alone or actin prebound to saturating levels (20 μM) of Tpm1p or Tpm2p. Asterisk indicates a significant reduction in the rate of actin filament gliding in the presence of saturating Tpm2p (P < 0.05). Error bars represent the SD. n > 40 for each condition. (D) Isoform-specific effects of Tpm2p on binding of F-actin to Myo1p. 20 μM phalloidin-stabilized yeast F-actin was pretreated with 10 μM purified Tpm1p or Tpm2p at 4°C for 15 min. 30 μl of the pretreated actin was mixed with magnetic beads containing immobilized Myo1p-GFP. Nucleotide (ATP or ADP) was added to a final concentration of 160 μM and the reaction mixture was incubated at 4°C for 75 s. Magnetic beads were separated from the reaction mixture and solubilized in SDS sample buffer. A 20-μl aliquot of the supernatant was also solubilized in SDS sample buffer. Solubilized material was analysis by SDS-PAGE and Western blots. Purification of actin, Tpm1p, and Tpm2p was performed as described in Materials and methods. Immobilization of Myo1p-GFP on magnetic beads, separation of magnetic beads from the binding reaction mixture, and solubilization and analysis of magnetic beads and supernatant after binding were all performed as for Fig. 4. Asterisks mark IgG heavy and light chains. The image shown is a single blot that was exposed at two different times to produce similar density for all Myo1p-GFP bands. Lanes 1–4 (left) and lanes 5–8 (right) were from different exposures.
To determine whether tropomyosins exert isoform-specific effects on this process, we used established protocols to purify Tpm1p from yeast carrying a deletion in TPM2, and Tpm2p from yeast carrying a deletion in TPM1. Pull-down assays, which measured binding of the yeast tropomyosins to yeast actin, revealed that purified Tpm1p and Tpm2p retained biological activity (Fig. 7 B). The affinity of purified Tpm1p and Tpm2p for yeast actin is similar to that reported previously for rabbit skeletal muscle actin (Maytum et al., 2000).
Finally, we tested the effects of Tpm1p and Tpm2p on type II myosin–driven microfilament sliding and on binding of F-actin to type II myosin. To do so, we compared the rates of type II myosin–driven sliding of F-actin that was either untreated or pretreated with saturating amounts of Tpm1p or Tpm2p (Fig. 7 C). We found that the rate of microfilament gliding of F-actin containing bound Tpm1p (0.64 ± 0.10 μm/s) was similar to that observed for untreated F-actin without tropomyosin (0.63 ± 0.10 μm/s). In contrast, the rate of sliding of actin filaments containing bound Tpm2p (0.41 ± 0.12 μm/s) was significantly lower than that observed for either untreated or Tpm1p-treated F-actin. Interestingly, Tpm2p exhibits isoform-specific effects on type II myosin–driven microfilament sliding in vitro that is equal and opposite to the observed effect of deletion of TPM2 on type II myosin–mediated retrograde actin cable flow in vivo. This finding is consistent with the interpretation that Tpm2p exerts an isoform-specific effect on inhibition of binding of F-actin to yeast type II myosin. To test this hypothesis directly, phalloidin-stabilized yeast F-actin was pretreated with equal amounts of purified Tpm1p or Tpm2p, and tested for binding to immobilized type II myosin (Fig. 7 D). At the concentrations tested, we found that Tpm2p, but not Tpm1p, inhibited the binding of actin filaments to type II myosin of budding yeast.
Discussion
Retrograde actin flow occurs in cortical actin networks in both motile and nonmotile cells, as well as in actin bundles in apical protrusions including microvilli, filopodia, and stereocilia. Several studies support a role for type II myosin and a tropomyosin in this process. First, both proteins localize to the lamella, an actin network that is proximal to the lamellipodium and exhibits retrograde flow in migrating cells (Ponti et al., 2004). Second, retrograde flow of actin lamella and of actin bundles in neuronal filopodia is perturbed by 2,3-butanedione monoxime and blebbistatin, agents that inhibit type II myosin ATPase activity (Lin et al., 1996; Waterman-Storer and Salmon, 1997; Salmon et al., 2002; Ponti et al., 2004; Gupton et al., 2005; Medeiros et al., 2006). Third, microinjection of the skeletal muscle tropomyosin, but not endogenous tropomyosin, into PtK1 cells produces an altered rate of retrograde actin flow (Gupton et al., 2005).
Although type II myosins and specific tropomyosin isoforms have been implicated in retrograde actin flow in animal cells, there was no evidence for a role of these proteins in actin cable dynamics in budding yeast. Moreover, the precise function of tropomyosin in retrograde actin flow in animal cells has been unclear. We provide evidence that type II myosin and a specific tropomyosin contribute to the retrograde flow of actin cables in budding yeast. Our findings are consistent with a role for type II myosin in providing a pulling force on actin cables as they undergo retrograde flow. Our findings also indicate that type II myosin and Tpm2p may have antagonistic effects during retrograde actin cable flow. Tpm2p may exert its isoform-specific negative regulatory effect by inhibiting the binding of type II myosin with F-actin within actin cables. These findings indicate that retrograde actin flow is conserved from yeast to animal cells, and extend our understanding of the mechanism of action of type II myosin and tropomyosin in this process.
A new role for type II myosin in budding yeast
Although type II myosin has been implicated in cytokinesis in many cell types, the precise role of this protein in budding yeast remains enigmatic. Type II myosin is present at the bud neck during contractile ring closure (Bi et al., 1998; Lippincott and Li, 1998). However, localization studies have revealed that this type II myosin protein behaves differently compared with other actomyosin ring components. Specifically, actin and other resident contractile ring components localize to the bud neck at the G2–M transition in the cell cycle (Bi et al., 1998; Vallen et al., 2000; Tolliday et al., 2001). In contrast, Myo1p localizes to the bud neck earlier in the cell cycle, during G1 phase, immediately after commitment to a round of cell division. In addition, recent data suggests that the motor domain of type II myosin may not be required for cytokinesis in budding yeast. Specifically, Lord et al. (2005) have showed that budding yeast expressing the type II myosin tail in lieu of the wild-type protein exhibit relatively normal cytokinesis, with relatively little change in the rate of actin ring constriction and disassembly at the bud neck.
We find that deletion or mislocalization of type II myosin results in a reduction in the rate of retrograde actin cable flow. These observations indicate that localization of the protein at the bud neck is critical for its function in retrograde actin cable flow. Moreover, because retrograde actin cable flow occurs from the time of commitment to cell division until the end of cytokinesis in budding yeast, these observations explain why type II myosin localizes earlier to the bud neck during G1–M stages in the cell cycle, well before the actomyosin ring is assembled.
We also find that deletion or mutation of the type II myosin motor domain produces a decrease in the rate of retrograde actin cable flow that is similar to that observed upon deletion or mislocalization of full-length type II myosin. These observations indicate that type II myosin may exert a pulling force on actin cables as they move through the bud neck, a known “bottleneck” for intracellular transport in budding yeast. Additionally, because actin cables can assemble at the bud neck, type II myosin may pull newly assembled segments of actin cable away from the assembly site, providing greater access to the end of actin cables for the assembly machinery to continue cable assembly and elongation.
Tropomyosin function in budding yeast
We find that deletion of TPM2, but not TPM1, increases the rate of retrograde actin cable flow. These observations reveal a first isoform-specific function for Tpm2p as a negative regulator of retrograde actin cable flow in budding yeast. Moreover, we found that Tpm2p, but not Tpm1p, inhibits the binding of type II myosin to F-actin and microfilament gliding in vitro. Therefore, our evidence supports a model whereby Tpm2p exerts isoform-specific inhibition of retrograde actin cable flow by inhibiting the binding of type II myosin to F-actin present in dynamic actin cables.
Our studies also extend our understanding of the regulation of tropomyosin acetylation in budding yeast. Tropomyosins in yeast and other eukaryotes are modified by N-terminal acetylation. This cotranslational modification is essential for binding of tropomyosin to F-actin (Maytum et al., 2000). The mechanism for acetylation of Tpm2p is not known. However, recent studies indicate that Mdm20p functions with Nat3p to acetylate Tpm1p and regulate its actin-binding activity (Singer and Shaw, 2003). Because deletion of TPM2 increases the rate of retrograde actin cable flow, whereas deletion of either MDM20 or TPM1 has no effect on that process, we conclude that Mdm20p may not contribute to acetylation of Tpm2p.
Finally, these studies also extend our understanding of Tpm1p function in budding yeast. As described in the Introduction, one primary function of Tpm1p is to control actin cable stability and function. We find that Tpm1p binds to F-actin, but does not inhibit the binding of type II myosin to F-actin. Thus, the isoform-specific biochemical activity of Tpm1p is optimal for its function in budding yeast; it allows Tpm1p to stabilize actin cables without interfering with the ability of actin cables to serve as tracks for myosin-driven cargo transport during bud formation and growth.
The functions of retrograde flow
Because actin cables undergo retrograde flow, the track for anterograde movement is itself moving in the direction opposite that of transport. As a result, cargos that use actin cables as tracks are “swimming upstream” and must overcome the opposing force of retrograde actin cable flow. The rate of secretory vesicle movement driven by the essential type V myosin Myo2p (∼3 μm/s) is an order of magnitude higher than the rate of retrograde actin cable flow (Schott et al., 2002). In this case, retrograde actin cable flow may not have any significant effect on anterograde transport. In contrast, the rates of Arp2/3 complex–driven anterograde mitochondrial movement (0.3 μm/s) and of type V myosin (Myo4p)–driven mRNA movement (0.03 μm/s) are significantly lower (Beach et al., 1999; Fehrenbacher et al., 2004). In these cases, retrograde actin cable flow may have a significant effect on anterograde transport. In the case of mitochondria, we have proposed that actin cable dynamics favor bud-directed transport, and therefore would also favor inheritance of organelles that can harness the most robust anterograde force–generating machinery, i.e., inheritance of the “fittest” mitochondria (Boldogh et al., 2005). Thus, retrograde actin cable flow can affect the rate and efficacy of anterograde transport.
Work from this laboratory and others revealed a second function for retrograde actin flow—driving retrograde movement. Retrograde flow of actin networks in the leading edge of motile cells can drive movement of surface receptors and tubulin (Bray, 1970; Waterman-Storer and Salmon, 1997; Salmon et al., 2002). Retrograde actin flow has also been shown to drive movement of the nucleus, which results in reorientation and polarization of the microtubule-organizing center in migrating cells (Gomes et al., 2005). In budding yeast, mitochondria and actin patches bind to actin cables undergoing retrograde flow and use actin cables as “conveyor belts” for retrograde movement (Fehrenbacher et al., 2004; Huckaba et al., 2004). Indeed, because genes encoding pointed-end–directed myosins have not been identified in the yeast genome, retrograde actin flow may be the primary force for actin-dependent retrograde movement in this cell type. Thus, retrograde actin flow can drive retrograde movement in yeast and other eukaryotic cell types. Equally important, the organization and dynamics of actin cables in budding yeast allow for bidirectional movement using a single polarized cytoskeletal structure.
Finally, our finding that Tpm2p can regulate the rate of type II myosin–driven retrograde actin cable flow raises the question of why regulating retrograde flow rates would benefit the cell. A twofold increase in the rate of actin cable retrograde flow would have a significant impact on the rate of anterograde transport of mitochondria and mRNA. Thus, regulation of retrograde actin cable flow can affect mitochondrial inheritance and mRNA localization. Regulation of retrograde flow rates in budding yeast may also affect retrograde movement of mitochondria during inheritance, as well as retrograde movement of endosomes and their subsequent fusion with the sorting compartment. Indeed, our preliminary evidence indicates that the rate of retrograde endosome movement is reduced in yeast bearing mutations that inhibit retrograde actin cable flow. Studies designed to further evaluate the physiological role of altering the rate of actin cable retrograde flow are ongoing.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study are listed in Table II. With the exception of the myo3Δ myo5Δ strain, which is in the W303 genetic background, all strains used are S288C. Yeast cell growth and manipulations were performed according to established protocols (Sherman, 2002). To visualize actin cables in living cells, the C terminus of Abp140p was tagged with GFP (S65T) using PCR-based insertion of the GFP gene (Longtine et al., 1998) into the chromosomal copy of ABP140, as previously described (Yang and Pon, 2002). The cassette used to insert GFP into the ABP140 gene of wild-type and myosin mutant cells was amplified from pFA6a:GFP (S65T):HIS3 with the following primers: forward 5′- AAATATTGATAGTAACAATGCACAGAGTAAAATTTTCAGTCGGATCCCCGGGTTATTAA-3′ and reverse 5′-AAAGGATATAAAGTCTTCCAAATTTTTAAAAAAAAGTTCGGAATTCGAGCTCGTTAAAC-3′. Amplified DNA was transformed into the strains of interest, and cells that had integrated the GFP cassette into the ABP140 gene were selected by their ability to grow on synthetic media lacking histidine.
Table II.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YCY006 | MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 ABP140∷GFP∷KanMX6 | Yang and Pon, 2002 |
| YCY009 | MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 myo3Δ∷HIS3 myo5Δ∷TRP1
ABP140∷GFP∷KanMX6 |
this study |
| YCY010 | MATα leu2-3,112 his3-200 ura3-52 myo2-66∷HIS3 myo4Δ∷URA3 ABP140∷GFP∷KanMX6 | this study |
| YCY027 | MATα his3Δ leu2Δ met15Δ ura3Δ ABP140∷GFP∷KanMX6 | Huckaba et al., 2004 |
| THY114-2d | MATα his3Δ leu2Δ met15Δ lys2Δ ura3Δ mdm20Δ∷KanMX6 ABP140∷GFP∷KanMX6 | this study |
| THY118 | MATα his3Δ leu2Δ met15Δ ura3Δ tpm1Δ∷KanMX6 ABP140∷GFP∷HIS3 | this study |
| THY119 | MATα his3Δ leu2Δ met15Δ ura3Δ tpm2Δ∷KanMX6 ABP140∷GFP∷HIS3 | this study |
| THY120 | MATα leu2-3,112 ura3-52 ade1-101 myo1Δ∷URA3 ABP140∷GFP∷KanMX6 | this study |
| THY158 | MATα his3Δ leu2Δ met15Δ ura3Δ MYO1∷GFP∷HIS3 | this study |
| THY159 | MATα his3Δ leu2Δ met15Δ ura3Δ MYO1∷GFP∷HIS3 pTH12 (MYO1-tail)∷URA3 | this study |
| THY160 | MATα his3Δ leu2Δ met15Δ ura3Δ ABP140∷GFP∷KanMX6 pTH12 (pGal-MYO1-tail)∷URA3 | this study |
| THY192 | MATα his3Δ leu2Δ met15Δ ura3Δ ABP140∷GFP∷kanMX pTH15 (myo1-66∷GFP)∷URA3 | this study |
| THY193 | MATα his3Δ
leu2Δ
met15Δ
ura3Δ
ABP140∷GFP∷kanMX pTH15 (myo1-66∷GFP)∷URA3
myo1Δ∷HIS3 |
this study |
| THY194 | MATα his3Δ leu2Δ met15Δ ura3Δ p846 (MYO1-GFP)∷URA3 | this study |
| THY195 | MATα his3Δ leu2Δ met15Δ ura3Δ pTH14 (myo1-66)∷URA3 | this study |
| THY196 | MATα his3Δ leu2Δ met15Δ ura3Δ tpm2Δ∷kanMX pEH002 (GAL-TPM1)∷URA3 | this study |
| THY197 | MATα his3Δ leu2Δ met15Δ ura3Δ tpm1Δ∷kanMX pEH006 (GAL-TPM2)∷URA3 | this study |
| VSY21 | MATα leu2-3,112 his3-200 ura3-52 myo2-66∷HIS3 myo4Δ∷URA3 | Simon et al., 1995 |
| HA31-9c | MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 myo3Δ∷HIS3 myo5Δ∷TRP1 | Goodson et al., 1996 |
| BN4 | MATα leu2-3,112 ura3-52 ade1-101 myo1Δ∷URA3 | Negron et al., 1996 |
| BY4741 | MATα his3Δ leu2Δ met15Δ ura3Δ | Open Biosystems |
| RG7224 | MATα his3Δ leu2Δ met15Δ ura3Δ tpm1Δ∷KanMX6 | Open Biosystems |
| RG2297 | MATα his3Δ leu2Δ met15Δ ura3Δ tpm2Δ∷KanMX6 | Open Biosystems |
| RG11767 | MATα his3Δ leu2Δ lys2Δ ura3Δ mdm20Δ∷KanMX6 | Open Biosystems |
| TLY002 | MATα his3Δ leu2Δ met15Δ ura3Δ ABP140∷GFP∷kanMX pTL02 (MYO1∷GFP)∷LEU2 | this study |
| TLY003 | MATα his3Δ leu2Δ met15Δ ura3Δ ABP140∷GFP∷kanMX pTL03 (MYO1-Tail∷GFP)∷LEU2 | this study |
The strain containing a deletion in the MDM20 gene and a GFP tag on Abp140p (THY114-2d) was constructed by mating a haploid mdm20Δ cell (RG11767) with a wild-type cell expressing Abp140p-GFP (YCY027). The resulting diploids were sporulated in medium containing 1% KOAc + 0.025% glucose, and haploids produced from the diploids were selected using a visual screen for GFP-labeled actin cables and PCR to verify deletion of the gene of interest.
Plasmid pTH12 is a two-micrometer plasmid that encodes the 868 C-terminal amino acids of Myo1p under the control of the GAL1-10 promoter (similar to that described by Tolliday et al. [2003]). For the construction, DNA encoding the tail region of MYO1 was amplified from wild-type yeast genomic DNA using the following forward and reverse primers: 5′-TCAGGATCCTGACATAAAATTGGTCACTTTAGA-3′ and 5′-AGTTAAA AGCTTTTAACTGAAAATTTTACTCTGTGCATT-3′, respectively. The forward primer contains a BamHI restriction site and the reverse primer contains a HindIII restriction site. Restriction sites are underlined. The product was purified and ligated into the BamHI and HindIII sites of the plasmid pGAL68. The resulting vector contains an in-frame ATG from codon 1060 of MYO1 downstream of the GAL1 promoter. Induction of Myo1p tail overexpression was performed by shifting cells from synthetic media supplemented with raffinose into synthetic media containing both raffinose and galactose.
Plasmid pTH14 is a CEN-based plasmid that contains the MYO1 promoter within 555 bases of the upstream untranslated region of the gene followed by a GFP (S65T)-tagged MYO1 gene that carries an E528K mutation (myo1-66). To construct this plasmid, site-directed mutagenesis was performed on the plasmid p846 using the QuikChange Site-Directed Mutagenesis kit (Stratagene), which is a PCR-based method for mutagenesis. Primers were designed to mutate glutamate 528 to a lysine by a single nucleotide substitution (GAA→AAA). After PCR amplification, the mutated fragment was cleaved using AgeI and BclI and reinserted into p846. This new plasmid was sequenced to confirm the presence of the mutation. The plasma used to construct pTH14, which contains wild-type MYO1, was provided by E. Bi (University of Pennsylvania, Philadelphia, PA).
Plasmids pTL2 and pTL3 are centromere-based plasmids that contain full-length MYO1 and the MYO1 tail (aa 842–1,928) under control of the MYO1 promoter. These plasmids were produced from pGFP-MYO1 and pGFP-MYO1-Tail, which were generously provided by M. Lord (University of Vermont, Burlington, VT) and T. Pollard (Yale University, New Haven, CT; Lord et al., 2005). To allow for transformation into strains of interest, the TRP1 marker on pGFP-MYO1 and pGFP-MYO1-Tail was replaced with LEU2. To do so, the TRP1 marker was excised from pGFP-MYO1 and pGFP-MYO1-Tail by digestion with PvuI. The LEU2 marker, which was amplified from pRS415, was then inserted into the plasmid at the PvuI site.
Microscopy and image analysis
For time-lapse fluorescence imaging of Abp140p-GFP, cells were grown in synthetic complete or lactate medium (Yang and Pon, 2002) until early log phase at 25°C. 3 μl of cell suspension was applied to a microscope slide and covered with a coverslip. Microscopy was performed using a microscope (E600; Nikon) equipped with a Plan Apo ×100/1.4 NA objective and a cooled charge-coupled device camera (Orca-ER; Hamamatsu). Illumination with a 100 W mercury arc lamp was controlled with a shutter (Uniblitz D122; Vincent Associates). The temperature of the objective lens was controlled using an objective heater (Bioptechs). Images were collected and analyzed using Openlab 3.0.8 software (Improvision) and ImageJ 1.28 (National Institutes of Health), respectively. The exposure time and time interval between successive image acquisitions were 400 and ∼500 ms, respectively. The total imaging time for the time-lapse imaging was 20 s. For determination of the velocity of elongating actin cables, the change in position of fluorescent fiduciary marks on elongating cables was measured as a function of time, as previously described (Yang and Pon, 2002).
Time-lapse imaging of fluorescently labeled actin filaments in actin- gliding assays was performed as described in the previous paragraph. The exposure time and time interval between successive image acquisitions were 400 and 500 ms, respectively. The total imaging time for the time-lapse imaging was 50 s. For determination of the velocity of microfilament gliding, the change in position of the tips of gliding actin filaments was measured as a function of time, as previously described (Yang and Pon, 2002).
For analysis of the localization of GFP-tagged wild-type and myo1-66 mutant proteins, 25 z sections were obtained at 0.2-μm intervals through the entire cell. Z sectioning for 3D imaging was performed using a piezoelectric focus motor mounted on the objective lens of the microscope (Polytech PI). Out-of-focus light was removed by digital deconvolution, and each series of deconvolved images was projected and rendered with Volocity software (Improvision, Inc.).
Protein purification
Yeast actin was isolated from S. cerevisiae as previously described (Boldogh and Pon, 2001). In brief, actin was purified from cell lysate by DNase affinity chromatography, followed by ion-exchange chromatography using DE52. Peak fractions were pooled and subjected to multiple rounds of polymerization/depolymerization to ensure polymerization-competent actin. Yeast tropomyosins (Tpm1p and Tpm2p) were purified as previously described (Maytum et al., 2000). Tpm1p was purified from a strain that carries a deletion in the TPM2 gene and is transformed with a plasmid bearing TPM1 under control of the galactose-inducible GAL1 promoter (THY196). Tpm2p was purified from a strain that carries a deletion in the TPM1 gene and is transformed with a plasmid bearing TPM2 under control of the galactose-inducible GAL1 promoter (THY197). Tropomyosin constructs were provided by A. Bretscher and D. Pruyne (Cornell University, Ithaca, NY). Protein concentrations were determined using the bicinchoninic acid protein assay reagent (Pierce Chemical Co.) according to the manufacturer's protocol.
Preparation of yeast cell lysate
For purification of actin and the GFP-tagged wild-type and mutant Myo1p proteins from yeast, cells were grown to mid-log phase, concentrated by centrifugation (1,500 g for 10 min), and suspended in cell lysate buffer (25 mM imidazole-HCl, pH 7.4, 25 mM KCl, 4 mM MgCl2, 200 μM ATP, 2 mM DTT, 1 mM EGTA, 1 mM PMSF, and a previously described protease inhibitor cocktail [Lazzarino et al., 1994]). The cell suspension was then added drop-wise into liquid nitrogen. The resulting frozen pellets were ground in a liquid nitrogen–cooled coffee grinder for 5–10 s, and the ground powder was stored at –80°C. The ground powder was thawed at 4°C and clarified by centrifugation at 1,500 g for 4 min at 4°C. The pellet was discarded and the supernatant was used as in whole-cell lysate for microfilament gliding and myosin–actin–binding assays.
For quantitation of the steady-state level of Myo1p and myo1-66p, cells were grown to mid-log phase (OD600 = 0.7) in 10 ml of synthetic complete–ura liquid medium. Cells were concentrated by centrifugation, washed with distilled water, and resuspended in 300 μl lysis buffer (50 mM imidazole, pH 7.4, 1% Triton X-100, 2 mM PMSF, and protein inhibitor cocktails 1 and 2). 1 ml of 0.5-mm glass beads (Biospec Products, Inc.) were washed in lysis buffer and added to the cell suspension. The mixture was vortexed vigorously for 6 min at 4°C. Thereafter, 200 μl of the lysate was removed and mixed with 66.6 μl of 4× SDS sample buffer. The mixture was incubated at 100°C for 3–4 min. Proteins in the solubilized lysate were analyzed by SDS-PAGE, followed by Western blots.
Tropomyosin–actin–binding assays
Purified yeast tropomyosins at the indicated concentrations were added to 0.4 mg/ml F-actin in tropomyosin-binding buffer (150 mM KCl, 5 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 1 mM EGTA, and 1 mM DTT). Samples were incubated for 20 min at 4°C. Samples were then subjected to ultracentrifugation for 20 min at 100,000 g at 4°C in a fixed angle rotor (TA 100.3) using a ultracentrifuge (TL-100; both Beckman Coulter). Supernatant was removed, and the pellet was resuspended in an equal volume of SDS sample buffer. Equal volumes were loaded in wells of a polyacrylamide gel and subjected to electrophoresis, followed by Coomassie blue staining. Quantitative densitometry was performed to assess the cosedimentation of tropomyosin with actin (bound vs. unbound fractions).
Microfilament gliding assays
Construction of nitrocellulose-coated flow chambers.
Ethanol- washed coverslips were coated with 0.5% nitrocellulose and allowed to air-dry. The coverslip was then applied onto a microscope slide with the coated side facing down, using double-stick tape as adherent spacers. The resultant flow chamber volume was ∼30 μl.
Immobilization of GFP-tagged type II myosin in the flow chamber.
0.5 mg/ml protein A in TBS was added to the flow chamber and allowed to incubate for 30 min at RT. Unbound protein A was removed by three washes with TBS. 50 μg/ml anti-GFP antibody (Roche) was added to the flow chamber and allowed to incubate for 30 min at RT. Unbound antibody was removed by three washes with PBS. To prevent nonspecific binding, 5 mg/ml BSA in TBS was added, and after a 30-min incubation at RT, unbound material was removed by two washes with TBS and two washes with cell lysate buffer. Finally, cell lysate from THY158 (wild-type cells expressing C-terminally GFP-tagged Myo1p) was added to the flow chamber and incubated for 2–4 h at 4°C. Unbound material was removed by two washes with motility buffer (25 mM imidazole HCl, pH 7.4, 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 600 mM sorbitol, 9.35 U/ml glucose oxidase, 3.6 μg/ml catalase, 3 mg/ml glucose, 1 mM PMSF, 10 mM DTT, and protease inhibitor cocktail as described in the previous sections) supplemented with 4 mg/ml BSA, followed by two washes in BSA-free motility buffer.
Pretreatment with unlabeled F-actin.
To block all rigor-binding sites of immobilized type II myosin, unlabeled phalloidin-stabilized yeast actin filaments (25 μg/ml in motility buffer) were added to the flow chamber and allowed to incubate for 2 min at 4°C. Unbound material was then removed by two washes with motility buffer supplemented with 10 μM ATP and an ATP-regenerating system (10 mM creatine phosphate and 100 μg/ml creatine phosphokinase). Unbound material was removed by three washes with motility buffer.
Visualization of microfilament gliding.
Yeast F-actin that was fluorescently labeled and stabilized with rhodamine phalloidin was suspended in ATP-free motility buffer to a final concentration of 10 μg/ml. This solution was added to the flow chamber containing immobilized type II myosin. After incubation at RT for 2 min, unbound material was removed by two washes with motility buffer. The microscope slide flow chamber was placed on the stage of an epifluorescence microscope (see above). Motility buffer containing ATP and an ATP-regenerating system was added and microfilament gliding was monitored by time-lapse fluorescence imaging as described in Microscopy and image analysis.
Myosin–actin–binding assays
Goat anti–mouse IgG magnetic beads (New England Biolabs; 10 μl per reaction) were washed three times with sterile-filtered TBS, pH 8.0, and incubated with 5 μg of mouse anti-GFP IgGs (Roche) for 1 h at 4°C. Beads were washed two times in sterile-filtered TBS and two times in lysate buffer, and then incubated with 100 μl of either 1 mg/ml BSA, lysate from cells expressing GFP-tagged Myo1p, or lysate from cells expressing GFP-tagged myo1-66p for 2 h at 4°C. Beads were washed twice in lysate buffer and twice with motility buffer to remove unbound material. Phalloidin-stabilized yeast F-actin was added at the concentrations indicated in the presence of 1 mM ATP or ADP, pH 7.4. After incubation at 4°C for 90 s, beads were separated from the supernatant. Bound material (beads) and unbound material were solubilized in SDS sample buffer and subjected to PAGE and Western blot analysis using antibodies that recognized GFP and actin. Quantitative densitometry was performed to assess the fraction of actin that was bound to myosin or released to the supernatant.
Online supplemental material
Video 1 shows retrograde actin cable flow in wild-type cells. Video 2 shows retrograde actin cable flow in myo1Δ cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200609155/DC1.
Supplementary Material
Acknowledgments
We thank A. Bretscher and D. Pruyne for tropomyosin expression constructs, E. Bi for the Myo1p-GFP plasmid, M. Lord and T. Pollard for the MYO1 constructs, J. Rodriguez-Medina for the myo1Δ strain, J. Lessard for C4D6 antibody, G. Schatz for the anti-hexokinase antibody, I. Boldogh for guidance in protein purification, and members of the Pon laboratory for critical reading of this manuscript.
This work was supported by research grants from the National Institutes of Health to L. Pon (R01 GM066307) and to T. Huckaba (DDK07786).
References
- Adams, A.E., and J.R. Pringle. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D.C. 1998. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol. Biol. Cell. 9:3259–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura, T., T. Sasaki, F. Nagano, A. Satoh, H. Obaishi, H. Nishioka, H. Imamura, K. Hotta, K. Tanaka, H. Nakanishi, and Y. Takai. 1998. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 16:121–130. [DOI] [PubMed] [Google Scholar]
- Beach, D.L., E.D. Salmon, and K. Bloom. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9:569–578. [DOI] [PubMed] [Google Scholar]
- Bi, E., P. Maddox, D.J. Lew, E.D. Salmon, J.N. McMillan, E. Yeh, and J.R. Pringle. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I.R., and L.A. Pon. 2001. Assaying actin-binding activity of mitochondria in yeast. Methods Cell Biol. 65:159–173. [DOI] [PubMed] [Google Scholar]
- Boldogh, I.R., H.C. Yang, W.D. Nowakowski, S.L. Karmon, L.G. Hays, J.R. Yates III, and L.A. Pon. 2001. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc. Natl. Acad. Sci. USA. 98:3162–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I.R., K.L. Fehrenbacher, H.C. Yang, and L.A. Pon. 2005. Mitochondrial movement and inheritance in budding yeast. Gene. 354:28–36. [DOI] [PubMed] [Google Scholar]
- Bray, D. 1970. Surface movements during the growth of single explanted neurons. Proc. Natl. Acad. Sci. USA. 65:905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J., and P.C. Bridgman. 2003. Role of myosin II in axon outgrowth. J. Histochem. Cytochem. 51:421–428. [DOI] [PubMed] [Google Scholar]
- Dong, Y., D. Pruyne, and A. Bretscher. 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B., C. Brown, B.G. Barrell, and A. Bretscher. 1995. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J. Cell Biol. 128:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D.G., K.G. Miller, and D. Botstein. 1988. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107:2551–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., B.M. Klebl, A.H. Tong, B.A. Webb, T. Leeuw, E. Leberer, M. Whiteway, D.Y. Thomas, and C. Boone. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., D. Pruyne, D.C. Amberg, C. Boone, and A. Bretscher. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:32–41. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher, K.L., H.C. Yang, A.C. Gay, T.M. Huckaba, and L.A. Pon. 2004. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 14:1996–2004. [DOI] [PubMed] [Google Scholar]
- Forscher, P., and S.J. Smith. 1988. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107:1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, E.R., S. Jani, and G.G. Gundersen. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. [DOI] [PubMed] [Google Scholar]
- Goodson, H.V., and J.A. Spudich. 1995. Identification and molecular characterization of a yeast myosin I. Cell Motil. Cytoskeleton. 30:73–84. [DOI] [PubMed] [Google Scholar]
- Goodson, H.V., B.L. Anderson, H.M. Warrick, L.A. Pon, and J.A. Spudich. 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133:1277–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans, B.L., H. Grotsch, D. Mukhopadhyay, I.M. Fernandez, J. Pannsfield, F.Z. Idrissi, J. Lechner, H. Riezman, and M.I. Geli. 2006. TEDS site phosphorylation of the yeast myosins I is required for ligand-induced but not for constitutive endocytosis of the G protein-coupled receptor Ste2p. J. Biol. Chem. 281:11104–11114. [DOI] [PubMed] [Google Scholar]
- Gupton, S.L., W.C. Salmon, and C.M. Waterman-Storer. 2002. Converging populations of f-actin promote breakage of associated microtubules to spatially regulate microtubule turnover in migrating cells. Curr. Biol. 12:1891–1899. [DOI] [PubMed] [Google Scholar]
- Gupton, S.L., K.L. Anderson, T.P. Kole, R.S. Fischer, A. Ponti, S.E. Hitchcock-DeGregori, G. Danuser, V.M. Fowler, D. Wirtz, D. Hanein, and C.M. Waterman-Storer. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer, B.K., A. Petzold, S.H. Lillie, and S.S. Brown. 1994. Identification of MYO4, a second class V myosin gene in yeast. J. Cell Sci. 107:1055–1064. [DOI] [PubMed] [Google Scholar]
- Heath, J.P. 1983. Behaviour and structure of the leading lamella in moving fibroblasts. I. Occurrence and centripetal movement of arc-shaped microfilament bundles beneath the dorsal cell surface. J. Cell Sci. 60:331–354. [DOI] [PubMed] [Google Scholar]
- Henson, J.H., T.M. Svitkina, A.R. Burns, H.E. Hughes, K.J. MacPartland, R. Nazarian, and G.G. Borisy. 1999. Two components of actin-based retrograde flow in sea urchin coelomocytes. Mol. Biol. Cell. 10:4075–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G.J., E.J. King, and J.M. Shaw. 1997. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J. Cell Biol. 137:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K.L., N.L. Catlett, and L.S. Weisman. 1996. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 135:1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba, T.M., A.C. Gay, L.F. Pantalena, H.C. Yang, and L.A. Pon. 2004. Live cell imaging of the assembly, disassembly, and actin cable– dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 167:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, E., J. Kusch, Y. Barral, and T.C. Huffaker. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi, F.Z., B.L. Wolf, and M.I. Geli. 2002. Cofilin, but not profilin, is required for myosin-I-induced actin polymerization and the endocytic uptake in yeast. Mol. Biol. Cell. 13:4074–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, H., K. Tanaka, T. Hihara, M. Umikawa, T. Kamei, K. Takahashi, T. Sasaki, and Y. Takai. 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16:2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, G.C., J.A. Prendergast, and R.A. Singer. 1991. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113:539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado, C., J.R. Haserick, and J. Lee. 2005. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol. Biol. Cell. 16:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, H., K. Tanaka, A. Mino, M. Umikawa, H. Imamura, T. Fujiwara, Y. Fujita, K. Hotta, H. Qadota, T. Watanabe, et al. 1996. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Lazzarino, D.A., I. Boldogh, M.G. Smith, J. Rosand, and L.A. Pon. 1994. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol. Biol. Cell. 5:807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie, S.H., and S.S. Brown. 1994. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 125:825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.H., E.M. Espreafico, M.S. Mooseker, and P. Forscher. 1996. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 16:769–782. [DOI] [PubMed] [Google Scholar]
- Lin, H.W., M.E. Schneider, and B. Kachar. 2005. When size matters: the dynamic regulation of stereocilia lengths. Curr. Opin. Cell Biol. 17:55–61. [DOI] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li. 1998. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143:1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.P., and A. Bretscher. 1989. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 57:233–242. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Lord, M., and T.D. Pollard. 2004. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J. Cell Biol. 167:315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, M., E. Laves, and T.D. Pollard. 2005. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell. 16:5346–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu, A., and T. Mitchison. 1999. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 146:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytum, R., M.A. Geeves, and M. Konrad. 2000. Actomyosin regulatory properties of yeast tropomyosin are dependent upon N-terminal modification. Biochemistry. 39:11913–11920. [DOI] [PubMed] [Google Scholar]
- Medeiros, N.A., D.T. Burnette, and P. Forscher. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:215–226. [DOI] [PubMed] [Google Scholar]
- Negron, J.A., S. Gonzalez and J.R. Rodriguez-Medina. 1996. In vivo phosphorylation of type II myosin in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 221:515–520. [DOI] [PubMed] [Google Scholar]
- Ponti, A., M. Machacek, S.L. Gupton, C.M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615. [DOI] [PubMed] [Google Scholar]
- Pruyne, D.W., D.H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931–1945. [DOI] [PubMed] [Google Scholar]
- Rayment, I., W.R. Rypniewski, K. Schmidt-Base, R. Smith, D.R. Tomchick, M.M. Benning, D.A. Winkelmann, G. Wesenberg, and H.M. Holden. 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 261:50–58. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., M.J. Tyska, P.J. Novick, and M.S. Mooseker. 2001. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153:1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese, O.W., C.A. Reinke, B.J. Bevis, A.T. Hammond, I.B. Sears, J. O'Connor, and B.S. Glick. 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153:47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzadzinska, A.K., M.E. Schneider, C. Davies, G.P. Riordan, and B. Kachar. 2004. An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. J. Cell Biol. 164:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot, I., S.K. Klee, and D. Pellman. 2002. a. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42–50. [DOI] [PubMed] [Google Scholar]
- Sagot, I., A.A. Rodal, J. Moseley, B.L. Goode, and D. Pellman. 2002. b. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626–631. [DOI] [PubMed] [Google Scholar]
- Salmon, W.C., M.C. Adams, and C.M. Waterman-Storer. 2002. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 158:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, M.E., I.A. Belyantseva, R.B. Azevedo, and B. Kachar. 2002. Rapid renewal of auditory hair bundles. Nature. 418:837–838. [DOI] [PubMed] [Google Scholar]
- Schott, D.H., R.N. Collins, and A. Bretscher. 2002. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 156:35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41. [DOI] [PubMed] [Google Scholar]
- Simon, V., T.C. Swayne and L.A. Pon. 1995. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J. Cell Biol., 130:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, V.R., S.L. Karmon, and L.A. Pon. 1997. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton. 37:199–210. [DOI] [PubMed] [Google Scholar]
- Singer, J.M., and J.M. Shaw. 2003. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc. Natl. Acad. Sci. USA. 100:7644–7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., A. Sil, J.R. Swedlow, I. Herskowitz, and R.D. Vale. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 389:90–93. [DOI] [PubMed] [Google Scholar]
- Theriot, J.A., and T.J. Mitchison. 1991. Actin microfilament dynamics in locomoting cells. Nature. 352:126–131. [DOI] [PubMed] [Google Scholar]
- Tolliday, N., N. Bouquin, and R. Li. 2001. Assembly and regulation of the cytokinetic apparatus in budding yeast. Curr. Opin. Microbiol. 4:690–695. [DOI] [PubMed] [Google Scholar]
- Tolliday, N., M. Pitcher, and R. Li. 2003. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol. Biol. Cell. 14:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska, M.J., and M.S. Mooseker. 2002. MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys. J. 82:1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen, E.A., J. Caviston, and E. Bi. 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., and E.D. Salmon. 1997. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 139:417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, F.Z., G. Shiels, and E. Orr. 1987. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 6:3499–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K.M., and N.K. Wessells. 1973. Cytochalasin B: effects on membrane ruffling, growth cone and microspike activity, and microfilament structure not due to altered glucose transport. Dev. Biol. 31:413–420. [DOI] [PubMed] [Google Scholar]
- Yang, H.C., and L.A. Pon. 2002. Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA. 99:751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.