Abstract
Import of nucleus-encoded tRNAs into the mitochondria of the kinetoplastid protozoon Leishmania involves recognition of specific import signals by the membrane-bound import machinery. Multiple signals on different tRNA domains may be present, and further, importable RNAs interact positively (Type I) or negatively (Type II) with one another at the inner membrane in vitro. By co-transfection assays, it is shown here that tRNATyr (Type I) transiently stimulates the rate of entry of tRNAIle (Type II) into Leishmania mitochondria in transfected cells, and conversely, is inhibited by tRNAIle. Truncation and mutagenesis experiments led to the co-localization of the effector and import activities of tRNATyr to the D domain, and those of tRNAIle to the variable region–T domain (V-T region), indicating that both activities originate from a single RNA–receptor interaction. A third tRNA, human tRNALys, is imported into Leishmania mitochondria in vitro as well as in vivo. This tRNA has Type I and Type II motifs in the D domain and the V-T region, respectively, and shows both Type I and Type II effector activities. Such dual-type tRNAs may interact simultaneously with the Type I and Type II binding sites of the inner membrane import machinery.
INTRODUCTION
The mitochondria of a large number of species are deficient in one or several tRNA genes (1), and sustain translation of the organellar genes by specific import of the corresponding nucleus-encoded species from the cytoplasm (reviewed in 2). The number and identities of the tRNAs imported vary widely: from one in yeast (3) and marsupials (4), to about a dozen in plants (5), to at least 24 in kinetoplastid protozoa such as Leishmania and Trypanosoma, which lack all tRNA genes (6,7). Such diversity raises the questions of specificity, of the multiplicity of tRNA receptors and import mechanisms, and of the evolutionary origin of the import apparatus.
The recent development of in vitro import systems in yeast (8), Leishmania (9,10), trypanosomes (11,12) and potato (13) has enabled the interaction of tRNAs with mitochondria to be studied in detail. Two distinct import mechanisms are now recognized (2). The single-tRNA system in yeast appears to involve protein import channels and soluble carriers, including the cognate aminoacyl tRNA synthetase (14). This co-import model differs from the direct import mechanism apparently operating in kinetoplastid protozoa. Here soluble proteins are not required, and the tRNA import and protein import machineries are distinct (12,15). Specific sequence motifs and/or structures (import signals) on tRNAs are recognized (16,17). Moreover, by a selection-amplification protocol on Leishmania mitochondria, several import aptamers were isolated, suggesting the occurrence of multiple signals in different domains of various tRNAs (18). For example, tRNATyr(GUA) contains an import signal that includes the conserved motif YGGYAGAGC in the D domain (16,18), while tRNAIle(UAU) has an import signal in the variable region–T domain (V-T region) that contains the conserved motif UGGGU (18). Some tRNAs may have more than one signal; for example, human tRNALys(UUU), which is imported into trypanosome mitochondria (19), as well as its counterpart in Leishmania, contain both the D domain and the V-T region motifs (18) (see below). An additional level of complexity is that transfer of tRNA through the outer and inner mitochondrial membranes appear to be kinetically and biochemically separable processes (20). In particular, the effects of mutations within the import signal on these two steps are non-identical, suggesting the presence of distinct receptors at the two membranes (17). A 15 kDa tRNA binding protein is apparently required for outer membrane transfer (21); the inner membrane receptors are yet to be identified.
Of particular interest is the observation that in vitro, importable RNAs, including oligonucleotide aptamers and intact tRNAs, allosterically interact with one another at the inner membrane (18). Two types of RNAs have been observed: Type I RNAs, e.g. tRNATyr, can be imported into the matrix by themselves, while Type II RNAs, e.g. tRNAIle, require the presence of low levels of a Type I RNA. Moreover, Type II RNAs inhibit the transfer of Type I RNAs (18). Although the generality of this phenomenon with respect to all importable tRNAs is presently undetermined, the fact that known Type I and Type II aptamers are homologous to a large number of tRNAs (18) implies allosteric regulation to be widely applicable. This type of regulation may serve to create a balanced mitochondrial pool of tRNAs from a cytoplasmic pool containing tRNAs of widely different intrinsic import efficiencies.
In the present study, we have looked for evidence of such allosteric interactions in vivo, examined the relationship between the substrate and effector activities of tRNATyr and tRNAIle, and studied the interactions involving tRNALys. The results suggest the possibility of both sequential and simultaneous interactions between the Type I and Type II domains and their cognate binding sites on the inner membrane.
MATERIALS AND METHODS
Preparation of mitochondria
Mitochondria were prepared from lysates of Leishmania tropica strain UR6 promastigotes by Percoll gradient centrifugation as previously described (9). To selectively remove the outer membrane, mitochondria were treated with 320 µM digitonin (20). The resultant mitoplasts were subjected to freeze–thaw cycles to separate the soluble (matrix) and particulate (inner membrane) fractions (20). The purity of individual fractions was assessed by marker enzyme assays (20).
Import substrates and effectors
32P-labeled tRNATyr(GUA), tRNAGln(CUG) and tRNAIle (UAU) transcripts were prepared from cloned or PCR-amplified templates, and D domain RNAs from oligonucleotide templates bearing the stated mutations, using T7 RNA polymerase (16–18,20,21) (Fig. 1). Truncated tRNAIle[1–41] and tRNAIle[42–66] were obtained by run-off transcription of the appropriate subclones (18). The human cytoplasmic tRNALys1(UUU) gene (M.Sprinzl, K.S.Vassilenko, J.Emmerich and F.Bauer, http://www.uni-bayreuth.da/departments/biochemie/tRNA) was amplified from human genomic DNA by PCR using CGGGAATTCTAATACGACTCACTATAGCCCGGATAGCTCAGTCGG (sense) and TGGCGCCCGAACAGGGACTTG (antisense) as primers; the PCR product on transcription yields a 3′-CCA terminated transcript (Fig. 1). Import substrates were prepared using a high specific activity of [α-32P]UTP (∼100 c.p.m./fmol), while effector RNAs were synthesized at 1/250 of this specific activity (18).
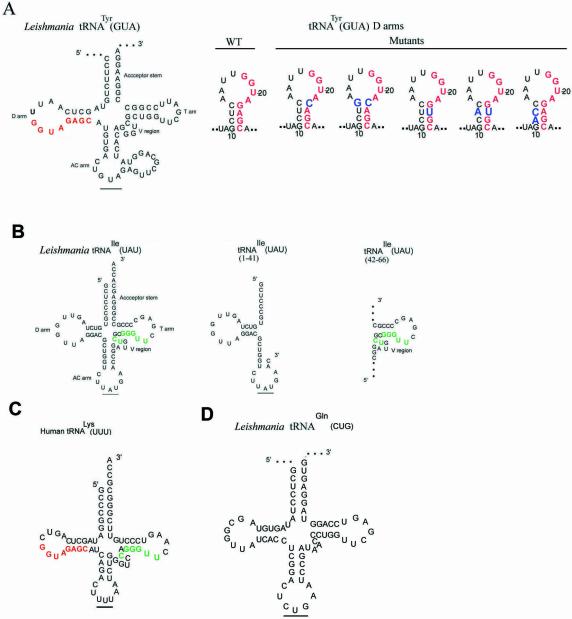
Figure 1.
Structures of tRNAs and their derivatives. (A) Left, Leishmania tRNATyr(GUA). The conserved D arm signal is shown in red. Right, D arm mini-helices, with mutations in blue. (B) Left, Leishmania tRNAIle(UAU); the conserved V-T region signal is shown in green. Right, truncated versions of tRNAIle[1–41] and tRNAIle[42–66]. (C) Human cytoplasmic tRNALys(UUU) with conserved D arm (red) and V-T (green) motifs. (D) tRNAGln(CUG).
In vitro import assays
Import into intact mitochondria or mitoplasts was assayed by RNase protection as previously described (17). Briefly, mitochondria (100 µg of protein) were incubated with 32P-labeled RNA (2.5 nM) in the presence of 4 mM ATP at 37°C for 15 min, then treated with RNases A plus T1, washed, and the RNase-resistant mitochondrial RNA recovered for gel electrophoretic analysis. To assess intra-mitochondrial distribution of imported RNA, RNase-treated mitochondria were fractionated as above, and RNA isolated from each fraction.
Binding assays
Mitochondria or mitoplasts were incubated with 32P-labeled RNA (1 nM) in the presence of 0.1 M NaCl on ice for 30 min, washed, and bound RNA recovered (21).
Transient transfection assays
Leishmania tropica promastigotes were electroporated in the presence of 32P-labeled RNA, the cells were incubated at 22°C for the indicated times, then lysed, and the particulate fraction containing mitochondria was treated with DNase and RNase before recovery of internalized RNA, as detailed previously (17). The cellular pool was isolated by RNase treatment of an aliquot of the transfected cells before lysis and preparation of total RNA. For time course experiments, import was terminated at the indicated times by the addition of 10 µM m-chlorocarbonylcyanide phenylhydrazone (CCCP).
RESULTS
Inter-tRNA interactions in transiently transfected cells
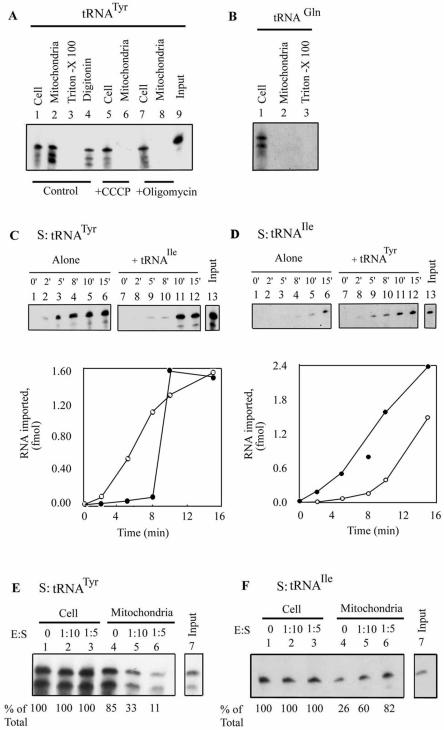
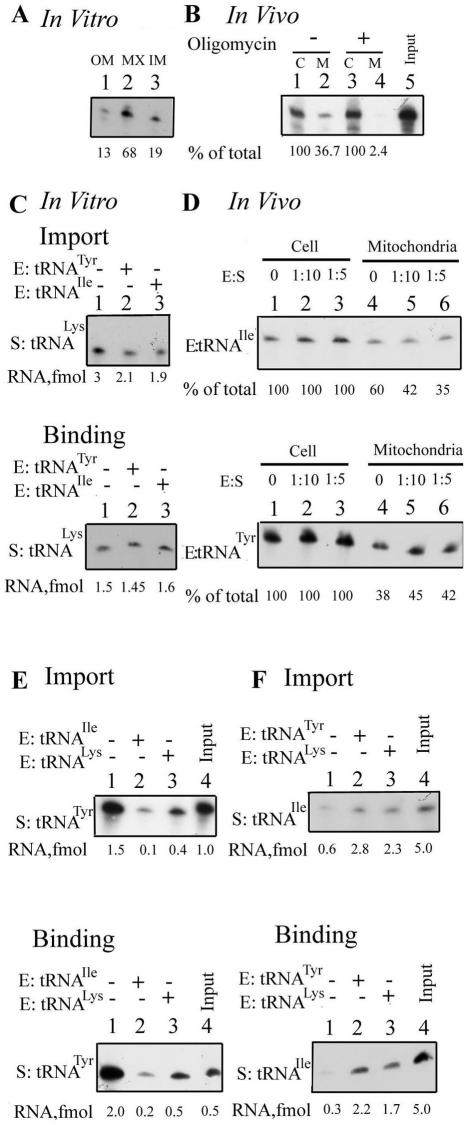
It was previously shown that, in vitro, tRNATyr (a Type I RNA) stimulates transfer of tRNAIle (a Type II RNA) across the inner membrane (18). To assess the physiological significance of this observation, a transient transfection assay was employed. Promastigotes were transfected with 32P-labeled tRNATyr, incubated at 22°C, and the total and mitochondrial RNAs were isolated. Using this approach, we have previously demonstrated specific, sequence-dependent mitochondrial uptake of oligoribonucleotides (17). As shown in Figure 2A, tRNATyr was recovered from the RNase-treated mitochondrial fraction. In independent experiments, an average of 69 ± 14% (n = 4) of the total intracellular transfecting RNA was present in mitochondria at 10 min post-transfection. Treatment of this fraction with Triton X-100 during or before RNase treatment resulted in degradation of the RNA, showing it to be membrane enclosed, but the RNA was partially resistant to treatment with digitonin, which selectively permeabilizes the outer mitochondrial membrane, indicating its presence in the matrix. In the presence of the protonophore uncoupler CCCP, which disrupts the proton gradient generated by actively respiring mitochondria, uptake was inhibited (Fig. 2A), showing the requirement of a membrane potential for import, as has been previously observed in vitro (17,20), as well as in vivo for import of oligonucleotides (17,18). Mitochondrial uptake was similarly inhibited by oligomycin, a specific inhibitor of the mitochondrial F1F0 ATPase (Fig. 2A). Moreover, uptake was specific with respect to tRNA, since tRNAGln(CUG), which is not imported in vivo (22), was not taken up by the mitochondrial fraction (Fig. 2B).
Figure 2.
Mitochondrial uptake of tRNAs in transfected cells. (A) Leishmania tropica promastigotes (4.3 × 108) were electroporated with 32P-labeled tRNATyr (500 fmol) in the absence (lanes 1–4) or presence of 10 µM CCCP (lanes 5 and 6) or 50 µM oligomycin (lanes 7 and 8). Aliquots of the transfected cells were lysed and mitochondrial fractions were treated with RNase and DNase in the absence (lanes 2, 6 and 8) or presence of 1% Triton X-100 (lane 3) or 320 µM digitonin (lane 4), before recovery and analysis of RNA in the particulate fraction. The corresponding total cellular pools (lanes 1, 5 and 7) were obtained by treating an identical aliquot of intact transfected cells with RNase before RNA isolation. (B) Transfection of promastigotes with 32P-labeled tRNAGln(CUG). Lane 1, total cellular pool; lane 2, mitochondrial fraction; lane 3, mitochondrial fraction treated with Triton X-100. (C) Kinetics of uptake of tRNATyr. Promastigotes were transfected with 500 fmol of high specific activity tRNATyr alone (lanes 1–6) or with 50 fmol of low specific activity tRNAIle (lanes 7–12). After incubation at 22°C for the indicated times, import was terminated by the addition of 10 µM CCCP, aliquots of transfected cells were lysed and RNase-resistant RNA in the mitochondrial fraction was recovered. Lane 13, input tRNATyr, 3 fmol. (D) Promastigotes were transfected with 500 fmol of high specific activity tRNAIle alone (lanes 1–6) or with 50 fmol of low specific activity tRNATyr (lanes 7–12) for the indicated times at 22°C, import terminated, and the mitochondrial uptake analyzed. Lane 13, input tRNAIle, 2 fmol. Quantitative data in (C) and (D) are graphically represented below the respective autoradiograms. Open circles, substrate alone; filled circles, substrate plus effector. (E) 32P-labeled high specific activity tRNATyr (S, 500 fmol) was co-transfected into promastigotes with low specific activity tRNAIle (E) at an E:S ratio of 0 (lanes 1 and 4), 1:10 (lanes 2 and 5) or 1:5 (lanes 3 and 6). Cells were incubated for 10 min at 22°C before analysis of total (lanes 1–3) or mitochondrial (lanes 4–6) RNA. Lane 7, input tRNATyr (1 fmol). (F) 32P-labeled high specific activity tRNAIle (S, 500 fmol) was co-transfected into promastigotes with low specific activity tRNATyr (E) at an E:S ratio of 0 (lanes 1 and 4), 1:10 (lanes 2 and 5) or 1:5 (lanes 3 and 6), and mitochondrial uptake was determined as in (E). Lane 7, input tRNAIle (1 fmol). Quantifications were performed by densitometry. The total cellular pool was taken as 100% in each case.
To examine inter-tRNA interactions, promastigotes were co-transfected with high specific activity, 32P-labeled tRNATyr (substrate) and low specific activity tRNAIle (effector) at a 10:1 molar ratio. In the absence of effector, tRNATyr was rapidly internalized by 10–15 min post-transfection with little or no lag (Fig. 2C). In the presence of tRNAIle, a pronounced lag of ∼8 min was observed, but the total amount imported was unaffected (Fig. 2C). In the reverse experiment, uptake of tRNAIle alone was noticeably slower than that of tRNATyr (lag of ∼8 min), but the rate was enhanced by co-transfection with tRNATyr effector, with little effect on the final yield (Fig. 2D). These effects were reproducible in three independent experiments, although the absolute value of the lag varied between 6 and 10 min, possibly a result of variations in the growth state of the unsychronized cell population used for transfection. Thus, effector activity in vivo is transient, being apparent only at early times post-transfection (within 10 min at 22°C). In a second set of experiments, early-time co-transfections were performed at different substrate:effector ratios. Dose- dependent inhibition of uptake of tRNATyr by tRNAIle (Fig. 2E), and stimulation of uptake of tRNAIle by tRNATyr (Fig. 2F), were observed.
Specificity of effector action
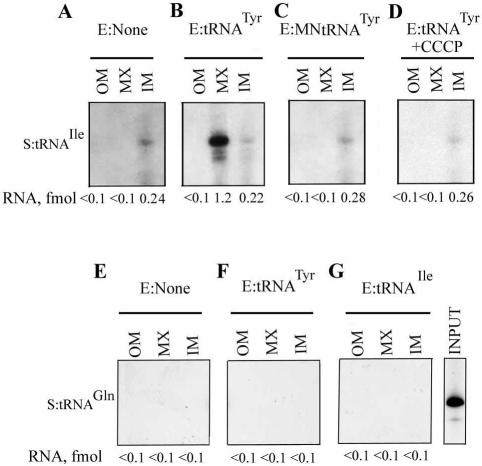
The intra-mitochondrial distribution of tRNAIle imported in vitro was assessed by fractionating post-import mitochondria into outer membrane plus intermembrane space, inner membrane and matrix compartments. Transfer of this Type II RNA into the matrix was strongly stimulated by the Type I tRNATyr (Fig. 3A and B), as previously observed (18). The effector activity of tRNATyr was sensitive to micrococcal nuclease (Fig. 3C), but not to DNase (data not shown), confirming that the effector is RNA in nature and not some adventitious contaminant of the RNA preparation. Effector-dependent import of the Type II RNA, like the effector-independent import of Type I RNA, was sensitive to CCCP (Fig. 3D). Moreover, import of tRNAGln(CUG), which is not imported in vivo, could not be induced by either Type I or Type II effector (Fig. 3E–G), implying that the effector does not cause a non-specific opening of import channels, but that the interactions are confined to importable RNAs.
Figure 3.
Specificity of effector action. The intra-mitochondrial distribution of tRNAIle(UAU) (A–D) or tRNAGln(CUG) (E–G) was assayed by sub- mitochondrial fractionation after import. OM, outer membrane plus intermembrane space; IM, inner membrane; MX, matrix. Import assays were performed with high specific activity substrate (S, 50 fmol) and low specific activity effector (E, 5 fmol) in the following combinations: (A and E) no effector; (B and F) E, tRNATyr(GUA); (C) E, micrococcal nuclease-treated tRNATyr; (D) E, tRNATyr, import in the presence of 50 µM CCCP; (G) E, tRNAIle.
Identification of effector domains in tRNATyr and tRNAIle
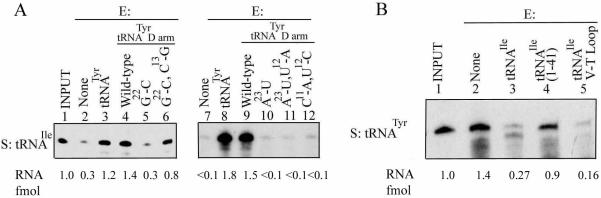
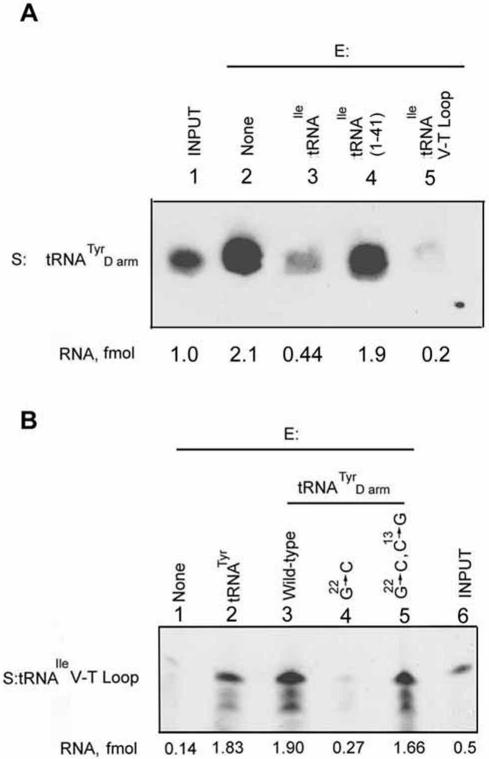
It is evident that each tRNA has two activities: as substrate for import, and as effector for the modulation of import of other tRNAs. It was necessary to determine whether the two activities are coincident in the molecule, or whether they reside in different domains. The import signal of tRNATyr has been localized to the D domain (16,17). In mitoplast import assays, an oligonucleotide containing the wild-type D domain (Fig. 1) could completely replace the intact molecule as positive effector for import of tRNAIle (Fig. 4A). Moreover, effector activity was sensitive to point mutations within the D domain (Fig. 4A). The mutation G22:C in the loop-closing base pair, which results in destabilization of the hairpin [ΔG of formation reduced from –9.2 to –2.2 kcal/mol (17)], resulted in complete loss of effector activity, but restoration of the base pair in the double mutant G22:C, C13:G partially restored activity, emphasizing the importance of secondary structure. Mutation of A23 to U similarly led to loss of effector activity, but this could not be restored in the double mutant A23:U, U12:A, indicating that at this position it is the identity of the base that is critical. The double mutation C11:A, U12:C outside the conserved motif, which disrupts two consecutive base pairs (ΔG –0.7 kcal/mol), also abolished activity. Importantly, the effect of each mutation on effector activity was identical to that on importability (17), indicating that these two phenomena are a consequence of the interaction of the Type I domain with a single binding site on the inner membrane.
Figure 4.
Localization of effector domains of tRNATyr and tRNAIle. High specific activity substrate (S, 50 fmol) tRNA was incubated with mitoplasts prepared from 100 µg of mitochondria either in the absence or presence of different low specific activity effectors (E, 5 fmol), and the imported RNA recovered. (A) S, tRNAIle. Lanes 2 and 7, no effector; lanes 3 and 8, E, tRNATyr; lanes 4 and 9, E, tRNATyr D arm, wild type; lane 5, E, G22:C mutant; lane 6, G22:C, C13:G; lane 10, A23:U; lane 11, A23:U, U12:A; lane 12, C11:A, U12:C. (B) S, tRNATyr. Lane 2, no effector; lane 3, E, tRNAIle; lane 4, E, tRNAIle[1–41]; lane 5, E, tRNAIle [42–66].
The Type II molecule tRNAIle has a strong import signal in the V-T region (18). By employing truncated versions of this tRNA, we sought to localize its effector domain. Whereas tRNAIle inhibited the inner membrane transfer of tRNATyr, tRNAIle[1–41], spanning the D domain and the anticodon loop, was only marginally inhibitory, while tRNAIle[42–66] was more effective than the intact molecule (Fig. 4B). Thus, as in the case of tRNATyr, the import signal and effector domains coincide.
From the above, it is clear that the interactions between tRNATyr and tRNAIle are effectively a reflection of the binding of their respective import signal-cum-effector domains to cognate binding sites on the inner membrane. Indeed, the import of the D arm of tRNATyr was specifically inhibited by the V-T region of tRNAIle, and vice versa (Fig. 5).
Figure 5.
Inter-domain interactions. Mitoplast import assays were performed with high specific activity substrate (50 fmol) and low specific activity effector (5 fmol) in the following combinations: (A) substrate tRNATyr D arm (wild type) in the absence (lane 2) or presence of effector: tRNAIle (lane 3), tRNAIle[1–41] (lane 4) or tRNAIle[42–66] (lane 5). (B) Substrate tRNAIle[42–66] in the absence (lane 1) or presence of effector: tRNATyr (lane 2), tRNATyr D arm, wild type (lane 3), G22:C mutant (lane 4) or G22:C, C13:G mutant (lane 5).
The dual activity of tRNALys
Previous experiments with import aptamers indicated that, in addition to tRNAs such as tRNATyr and tRNAIle, which contain single import signals, there may be tRNAs with more than one signal. One such molecule is tRNALys(UUU). Both the Leishmania (18) and the human (Fig. 1) varieties of this tRNA contain the conserved Type I motif YAGAGC in the D domain, as well as the conserved Type II motif UGGGU in the V-T region. Human tRNALys is imported into trypanosome mitochondria in vivo (19), indicating the recognition of a kinetoplastid-type import signal coincidentally present on this tRNA.
We first checked whether human tRNALys is imported into Leishmania mitochondria in vitro. Incubation of 32P-labeled tRNALys with intact mitochondria, followed by sub- mitochondrial fractionation, showed that a significant fraction of the tRNA was located in the matrix (Fig. 6A). Transient transfection of promastigotes resulted in oligomycin-sensitive mitochondrial internalization of tRNALys (Fig. 6B). The mean value of mitochondrial uptake in vivo was 45 ± 13% (n = 3) of the total intracellular pool under these conditions. Thus, human tRNALys is imported into Leishmania mitochondria in vitro as well as in vivo.
Figure 6.
Import of human tRNALys(UUU) into Leishmania mitochondria. (A) Intra-mitochondrial distribution assay. 32P-labeled tRNALys (100 fmol) was incubated with intact mitochondria (100 µg of protein), and its sub-mitochondrial distribution was analyzed. (B) Promastigotes were transfected with 500 fmol of high specific activity tRNALys in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 50 µM oligomycin. Total and mitochondrial pools were recovered as in Figure 2. (C) Effect of tRNATyr and tRNAIle on import (top) or binding (bottom) of tRNALys into or on mitoplasts in vitro. Lane 1, tRNALys substrate alone; lane 2, substrate plus tRNATyr effector; lane 3, substrate plus tRNAIle effector. (D) Effect of tRNATyr and tRNAIle on internalization of tRNALys into mitochondria of transfected cells. Co-transfection of promastigotes with high specific activity human tRNALys and low specific activity tRNAIle (top) or tRNATyr (bottom) was performed at effector:substrate ratios of 0 (lanes 1 and 4), 1:10 (lanes 2 and 5) and 1:5 (lanes 3 and 6), respectively. The total cellular pool (lanes 1–3) and mitochondrial pool (lanes 3–6) were recovered in each case. (E) Effect of tRNALys on import (top) or binding (bottom) of tRNATyr into or on mitoplasts. Import reactions contained 50 fmol of substrate and mitoplasts from 100 µg of mitochondria; corresponding values for binding assays were 10 fmol and 50 µg, respectively. Substrate:effector ratio was 10:1. Lane 1, tRNATyr alone; lane 2, tRNATyr plus tRNAIle effector; lane 3, tRNATyr plus tRNALys effector; lane 4, input tRNATyr, 1.0 and 0.5 fmol, respectively. (F) Effect of tRNALys on import (top) or binding (bottom) of tRNAIle. Assays were performed as in (E), except that tRNAIle was used as substrate. Lane 1, substrate alone; lane 2, substrate plus tRNATyr effector; lane 3, substrate plus tRNALys effector; lane 4, input tRNAIle, 5 fmol.
Next, the effector activity, if any, of tRNALys was assessed through its effect on known Type I and Type II tRNAs. tRNALys inhibited the inner membrane transfer, as well as binding, of tRNATyr (Fig. 6E), implying Type II activity, although the extent of inhibition was somewhat less, mole for mole, than that of the known Type II molecule tRNAIle. tRNALys also stimulated import and binding of tRNAIle (Fig. 6F); this is a characteristic of Type I RNA (18). In the converse experiment, the import or binding of tRNALys substrate in vitro was not appreciably affected by either tRNATyr (Type I) or tRNAIle (Type II) (Fig. 6C), and this was true at substrate:effector ratios ranging from 10:1 to 1:1 (data not shown). Similarly, neither tRNATyr nor tRNAIle had any effect on the internalization of tRNALys in vivo (Fig. 6D). It is evident from these observations that the effector activity of tRNALys is not a simple summation of Type I and Type II interactions.
DISCUSSION
In this study we show that the interactions between tRNAs at the inner membrane previously observed in vitro are also demonstrable in promastigotes by co-transfection assays, but that the effects are transient (Fig. 2). Relatively large amounts of tRNA, accounting for up to 80% or more of the total cellular pool, were localized in the mitochondrial fraction within a few minutes of transfection (Figs 2 and 6). This contrasts with the few per cent of the steady-state pool in mitochondria usually observed in vivo (2 and references therein). A possible explanation for the discrepancy is that the transfecting RNA enters through the flagellar pocket (23), with which the mitochondrion is in close proximity, and thus does not equilibrate with the cytoplasmic pool within the short time-span of experimental observation. The transience of effector action may be due to the presence of a complex cellular pool of endogenous tRNAs, some stimulatory and others inhibitory for import of a particular transfecting species. It was shown previously that the efficiencies of inner membrane transfer of individual import aptamers are significantly more variable in vitro than in vivo, and that the in vitro variance could be brought closer to the in vivo value by factoring in inter-RNA allosteric interactions (18). This is reflected in the present results: tRNATyr and tRNAIle are internalized to about the same extent in vivo (Fig. 2), whereas the corresponding intrinsic inner membrane transfer efficiencies in vitro are quite different (Fig. 4). The transient rate effect of the co-transfecting effector could be a consequence of a decline in its concentration in the cytoplasm, either due to degradation or, more likely, to sequestration through import into the mitocondrial matrix, allowing the endogenous effector pool to take over.
Secondly, our results demonstrate that the import and effector domains of a particular tRNA are coincident (Figs 4 and 5). The identical effects of mutations on import (17) and Type I effector activity of the D domain of tRNATyr probably reflect a common interaction with a binding site (receptor) on the inner membrane. Similarly, the Type II effector domain of tRNAIle co-localizes with its import signal. Since the Type I and Type II domains contain non-identical purine-rich sequences, their cognate biding sites are likely to be related but distinct, but they are presumed to be in close proximity to each other, as part of the same or different protein, and to have conformational flexibility. Thus, binding of a Type I domain to its cognate site would have two effects: import, via transfer to the import channel, and an allosteric conformation change in the Type II binding site, resulting in the opening up of this site for binding a Type II RNA (Fig. 7A). Type II domain binding would likewise lead to import of the Type II RNA, as well as to transmission of a negative allosteric effect on the Type I binding site (Fig. 7A).
Figure 7.
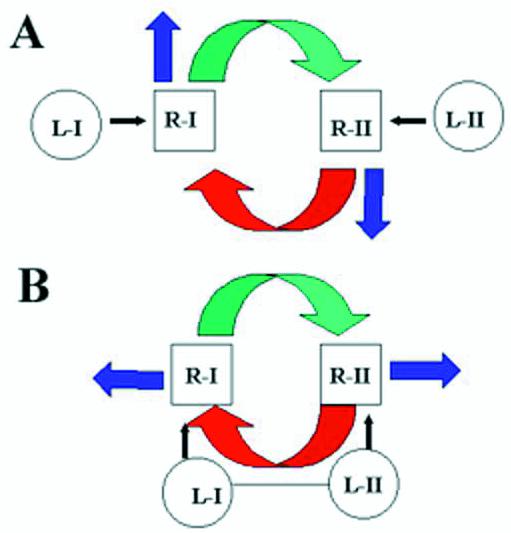
Type I and Type II interactions at the inner membrane. (A) The Type I RNA domain (L-I) binds to the cognate site R-I, leading to its transfer (blue arrow) and to positive regulation (green arrow) of the Type II site (R-II). The latter interaction results in binding of the Type II domain (L-II). Type II RNA is subsequently transferred (blue arrow) and also negatively regulates site R-I (red arrow). (B) The Type I and Type II domains are part of the same molecule. Sequential or simultaneous positive and negative effects occur on binding sites R-I and R-II.
We also show here that human tRNALys(UUU) is imported into Leishmania mitochondria in vitro as well as in vivo (Fig. 6). Recognition of a human cytoplasmic tRNA by the kinetoplastid import apparatus is of more than academic interest, since a number of human genetic diseases are caused by mutations in mitochondrial tRNA genes (24), and future therapeutic strategies might involve the introduction of critical kinetoplastid genes into mutant cells to induce tRNA import. Based on homology, it was predicted that tRNALys contains both Type I and Type II motifs (Fig. 1), and this was borne out by its ability to inhibit a Type I tRNA and stimulate a Type II tRNA (Fig. 6). This is understandable in terms of the two-site model in Figure 7A, in which one site is occupied by the single-domain tRNA, the other site being available for either domain of the double-domain tRNALys. However, the import of tRNALys substrate both in vivo and in vitro was not affected by either single-domain effector (Fig. 6). One explanation for this is that at high concentrations relative to the effector, the dual-domain tRNA can interact simultaneously or sequentially with both Type I and Type II binding sites, thus effectively precluding effector binding (Fig. 7B). It will be necessary to identify the corresponding tRNA binding proteins in order to study the details of these interactions.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a grant from the Department of Science and Technology, Government of India. S.G., S.N.B. and S.B. received fellowships from the Council of Scientific and Industrial Research and S.C. received a fellowship from the University Grants Commission.
REFERENCES
- 1.Gray M.W., Lang,B.F., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M., Brossard,N., Delage,E., Littlejohn,T.G., Plante,I., Rioux,P., Saint-Louis,D., Zhu,Y. and Burger,G. (1998) Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res., 26, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider A. and Marechal-Drouard,L. (2000) Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol., 10, 509–513. [DOI] [PubMed] [Google Scholar]
- 3.Martin R.P., Schneller,J.M., Stahl,A.J. and Dirheimer,G. (1979) Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry, 18, 4600–4605. [DOI] [PubMed] [Google Scholar]
- 4.Dörner M., Altmann,M., Pääbo,S. and Mörl,M. (2001) Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell, 12, 2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R., Marechal-Drouard,L., Akama,K. and Small,I. (1996) Striking differences in mitochondrial tRNA import between different plant species. Mol. Gen. Genet., 252, 404–411. [DOI] [PubMed] [Google Scholar]
- 6.Simpson A.M., Suyama,Y., Dewes,H., Campbell,D. and Simpson,L. (1989) Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res., 17, 5427–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock K. and Hajduk,S.L. (1990) The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem., 265, 19203–19215. [PubMed] [Google Scholar]
- 8.Tarrasov I. and Entelis,N.S. (1992) Mitochondrially-imported cytoplasmic tRNALys(CUU) of Saccharomyces cerevisiae: in vivo and in vitro targeting systems. Nucleic Acids Res., 20, 1277–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahapatra S., Ghosh,T. and Adhya,S. (1994) Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res., 22, 3381–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubio M.A., Liu,X., Yuzawa,H., Alonzo,J.D. and Simpson,L. (2000) Selective importation of RNA into isolated mitochondria from Leishmania tarentolae.RNA, 6, 988–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yermovsky-Kammerer A.E. and Hajduk,S.L. (1999) In vitro import of tRNA into the mitochondria of Trypanosoma brucei. Mol. Cell. Biol., 19, 6253–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nabholz C.E., Horn,E. and Schneider,A. (1999) tRNAs and proteins are imported into Leishmania mitochondria of Trypanosoma buucei by two distinct mechanisms. Mol. Biol. Cell, 10, 2747–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delage L., Dietrich,A., Cosset,A. and Marechal-Drouard,L. (2003) In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol., 23, 4000–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarassov I., Entelis,N. and Martin,R.P. (1995) An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J. Mol. Biol., 245, 315–323. [DOI] [PubMed] [Google Scholar]
- 15.Mahapatra S. and Adhya,S. (1996) Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J. Biol. Chem., 271, 20432–20437. [DOI] [PubMed] [Google Scholar]
- 16.Mahapatra S., Ghosh,S., Bera,S.K., Ghosh,T., Das,A. and Adhya,S. (1998) The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res., 26, 2037–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya S.N., Mukherjee,S. and Adhya,S. (2000) Mutations in a tRNA import signal define distinct receptors at the two membranes of Leishmania mitochondria. Mol. Cell. Biol., 20, 7410–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya S.N., Chatterjee,S. and Adhya,S. (2002) Mitochondrial RNA import in Leishmania tropica: aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Mol. Cell. Biol., 22, 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser R. and Schneider,A. (1995) tRNAs are imported into the mitochondria of Trypanosoma brucei independent of their genomic context and of their genetic origin. EMBO J., 14, 4212–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee S., Bhattacharyya,S.N. and Adhya,S. (1999) Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J. Biol. Chem., 274, 31249–31255. [DOI] [PubMed] [Google Scholar]
- 21.Adhya S., Ghosh,T., Das,A., Bera,S.K. and Mahapatra,S. (1997) Role of an RNA binding protein in import of tRNA into Leishmania mitochondria. J. Biol. Chem., 272, 21396–21402. [DOI] [PubMed] [Google Scholar]
- 22.Lye L.-F., Chen,D.-H.T. and Suyama,Y. (1993) Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol. Biochem. Parasitol., 58, 233–246. [DOI] [PubMed] [Google Scholar]
- 23.Landfear S.M. and Ignatushchenko,M. (2001) The flagellum and flagellar pocket of trypanosomatids. Mol. Biochem. Parasitol., 115, 1–17. [DOI] [PubMed] [Google Scholar]
- 24.Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1487. [DOI] [PubMed] [Google Scholar]