Abstract
Proper orientation and positioning of the mitotic spindle is essential for the correct segregation of fate determinants during asymmetric cell division. Although heterotrimeric G proteins and their regulators are essential for spindle positioning in many cell types, their mechanism of action remains unclear. In this study, we show that dyrb-1, which encodes a dynein light chain, provides a functional link between heterotrimeric G protein signaling and dynein activity during spindle positioning in Caenorhabditis elegans. Embryos depleted of dyrb-1 display phenotypes similar to a weak loss of function of dynein activity, indicating that DYRB-1 is a positive regulator of dynein. We find that the depletion of dyrb-1 enhances the spindle positioning defect of weak loss of function alleles of two regulators of G protein signaling, LIN-5 and GPR-1/2, and that DYRB-1 physically associates with these two proteins. These results indicate that dynein activity functions with regulators of G protein signaling to regulate common downstream effectors during spindle positioning in the early C. elegans embryo.
Introduction
Asymmetric cell division plays a crucial role in generating cell diversity. Proper positioning of the mitotic spindle is an essential step of an asymmetric division. In the early embryo of the nematode C. elegans, asymmetric positioning of the mitotic spindle depends on an imbalance in cortical force generators that act on astral microtubules and pull on spindle poles (Grill et al., 2001). Although the molecular nature of the force generators is not known, their spatial and temporal activation is controlled by heterotrimeric G protein signaling. Inactivation of two Gα subunits, GOA-1 and GPA-16, as well as the receptor-independent activators of G protein signaling GPR-1 and GPR-2 (two nearly identical proteins containing a Goloco domain, hereafter referred to as GPR-1/2) results in strongly reduced and symmetric pulling forces (Colombo et al., 2003). The coiled-coil protein LIN-5 also plays a crucial role in spindle positioning. LIN-5 interacts with GPR-1/2, and its inactivation results in a phenotype very similar to the phenotype of embryos lacking both Gα subunits or GPR-1/2 (Gotta et al., 2003; Srinivasan et al., 2003). The role of heterotrimeric G proteins in spindle positioning is conserved in other organisms, including flies and mammals (Bellaiche and Gotta, 2005; Sanada and Tsai, 2005). In Drosophila melanogaster neuroblasts, Gα and PINS, the functional homologue of GPR-1/2, are required for apical basal orientation of the mitotic spindle. In mammalian cells, Gα, the Goloco-containing protein LGN, and the microtubule-binding protein nuclear mitotic apparatus (NuMA) form a complex that has been suggested to regulate the interaction of astral microtubules with the cell cortex (Du and Macara, 2004). Interestingly, NuMA can bind to the dynein–dynactin complex (Merdes et al., 1996), and recent work has shown that Drosophila Mud and Caenorhabditis elegans LIN-5 are the homologues of NuMA (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). These results suggest a model in which the interaction of cortically localized NuMA/LIN-5/Mud with dynein results in the activation of this minus end–directed motor, locally increasing pulling forces and, thereby, resulting in posterior displacement of the mitotic spindle. To date, there is no evi dence for a dynein requirement in spindle positioning in Drosophila. Recent work in C. elegans using temperature-sensitive mutants has shown that partial dynein inactivation does not abolish spindle displacement to the posterior (Schmidt et al., 2005), but other studies have suggested a role for dynein in force generation during spindle positioning (Severson and Bowerman, 2003; Pecreaux et al., 2006).
In this study, we show that dyrb-1, which encodes a dynein light chain, plays a role in the regulation of spindle positioning. DYRB-1 is the only C. elegans member of the highly conserved roadblock/LC7 family (Koonin and Aravind, 2000). Mutations in the Drosophila robl gene result in defects in mitosis, the accumulation of vesicles in axons, and larval or pupal lethality (Bowman et al., 1999). Studies in Chlamydomonas reinhardtii have suggested that LC7 light chains are required for both motor assembly and regulation (DiBella et al., 2004). However, the exact function of roadblock/LC7 is unknown. We demonstrate that the depletion of dyrb-1 enhances the phenotype of gpr-1 and lin-5 temperature-sensitive mutants. Furthermore, we show that the forces pulling on the astral microtubules are reduced after the inactivation of dyrb-1. Interestingly, we find that DYRB-1 coimmunoprecipitates with GPR-1 and LIN-5. Based on these results, we propose a model in which GPR-1/2 and LIN-5 function with dynein to control spindle positioning.
Results and discussion
To identify additional genes that act with heterotrimeric G proteins in spindle positioning, we performed RNAi enhancer screens using lin-5 and gpr-1 temperature-sensitive alleles (see Materials and methods gpr-1(or574ts) and lin-5(ev571ts) enhancer screen section; unpublished data). In qualitative assays, we identified T24H10.6 as a candidate gene whose disruption by feeding bacteria that express double-stranded RNA (dsRNA) had no effect in wild-type animals but resulted in embryos that failed to hatch in lin-5(ev571ts) and gpr-1(or574ts) mutant backgrounds. This gene, which is named dyrb-1 (dynein light chain roadblock type-1), encodes a homologue of the roadbock/LC7 dynein light chain family (Bowman et al., 1999). The synthetic lethality was confirmed by feeding assays on solid media. In such assays, we found that the disruption of dyrb-1 in wild-type animals resulted in >99% embryonic viability, whereas viability was decreased considerably in both lin-5(ev571ts) and gpr-1(or574ts) mutants (Table I). Injection of dyrb-1 dsRNA resulted in ∼50% embryonic viability in wild-type animals, whereas viability was substantially decreased in lin-5(ev571ts) and gpr-1(or574ts) mutants (Table I). These results indicate that dyrb-1 genetically interacts with components of the heterotrimeric G protein pathway.
Table I.
Embryonic viability of dyrb-1(RNAi)
| N2 | gpr-1(or574ts) | lin-5(ev571ts) | |
|---|---|---|---|
| % | % | % | |
| Feeding | |||
| L4440 | 99.58 ± 0.32 | 98.75 ± 0.97 | 90.51 ± 3.75 |
| L4440 dyrb-1 | 99.33 ± 0.46 | 67.29 ± 9.13 | 20.47 ± 5.34 |
| Injections | |||
| Control (15°C) | 99.99 ± 0.23 | 99.22 ± 0.35 | 90.68 ± 2.65 |
| dyrb-1(RNAi) | 47.02 ± 15.93 | 3.80 ± 6.78 | 0 |
| Control (22°C) | 99.99 ± 0.3 | 96.30 ± 0.47 | 24.00 ± 12.0 |
| dyrb-1(RNAi) | 45.89 ± 24.49 | 0 | 0 |
The percentage of embryonic viability ± SD is shown after the depletion of dyrb-1 in wild-type, gpr-1(or574ts), and lin-5(ev571ts) strain backgrounds. Feeding was performed at 17°C. The progeny of 30 worms was analyzed for each genotype.
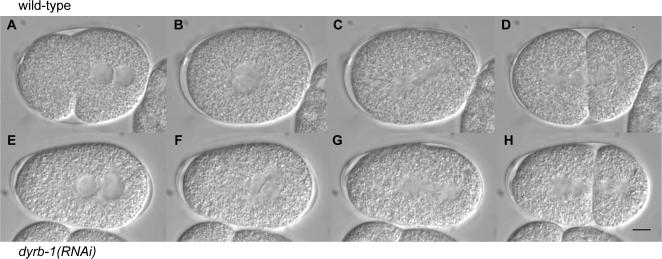
Because the injection of dyrb-1 dsRNA in wild-type animals results in reduced embryonic viability, we investigated by time-lapse differential interference contrast (DIC) microscopy whether dyrb-1(RNAi) embryos have early defects. The progression of events in wild-type embryos is illustrated in Fig. 1 (A–D). In wild type, the oocyte pronucleus migrates to the posterior to meet the sperm pronucleus, which also moves slightly to the center of the embryo. As a result, the two pronuclei meet at ∼68% of embryo length (0% anterior-most and 100% posterior-most; Table II). The two pronuclei and associated centrosomes then migrate toward the cell center while undergoing a 90° rotation (Fig. 1 B) that aligns the centrosomes along the anterior-posterior axis of the embryo. The spindle sets up in the center of the cell along this axis and is displaced toward the posterior by an imbalance of pulling forces at metaphase/anaphase. During this displacement, the posterior spindle pole undergoes transverse oscillations called rocking (Fig. 1 C). At telophase, the posterior spindle pole flattens, whereas the anterior remains round (Fig. 1 D). This asymmetric displacement of the spindle results in an asymmetry in cell size after cytokinesis, with a larger anterior cell (AB) and a smaller posterior cell (P1; Fig. 1 D).
Figure 1.
Early development of wild-type and dyrb-1(RNAi) embryos. Time-lapse images of wild-type (A–D) or dyrb-1(RNAi) (E–H) embryos during pronuclear meeting (A and E), nuclear envelope breakdown (B and F), anaphase (C and G), and after cytokinesis (D and H). Embryos were imaged at 22°C. Posterior is to the right. Bar, 10 μm.
Table II.
Disruption of dyrb-1 results in early defects
| Genotype | PNM | Position of NCC | Spindle length |
|---|---|---|---|
| % | % | % | |
| 15°C | |||
| Wild type (n = 18) | 69.62 ± 5.26 | 50.53 ± 1.21 | 43.44 ± 1.92 |
| gpr-1(or574ts) (n = 15) | 73.04 ± 9.02 | 50.57 ± 1.54 | 41.41 ± 3.24 |
| lin-5(ev571ts) (n = 22) | 71.39 ± 6.88 | 52.94 ± 2.30 | 44.04 ± 2.56 |
| dyrb-1(RNAi) (n = 23) | 70.58 ± 6.46 | 62.87 ± 4.66 | 40.29 ± 2.29a |
| gpr-1(or574ts); dyrb-1(RNAi) (n = 19) | 67.42 ± 5.81 | 64.53 ± 6.17 | 35.29 ± 3.18 |
| lin-5(ev571ts) dyrb-1(RNAi) (n = 17) | 73.85 ± 5.96 | 69.49 ± 6.49 | 36.95 ± 5.01 |
| 22°C | |||
| Wild type (n = 27) | 68.35 ± 3.89 | 51.08 ± 2.25 | 44.32 ± 3.01 |
| gpr-1(or574ts) (n = 17) | 69.69 ± 6.11 | 50.41 ± 1.85 | 41.89 ± 2.11 |
| lin-5(ev571ts) (n = 26) | 67.10 ± 6.10 | 50.08 ± 3.81 | 42.44 ± 2.15 |
| dyrb-1(RNAi) (n = 22) | 66.37 ± 1.85 | 63.31 ± 5.58 | 38.39 ± 3.49a |
| gpr-1(or574ts); dyrb-1(RNAi) (n = 16) | 67.33 ± 3.74 | 68.33 ± 5.77 | 37.73 ± 4.55 |
| lin-5(ev571ts) dyrb-1(RNAi) (n = 17) | 72.47 ± 7.46 | 75.65 ± 12.16 | 36.08 ± 4.03 |
| gpr-1/2(RNAi) (n = 19) | 62.29 ± 5.93 | 50.28 ± 1.60 | 38.84 ± 1.96 |
| lin-5(RNAi) (n = 23) | 64.44 ± 4.19 | 52.31 ± 3.46 | 38.42 ± 3.53 |
All events were calculated as the percentage of egg length ± SD where 0% is the anterior-most point and 100% is the posterior-most point in the embryo. n is the number of embryos analyzed. PNM, pronuclear meeting. Pronuclear meeting corresponds to the position of pronuclear meeting along the a-p axis. The difference of pronuclear meeting position at 22°C between gpr-1/2(RNAi) and wild type and between lin-5(RNAi) and wild type are significant (P = 0.0006 and P = 0.002, respectively; t test). NCC, nuclear centrosomal complex. The position of the nuclear centrosomal complex was calculated at the onset of nuclear envelope breakdown. Spindle pole separation was calculated at anaphase (by dividing spindle length by embryo length).
The difference in spindle length of dyrb-1(RNAi) embryos compared with gpr-1(or574ts); dyrb-1(RNAi), lin-5(ev571ts) dyrb-1(RNAi), and wild-type embryos is significant (P < 0.00001; t test).
We found that many of these processes are affected in dyrb-1(RNAi) embryos. dyrb-1(RNAi) embryos display delayed migration of the oocyte pronucleus (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200707085/DC1), failure in pronuclear centration (Table II), and delay in rotation of the pronucleus–centrosome complex (Fig. 1 F), although in all embryos, the mitotic spindle is aligned along the anterior-posterior axis at cytokinesis (Fig. 2 G). In addition, the first mitotic spindle is significantly shorter than in wild type (Table II), spindle rocking is absent in all embryos, and the posterior aster does not flatten (Fig. 1 H). These phenotypes are highly penetrant (as observed in nearly 100% of the embryo) and, therefore, cannot solely be the cause of embryonic lethality because 50% of the dyrb-1(RNAi) embryos are viable in these conditions.
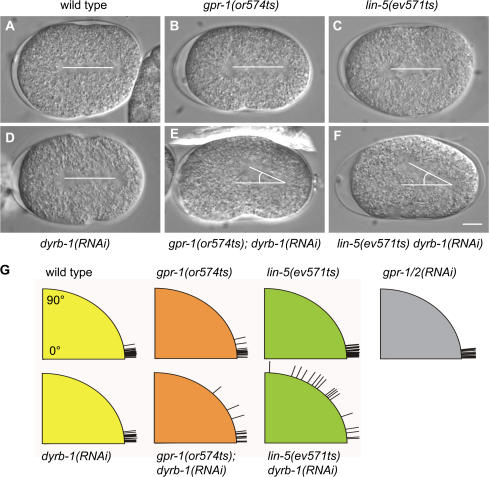
Figure 2.
Inactivation of DYRB-1 in lin-5(ev571) and gpr-1(or574) mutants results in aberrant spindle orientation in the one-cell embryo before cytokinesis. (A–F) One-cell embryos of the indicated genotypes before cytokinesis. Embryos were imaged at 22°C. Posterior is to the right. (G) Spindle orientation at cytokinesis in wild-type and mutant embryos of the indicated genotype. Alignment along the anterior-posterior axis corresponds to 0°C. Bar, 10 μm.
The phenotypes of dyrb-1(RNAi)–depleted embryos are similar to the phenotypes observed upon weak RNAi depletion of dhc-1, which encodes a dynein heavy chain (Gonczy et al., 1999), or in animals bearing temperature-sensitive alleles of dhc-1 (Schmidt et al., 2005). Strong RNAi depletion of dhc-1 results in more severe phenotypes, including defects in centrosome separation, bipolar spindle formation, and cytokinesis (Gonczy et al., 1999). We did not observe such phenotypes upon the RNAi depletion of dyrb-1. Although transgenically expressed GFP–DYRB-1 is strongly depleted under our RNAi conditions (Fig. 3), we cannot exclude that there is remaining endogenous DYRB-1 in the depleted animals, and attempts to obtain anti–DYRB-1 antibodies were unsuccessful. However, embryos from dyrb-1(tm2645) homozygote mutant mothers, which are predicted to produce a truncated protein that only contains the first 29 amino acids of DYRB-1, show phenotypes identical to dyrb-1(RNAi) embryos (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200707085/DC1). Because there are at least six other dynein light chains predicted in the C. elegans genome, it is possible that one or more of these partially compensate for the loss of dyrb-1 in the early embryo. Collectively, these results suggest that DYRB-1 is required for several dynein-dependent processes in the one-cell embryo, including proper spindle orientation, and are consistent with DYRB-1 regulating dynein activity.
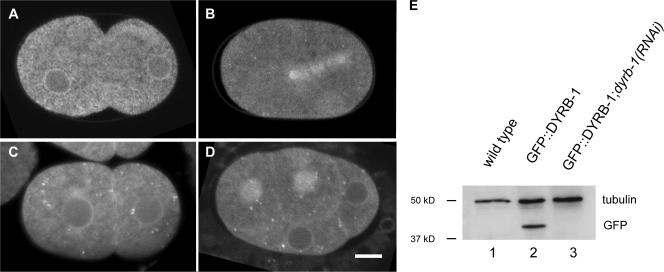
Figure 3.
Localization pattern of GFP–DYRB-1 in early embryos. (A–E) Localization pattern of GFP–DYRB-1 at different cell cycle stages. (A) One-cell embryo during pronuclear migration. (B) One-cell embryo at anaphase. (C) Two-cell embryo. (D) Four-cell embryo. Posterior is to the right. (E) Western blot analysis of C. elegans embryos probed with anti-GFP antibodies and antitubulin antibodies. Anti-GFP antibodies recognize a band of ∼40 kD (lane 2) that is absent in the extract from wild-type embryos (lane 1). GFP–DYRB-1 is fully depleted in extract from dyrb-1(RNAi) embryos (lane 3). Tubulin is used as a loading control. Bar, 10 μm.
To further investigate the genetic interaction between dyrb-1 and genes in the heterotrimeric G protein pathway, we analyzed the phenotype resulting from dyrb-1 depletion in lin-5 or gpr-1 mutants. Codisruption of dyrb-1(RNAi) and gpr-1(or574ts) or lin-5(ev571ts) at permissive temperature for both mutants resulted in a complete loss of embryonic viability (Table I) and a strong synthetic phenotype in early events. The male pronuclear envelope broke down before the pronuclei met in 9/17 cases for gpr-1(or574ts); dyrb-1(RNAi) and in 9/16 cases for lin-5(ev571ts) dyrb-1(RNAi) embryos. During prophase in wild type, gpr-1(or574ts), lin-5(RNAi), and dyrb-1(RNAi) one-cell embryos, the two centrosomes localized to opposite sides of the male pronucleus. In 7/17 embryos codisrupted for lin-5 dyrb-1 and in 6/10 embryos codisrupted for gpr-1; dyrb-1, this separation failed, and centrosomes remained in close proximity to each other (Videos 1–5, available at http://www.jcb.org/cgi/content/full/jcb.200707085/DC1). This phenotype is similar to embryos strongly depleted of DHC-1 (Gonczy et al., 1999). Furthermore, although in dyrb-1(RNAi), gpr-1(or574ts), and lin-5(ev571ts) embryos, the mitotic spindle was oriented along the anterior-posterior axis at the onset of cytokinesis, rotation was further delayed in the double mutants, and the spindle was misaligned in 13/17 lin-5(ev571) dyrb-1(RNAi) embryos and 3/16 gpr-1(or574); dyrb-1(RNAi) embryos (Fig. 2). This phenotype is independent from the centrosome separation defect because we observed misaligned spindles in embryos in which the centrosomes had correctly separated.
These results indicate that lin-5(ev571) dyrb-1(RNAi) and gpr-1(or574); dyrb-1(RNAi) double mutant embryos have a stronger phenotype than any single mutant, which is consistent with GPR-1, LIN-5, and DYRB-1 regulating common processes in the one-cell embryo. Interestingly, the pronuclear migration defect was enhanced in both double mutant combinations, suggesting that LIN-5 and GPR-1/2 play a role in this process. Therefore, we quantified the position of pronuclear meeting in gpr-1/2(RNAi) and lin-5(RNAi) embryos and found that they meet more anterior when compared with wild-type embryos (Table II). This indicates a novel role for the heterotrimeric G protein pathway in the regulation of pronuclear migration.
Altogether, the phenotypes of dyrb-1(RNAi) embryos suggest that DYRB-1 may regulate dynein activity. To address this, we generated transgenic animals expressing DYRB-1 fused to GFP in the early embryo. We found that DYRB-1 localizes in a punctate manner in the cytoplasm and is found at the periphery of pronuclei and nuclei (Fig. 3 A). During anaphase, GFP–DYRB-1 localizes to the mitotic spindle, the two centrosomes, and around the chromosomes (Fig. 3 B). We also observed a weak localization to the cortex that is more apparent in two- and four-cell embryos as well as in older embryos (Fig. 3, C and D). The depletion of GPR-1/2 or LIN-5 did not affect this localization pattern at any stage (unpublished data). Therefore, DYRB-1 localization appears similar to that of DHC-1 (Gonczy et al., 1999), which is consistent with a role in regulating dynein activity in the early embryo. To further test this possibility, we depleted dyrb-1 in let-99(or204ts) mutants, which exhibit hyperactive nuclear movements during centration (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200707085/DC1). This phenotype results from an excess of Gα signaling and dynein activity because it is suppressed in let-99; Gα(RNAi) embryos and in let-99 embryos in which dhc-1 has been weakly depleted by RNAi (Tsou et al., 2002, 2003). We found that the depletion of dyrb-1 also suppresses the nuclear hyperactive movement of let-99 embryos (Fig. S2, M–P; and Video 6). These results indicate that DYRB-1 plays a positive role in the regulation of dynein activity.
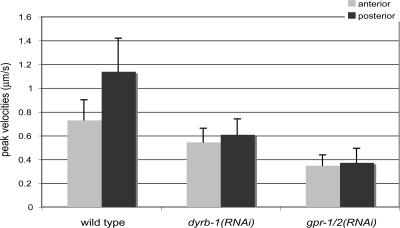
Recent work in C. elegans has shown that dynein activity may contribute to posterior spindle displacement, although its function is not strictly essential for this process (Severson and Bowerman, 2003; Schmidt et al., 2005; Pecreaux et al., 2006). Several observations indicate that the pulling forces that regulate spindle positioning are weaker in dyrb-1(RNAi) embryos: the spindle is significantly shorter than in wild type, spindle rocking does not occur, and the posterior aster does not flatten (Table II). Therefore, we tested whether dyrb-1 regulates spindle positioning by investigating whether pulling forces are compromised in dyrb-1(RNAi) embryos when the mitotic spindle is severed by a laser microbeam (Grill et al., 2001). Because the mitotic spindle forms at the posterior of dyrb-1(RNAi) embryos, comparisons were made with wild-type embryos in which the spindle was also severed when it was more posterior at the onset of anaphase B (see Materials and methods RNAi, microscopy, and spindle severing section). In wild-type embryos, forces on each side of the spindle are asymmetric, and the mean peak velocity of the posterior centrosome is ∼1.6 times higher than the one of the anterior centrosome (Fig. 4; Grill et al., 2001). The mean peak velocity of both anterior and posterior centrosomes in dyrb-1(RNAi) embryos after spindle severing is significantly reduced compared with wild type (P < 0.001; t test), and the asymmetry is lost (Fig. 4 and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200707085/DC1). The mean peak velocity of both centrosomes in dyrb-1(RNAi) embryos was faster than that in gpr-1/2(RNAi) embryos, indicating that RNAi disruption of dyrb-1 does not completely inactivate the force generators, as is the case for gpr-1/2(RNAi). These reduced forces are not a consequence of polarity defects because PAR proteins are properly localized in dyrb-1(RNAi) embryos (Fig. S3). Likewise, the depletion of dyrb-1 did not affect the localization pattern of GPR-1/2 or LIN-5 (Fig. S3 and not depicted). These results indicate that dyrb-1 plays an important role in regulating the asymmetry in forces that pull on astral microtubules and that it functions downstream or in parallel to polarity cues and LIN-5/GPR-1/2.
Figure 4.
The mean peak velocities of anterior and posterior asters are reduced in dyrb-1(RNAi) embryos. Mean peak velocities (micrometer/second) of anterior (light gray) and posterior (dark gray) spindle poles measured after spindle severing in one-cell stage embryos of the indicated genotypes (wild type, n = 24; dyrb-1(RNAi), n = 26; gpr-1/2(RNAi), n = 15). Error bars correspond to SD.
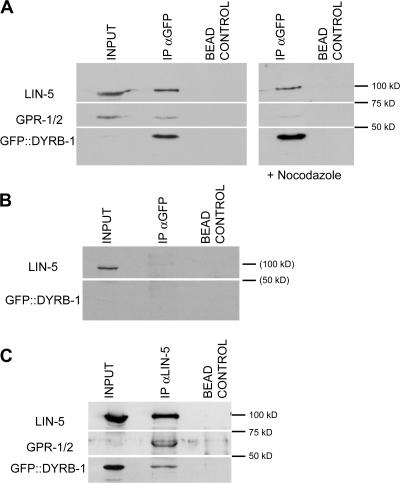
Our results suggest that DYRB-1 could function together with the heterotrimeric G protein pathway to regulate microtubule-dependent events. In mammalian cells, NuMA was shown to physically interact with both dynein and LGN (Merdes et al., 1996; Du et al., 2001). This suggested the possibility that DYRB-1 could molecularly interact with components of the heterotrimeric G protein pathway. Thus, we investigated whether DYRB-1 can be recovered in a complex with LIN-5 and/or GPR-1/2 using embryonic extracts made from the transgenic strain expressing GFP–DYRB-1. We found that both LIN-5 and GPR-1/2 could be coimmunoprecipitated with anti-GFP antibodies (Fig. 5). Conversely, both GPR-1/2 and GFP–DYRB-1 could be coimmunoprecipitated with anti–LIN-5 antibodies. These interactions are specific because they are not detected in control immunoprecipitations in which anti-GFP antibodies were incubated with extracts made from dyrb-1(RNAi)-depleted animals. GFP–DYRB-1 could still be coimmunoprecipitated with LIN-5 and GPR-1/2 in the presence of 50 μM of the microtubule-depolymerizing drug nocodazole, indicating that this interaction is microtubule independent. Unfortunately, the depletion of lin-5 or gpr-1/2 in the GFP–DYRB-1–expressing strain resulted in complete sterility, thus precluding us from assessing whether this interaction depends on LIN-5 or GPR-1/2 (see Materials and methods Preparation of extracts and Western blot analyses section). These results suggest that LIN-5, GPR-1/2, and DYRB-1 are parts of a common protein complex, which is consistent with these three proteins functioning in the same pathway.
Figure 5.
DYRB-1, LIN-5, and GPR-1/2 can coimmunoprecipitate. Immunoprecipitations from embryonic extracts expressing a GFP–DYRB-1 transgene using anti-GFP antibodies (A and B) or LIN-5 antibodies (C). All three proteins can be coimmunoprecipitated in each condition as well as in the presence of 50 μM nocodazole (A). (B) Coimmunoprecipitation using anti-GFP antibodies is not observed when DYRB-1 is depleted by RNAi. Input is 1/20 of the total amount used in all blots.
In conclusion, we have shown that the embryonic loss of dyrb-1, which encodes a dynein light chain subunit of the roadblock family, phenocopies a weak loss of dynein activity. Furthermore, we have demonstrated that DYRB-1 genetically and physically interacts with LIN-5 and GPR-1/2, which are two positive regulators of the heterotrimeric G protein pathway. This suggests that DYRB-1 or another component of the dynein complex is an effector of the heterotrimeric G protein pathway. Interestingly, spindle-positioning forces are reduced, and the asymmetry in pulling forces between anterior and posterior poles is lost in dyrb-1–depleted embryos. Collectively, these results suggest a model in which heterotrimeric G protein signaling controls spindle positioning, at least in part, by regulating dynein activity. One possibility is that LIN-5 and GPR-1/2 could asymmetrically activate cortically anchored dynein, which would then promote pulling of the mitotic spindle toward the posterior pole of the embryo. Recent results by Pecreaux et al. (2006) showed that a weak depletion of dynein results in a loss of pole oscillations during spindle positioning, which is consistent with our observation that such oscillations are lost in dyrb-1(RNAi) embryos. However, their results and results from Grill et al. (2003) predicted that reducing dynein activity leads to an overall reduction in pulling forces rather than a loss in asymmetry, which is inconsistent with our observation that forces are weaker and symmetric in dyrb-1(RNAi) embryos. One possibility to reconcile these results is to suggest that DYRB-1 regulates all asymmetries in pulling forces and that the remaining forces in dyrb-1(RNAi) embryos are not DYRB-1 dependent. These remaining forces could depend on a variety of other potential regulators of force generators (e.g., another dynein light chain, the dynactin complex, or microtubule–cortex interactions).
Spindle positioning was recently proposed to be mainly controlled by the regulation of microtubule dynamics (Kozlowski et al., 2007) based on computer simulations. It is unclear at present how dynein regulates spindle positioning, but previous studies in other cell types have shown that its motor activity can regulate microtubule dynamics (Carminati and Stearns, 1997; Yamamoto et al., 2001). Therefore, spindle positioning in C. elegans embryos could occur through a regulation of dynein motor activity by DYRB-1 and the heterotrimeric G protein signaling pathway, which could, in turn, modulate microtubule dynamics. These two events need not be mutually exclusive.
Materials and methods
Strains and alleles
C. elegans Bristol strain N2 was used as the standard wild-type strain. Nematode culturing was performed as described previously (Brenner, 1974). The alleles used in this study are gpr-1(or574ts), lin-5(ev571ts) (Lorson et al., 2000), let-99(or204ts), and dyrb-1(tm2645). Temperature-sensitive mutant strains were grown at a permissive temperature of 15°C and shifted to 22°C 24 h before the embryos were examined.
gpr-1(or574ts) was isolated in a screen for temperature-sensitive embryonic lethal mutations (Encalada et al., 2000) and was outcrossed six times with N2 males. The mutation mapped between dpy-17 and unc-50, close to gpr-1 and its paralogous locus gpr-2. These two closely linked paralogues are 97% identical in coding DNA and cannot be distinguished by expression analysis or RNAi. Because embryos from or574ts homozygotes show a mild form of the cytological defects resulting from RNAi inactivation of gpr-1/gpr-2, we sequenced both loci in the mutant. Genomic DNA fragments encompassing the F22B7.13 (gpr-1) and C38C10.4 (gpr-2) loci were amplified from the mutant by PCR using primer sequences unique to each locus. Pooled PCR products were sequenced using a sequencer (CEQ 800; Beckman Coulter) at the University of Oregon Sequencing Facility. or574ts was found to carry a missense mutation in gpr-1 that was predicted to convert glycine 289 to arginine. This mutation was not present in the background strain used for mutagenesis (CB1309), and no other mutations were found in or574ts within 0.5-kb gpr-1 or gpr-2 coding regions. Eggs laid by or574ts homozygotes were >99% viable at 23°C, 22% viable at 25°C, and <1% viable at 26°C. Eggs laid by heterozygotes were >99% viable at 26°C, indicating that or574ts is fully recessive. However, trans-heterozygotes for or574ts and nDf20, a deficiency predicted to uncover gpr-1 but not gpr-2, produced 33% viable eggs at 26°C, suggesting that the or574ts mutant GPR-1 protein acts in a dominant-negative fashion for both GPR-1 and -2.
For the GFP–DYRB-1 strain, full-length DYRB-1 cDNA was cloned into pID3.01 (Pellettieri et al., 2003) using Gateway technology (Invitrogen). GFP lines were created using the microparticle bombardment technique as described previously (Praitis et al., 2001). The transgene analyzed fully rescues the embryonic lethality of the dyrb-1(tm2645) allele, indicating that the GFP fusion is functional.
gpr-1(or574ts) and lin-5(ev571ts) enhancer screen
The screen was performed as described previously (Labbé et al., 2006) with some modifications. In brief, lin-5(ev571ts) and gpr-1(or574ts) mutant animals were grown in large quantities on solid media at 15°C and were bleached to collect embryos. These embryos were incubated at 15°C with rocking in M9 buffer to allow hatching of L1 larvae. Assays were performed in plates containing 96 wells. RNAi clones from the available collection (Kamath et al., 2003) were seeded in individual wells and grown overnight at 37°C in Luria Broth medium containing 100 μg/ml carbenicillin. Each well of fresh 96-well plates was then filled with 75 μl 3x nematode growth medium (NGM; regular NGM with a triple amount of peptone), and 2 μl of overnight bacterial culture was added. The plates were incubated at 37°C for 2.5 h without shaking. 25 μl 3xNGM containing 24 mM IPTG was then added to each well (6 mM IPTG final), and the plates were incubated at 37°C for 5 h without shaking. 5–10 L1 worms of each genotype were then added to each well (15 μl from a suspension containing approximately eight worms per 15 μl M9 buffer). The plates were incubated for 6–7 d at the semirestrictive temperature of 17°C without agitation until food was depleted and F1 progeny had hatched. Enhancement of lethality was estimated by visual inspection under a dissecting scope, and the relative decrease of swimming L1 larvae compared with the control was scored as positive. Liquid handling was performed using a robotic system (Biomek FX; Beckman Coulter) equipped with a 96-channel pipetting head.
RNAi, microscopy, and spindle severing
dsRNA was produced as described previously (Zipperlen et al., 2001). For live imaging of embryos, dsRNA was injected into young adult hermaphrodites. Animals were dissected, and embryos were analyzed 24 h after injection.
For DIC analysis of living embryos, embryos were mounted as described previously (Gotta et al., 2003). The first cell cycle of embryos was visualized with a camera (Orca ER; Hamamatsu) mounted on an inverted microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.), and the acquisition system was controlled by Openlab software (Improvision). Images were captured at 5-s intervals using a plan Apochromat 63× 1.4 NA objective. For immunofluorescence experiments, a microscope system (DeltaVision 3000; Olympus) was used for capturing and deconvolving images of embryos.
Spindle-severing experiments were performed similar to Grill et al. (2001) using an inverted microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) equipped with a PALM laser system (Mikrolaser Technologie). The pulsed laser was focused to an ∼1-μm-thick spot in the focal plane. C. elegans embryos were mounted on the inverted microscope. Because the spindle does not go to the center of the cell in dyrb-1(RNAi) embryos, the spindle was cut along the midzone when at a posterior position in both wild-type and dyrb-1(RNAi) animals, which corresponds to the onset of anaphase B. The wild-type centrosome speeds that were measured in these experiments are comparable with those measured previously (Grill et al., 2001), indicating that severing the spindle when it is at a posterior position does not preclude the accurate measurement of centrosome velocity. For monitoring spindle severing, one DIC image was captured every second. Measurements of peak velocities were performed by manual tracking with ImageJ (tracking the center of the aster; National Institutes of Health).
Indirect immunofluorescence
Indirect immunofluorescence of embryos was performed as described previously (Gotta and Ahringer, 2001). We used rabbit anti–GPR-1/2 (1:80; Couwenbergs et al., 2004), mouse anti–α-tubulin (1:1,000; DM1A; Sigma-Aldrich), and rabbit anti-GFP (1:150; Abcam) as primary antibodies. Secondary antibodies were anti–rabbit AlexaFluor488, anti–rabbit AlexaFluor568, and anti–mouse AlexaFluor568 (1:500 each). DNA was visualized with DAPI.
Preparation of extracts and Western blot analyses
To prepare dyrb-1(RNAi) embryonic extracts, wild-type embryos were obtained by hypochlorite treatment, and newly hatched synchronized L1 larvae were grown on bacteria expressing dyrb-1 dsRNA until adulthood. Adult worms were collected and treated with hypochlorite to recover embryos. These embryos were resuspended in 1 vol of 4× Laemmli buffer and boiled for 10 min at 95°C before Western blot analysis. For immunoprecipitation experiments, the embryos were washed in immunoprecipitation lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 1% Triton X-100, and protease inhibitor cocktail; Afshar et al., 2004), resuspended in an equal volume of buffer, and frozen at −80°C. They were then broken with glass beads (Lysing Matrix C; MP Biomedicals) in a bead beater (3 × 30 s with a 1-min interval at 4°C). The lysate was spun down at 14,000 rpm for 15 min at 4°C. The extract was incubated with 3 mg anti-GFP antibodies (Roche) or 2 mg anti–LIN-5 antibodies (Lorson et al., 2000). Antibody and protein A–Sepharose bead binding was performed as described previously (Afshar et al., 2004). For SDS-PAGE and Western blotting, standard procedures were used. The GPR-1/2 antibody used for blotting was described previously (Couwenbergs et al., 2004). Although both lin-5 or gpr-1/2 could be depleted by dsRNA injection in adult hermaphrodites expressing GFP–DYRB-1, the depletion of either lin-5 or gpr-1/2 by feeding dsRNA to GFP–DYRB-1 L1 or L3/L4 animals (in liquid or on solid media) resulted in strong sterility, thereby preventing us from preparing embryonic extracts for immunoprecipitation experiments. We note that the amount of GPR-1/2 that was coimmunoprecipitated with anti–LIN-5 antibodies in Fig. 5 C appears proportionally high compared with the loading control. This could indicate that the anti–LIN-5 antibodies have a higher affinity for the pool of LIN-5 protein that is in a complex with GPR-1/2.
Online supplemental material
Fig. S1 shows the early development of dyrb-1(tm2645) mutant embryos. Fig. S2 shows that the hyperactive nuclear movement of let-99 mutant embryos is suppressed in let-99(or204ts); dyrb-1(RNAi) embryos. Fig. S3 shows that DYRB-1–depleted embryos have normal polarity. Videos 1 and 2 show wild-type (Video 1) and gpr-1(or574) (Video 2) embryos expressing GFP-tubulin and imaged at 22°C. Videos 3 and 4 show lin-5(RNAi) (Video 3) and gpr-1(or574); dyrb-1(RNAi) (Video 4) embryos expressing GFP-tubulin and imaged at 22°C. Video 5 shows lin-5(RNAi) dyrb-1(RNAi) embryos expressing GFP-tubulin and imaged at 22°C. Video 6 shows a combined video with let-99(or204ts), dyrb-1(RNAi), and dyrb-1(RNAi); let-99(or204ts) embryos. Video 7 shows a combined video with spindle-cutting experiments in a wild-type embryo, a dyrb-1(RNAi) embryo, and a gpr-1/2(RNAi) embryo. Table S1 shows that pronuclear migration is delayed in dyrb-1–depleted embryos. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707085/DC1.
Supplementary Material FROM ROCKUP!
Acknowledgments
We are indebted to Matthias Peter for insightful discussions and for providing us with access to the robotic equipment. We are also grateful to Ulrike Margelisch, Caroline Zbinden, and Anton Lehmann for technical assistance as well as to Patrick Meraldi, Matthias Peter, and the Gotta and Peter laboratories for helpful advice and stimulating discussions. We also thank the Light Microscopy Center of the ETH for providing state of the art microscopes and Gabor Csucs for technical help with image acquisition. We thank Sander van den Heuvel, Shohei Mitani (Japanese National Bioresource Project for C. elegans), and the Caenorhabditis Genetics Center (which is funded by the National Center for Research Resources of the National Institutes of Health) for strains and reagents.
This work was supported by a European Molecular Biology Organization postdoctoral fellowship to J.-C. Labbé, a Human Frontier Science Program postdoctoral fellowship to T. Marty, as well as grants from the Swiss National Foundation, Novartis, and the ETH to M. Gotta.
C. Couwenbergs and J.-C. Labbé contributed equally to this paper.
J.-C. Labbé's present address is Institut de recherche en immunologie et cancérologie, Université de Montréal, Succursale Centre-ville, Montréal, Quebec H3C 3J7, Canada.
T. Marty's present address is Swiss Contact Office for Research and Higher Education, B-1050 Brussels, Belgium.
Abbreviations used in this paper: DIC, differential interference contrast; dsRNA, double-stranded RNA; NGM, nematode growth medium; NuMA, nuclear mitotic apparatus.
References
- Afshar, K., F.S. Willard, K. Colombo, C.A. Johnston, C.R. McCudden, D.P. Siderovski, and P. Gonczy. 2004. RIC-8 is required for GPR-1/2-dependent Galpha function during asymmetric division of C. elegans embryos. Cell. 119:219–230. [DOI] [PubMed] [Google Scholar]
- Bellaiche, Y., and M. Gotta. 2005. Heterotrimeric G proteins and regulation of size asymmetry during cell division. Curr. Opin. Cell Biol. 17:658–663. [DOI] [PubMed] [Google Scholar]
- Bowman, A.B., R.S. Patel-King, S.E. Benashski, J.M. McCaffery, L.S. Goldstein, and S.M. King. 1999. Drosophila roadblock and Chlamydomonas LC7: a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 146:165–180. [PMC free article] [PubMed] [Google Scholar]
- Bowman, S.K., R.A. Neumuller, M. Novatchkova, Q. Du, and J.A. Knoblich. 2006. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 10:731–742. [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, J.L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, K., S.W. Grill, R.J. Kimple, F.S. Willard, D.P. Siderovski, and P. Gonczy. 2003. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 300:1957–1961. [DOI] [PubMed] [Google Scholar]
- Couwenbergs, C., A.C. Spilker, and M. Gotta. 2004. Control of embryonic spindle positioning and Galpha activity by C. elegans RIC-8. Curr. Biol. 14:1871–1876. [DOI] [PubMed] [Google Scholar]
- DiBella, L.M., M. Sakato, R.S. Patel-King, G.J. Pazour, and S.M. King. 2004. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol. Biol. Cell. 15:4633–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Q., and I.G. Macara. 2004. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 119:503–516. [DOI] [PubMed] [Google Scholar]
- Du, Q., P.T. Stukenberg, and I.G. Macara. 2001. A mammalian partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 3:1069–1075. [DOI] [PubMed] [Google Scholar]
- Encalada, S.E., P.R. Martin, J.B. Phillips, R. Lyczak, D.R. Hamill, K.A. Swan, and B. Bowerman. 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228:225–238. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., S. Pichler, M. Kirkham, and A.A. Hyman. 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147:135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta, M., and J. Ahringer. 2001. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell Biol. 3:297–300. [DOI] [PubMed] [Google Scholar]
- Gotta, M., Y. Dong, Y.K. Peterson, S.M. Lanier, and J. Ahringer. 2003. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 13:1029–1037. [DOI] [PubMed] [Google Scholar]
- Grill, S.W., P. Gonczy, E.H. Stelzer, and A.A. Hyman. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633. [DOI] [PubMed] [Google Scholar]
- Grill, S.W., J. Howard, E. Schaffer, E.H. Stelzer, and A.A. Hyman. 2003. The distribution of active force generators controls mitotic spindle position. Science. 301:518–521. [DOI] [PubMed] [Google Scholar]
- Izumi, Y., N. Ohta, K. Hisata, T. Raabe, and F. Matsuzaki. 2006. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8:586–593. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., A.G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., and L. Aravind. 2000. Dynein light chains of the Roadblock/LC7 group belong to an ancient protein superfamily implicated in NTPase regulation. Curr. Biol. 10:R774–R776. [DOI] [PubMed] [Google Scholar]
- Kozlowski, C., M. Srayko, and F. Nedelec. 2007. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 129:499–510. [DOI] [PubMed] [Google Scholar]
- Labbé, J.C., A. Pacquelet, T. Marty, and M. Gotta. 2006. A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR protein-dependent cell polarity in Caenorhabditis elegans. Genetics. 174:285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson, M.A., H.R. Horvitz, and S. van den Heuvel. 2000. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J. Cell Biol. 148:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., K. Ramyar, J.D. Vechio, and D.W. Cleveland. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. [DOI] [PubMed] [Google Scholar]
- Pecreaux, J., J.C. Roper, K. Kruse, F. Julicher, A.A. Hyman, S.W. Grill, and J. Howard. 2006. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 16:2111–2122. [DOI] [PubMed] [Google Scholar]
- Pellettieri, J., V. Reinke, S.K. Kim, and G. Seydoux. 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C elegans. Dev. Cell. 5:451–462. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada, K., and L.H. Tsai. 2005. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 122:119–131. [DOI] [PubMed] [Google Scholar]
- Schmidt, D.J., D.J. Rose, W.M. Saxton, and S. Strome. 2005. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell. 16:1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, A.F., and B. Bowerman. 2003. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 161:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller, K.H., C. Cabernard, and C.Q. Doe. 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8:594–600. [DOI] [PubMed] [Google Scholar]
- Srinivasan, D.G., R.M. Fisk, H. Xu, and S. van den Heuvel. 2003. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 17:1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou, M.F., A. Hayashi, L.R. DeBella, G. McGrath, and L.S. Rose. 2002. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 129:4469–4481. [DOI] [PubMed] [Google Scholar]
- Tsou, M.F., A. Hayashi, and L.S. Rose. 2003. LET-99 opposes Galpha/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development. 130:5717–5730. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., C. Tsutsumi, H. Kojima, K. Oiwa, and Y. Hiraoka. 2001. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol. Biol. Cell. 12:3933–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperlen, P., A.G. Fraser, R.S. Kamath, M. Martinez-Campos, and J. Ahringer. 2001. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20:3984–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.