Abstract
Tissue damage is usually followed by healing, as both differentiated and stem cells migrate to replace dead or damaged cells. Mesoangioblasts (vessel-associated stem cells that can repair muscles) and fibroblasts migrate toward soluble factors released by damaged tissue. Two such factors are high mobility group box 1 (HMGB1), a nuclear protein that is released by cells undergoing unscheduled death (necrosis) but not by apoptotic cells, and stromal derived factor (SDF)–1/CXCL12. We find that HMGB1 activates the canonical nuclear factor κB (NF-κB) pathway via extracellular signal-regulated kinase phosphorylation. NF-κB signaling is necessary for chemotaxis toward HMGB1 and SDF-1/CXCL12, but not toward growth factor platelet-derived growth factor, formyl-met-leu-phe (a peptide that mimics bacterial invasion), or the archetypal NF-κB–activating signal tumor necrosis factor α. In dystrophic mice, mesoangioblasts injected into the general circulation ingress inefficiently into muscles if their NF-κB signaling pathway is disabled. These findings suggest that NF-κB signaling controls tissue regeneration in addition to early events in inflammation.
Introduction
Damage to tissues and organs is frequent in the life of vertebrates: tissues can be ripped, squashed, or wounded by mechanical forces, mishaps, or predators. Freezing or burns, chemical insults (strong acids or bases or cytotoxic poisons produced by invading bacteria), radiation, or the withdrawal of oxygen and/or nutrients can also kill cells. Thus, the ability to repair damaged tissues is essential for evolutionary success. Very often the new cells that replace the dead ones migrate from specific niches within the tissue or from distant districts such as the bone marrow. Although the mechanism of cell migration has been intensely studied, the orchestration of the physiological responses that bring the relevant cells to the required sites is much less understood.
We and others have found that high mobility group box 1 (HMGB1), an abundant component of the cell nucleus, when present in the extracellular space, signals tissue damage (Bianchi, 2007). HMGB1 is released by cells undergoing necrosis (accidental cell death) but not by cells undergoing apoptosis (Scaffidi et al., 2002). Extracellular HMGB1 then promotes the ingression of inflammatory cells (Scaffidi et al., 2002), but also the migration and proliferation of stem cells (Palumbo et al., 2004; Limana et al., 2005). Thus, HMGB1 has the expected characteristics of a signal that can orchestrate tissue regeneration, although it is not expected to be the only one (Bianchi, 2007).
In particular, we previously described that extracellular HMGB1 can attract mesoangioblasts, both in vitro and in vivo (Palumbo et al., 2004). Mesoangioblasts are a specific population of mesodermal stem cells that are associated with the walls of fetal and postnatal vessels (Minasi et al., 2002). They grow extensively in culture and can differentiate into most mesodermal cell types. When injected into the general circulation of dystrophic mice and dogs, they migrate into muscles and contribute to their regeneration and functional recovery (Sampaolesi et al., 2003, 2006).
Here, we have investigated the signaling pathways that activate cell migration toward extracellular HMGB1 and allow mesoangioblasts to navigate to damaged muscles. HMGB1 is known to activate MAPKs and nuclear factor κB (NF-κB); we show that NF-κB activation proceeds via extracellular signal-regulated kinase (ERK) phosphorylation. Surprisingly, mesoangioblasts and fibroblasts do not migrate toward HMGB1 if NF-κB activation is blocked. This same NF-κB dependency applies to stromal derived factor (SDF)–1/CXCL12, which also directs the migration of stem cells, but not to TNF-α, the archetypal NF-κB activating signal.
Results and discussion
Fibroblasts respond chemotactically to HMGB1
Mesoangioblasts provide an excellent model to investigate cell navigation to damaged tissues in living animals; however, embryonic fibroblasts from genetically modified mice allow the unequivocal identification of the components of the signaling pathways activated by individual chemoattractants.
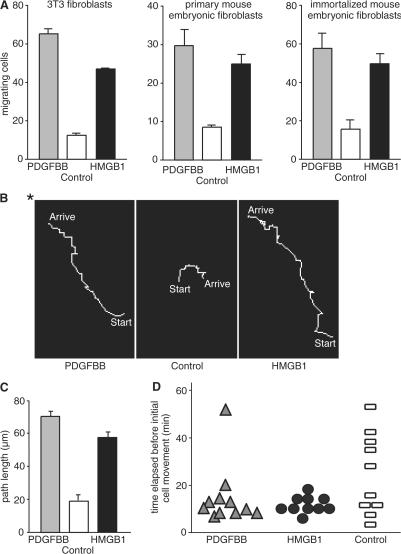
Fibroblast cell lines such as 3T3 and wild-type (wt) mouse embryonic fibroblasts (MEFs), either primary or immortalized with polyoma large T antigen (Calogero et al., 1999), respond chemotactically to HMGB1 in Boyden chambers (Fig. 1 A). The migration is directional, as shown by the tracking of living 3T3 fibroblasts in chemoattractant gradients formed between the inner well and the external ring chamber of a Dunn chemotaxis apparatus (Fig. 1 B). Most cells migrated toward HMGB1 or PDGF, with mean paths of ∼70 and 55 μm, respectively, but were immobile or moved randomly (mean path of 20 μm) in the absence of chemoattractants (Fig. 1 C). Movement occurred within ∼10, 15, and 25 min in the presence of HMGB1, PDGF, and serum-free medium, respectively (Fig. 1 D). Similar results were obtained with primary and immortalized MEFs (unpublished data).
Figure 1.
Fibroblasts migrate in response to HMGB1. (A) Cell migration toward 10 ng/ml PDGFBB and 30 ng/ml HMGB1 was tested in Boyden chambers for 3 h for 3T3 fibroblasts, primary fibroblasts derived from wt embryonic day 14.5 mouse embryos, and fibroblasts immortalized with large T antigen. Each error bar represents the mean ± SD of triplicate samples. The difference between control and chemoattractants is statistically significant (P < 0.01). Each cell type was tested at least three times. (B) In Dunn chambers, cells migrate directionally toward the source of HMGB1 and PDGFBB (asterisk) and move randomly in the absence of chemoattractant. (C) Length of the paths of migrating cells under different conditions. Each error bar represents the mean ± SD of triplicate samples. The difference between control and chemoattractants is statistically significant (P < 0.01). This experiment was repeated three times. (D) Time elapsed before cell movement. Each point represents one cell.
NF-κB is activated in response to HMGB1 via an ERK-dependent mechanism
Extracellular HMGB1 has been reported to engage multiple receptors, including the receptor for advanced glycation end products (RAGE; Hori et al., 1995) and Toll-like receptors 2 and 4 (Park et al., 2004). RAGE has been reported to activate MAPKs; both RAGE and Toll-like receptors activate NF-κB (Bonizzi and Karin, 2004; Bierhaus et al., 2005).
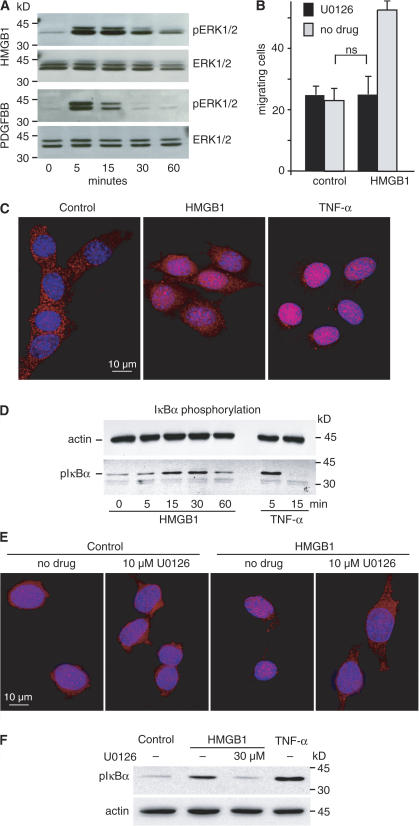
We had previously shown that U0126, a specific inhibitor of MAPK/ERK kinase (MEK) 1/2, which phosphorylates ERKs, abrogates the migration of smooth muscle cells in response to HMGB1 (Degryse et al., 2001). Likewise, HMGB1 induced the rapid phosphorylation of ERK1/2 in 3T3 fibroblasts (Fig. 2 A) and U0126 inhibited the HMGB1-induced migration (Fig. 2 B).
Figure 2.
HMGB1 induces NF-κB activation via an ERK- dependent mechanism. (A) 3T3 fibroblasts were stimulated with 100 ng/ml HMGB1 or 10 ng/ml PDGFBB for 5, 15, 30, or 60 min, and total cell proteins were analyzed by Western blot. ERK1/2 activation was visualized with anti–phospho- ERK antibody (pERK1/2). Stripped blots were reprobed with antibodies against total ERK (ERK1/2). Results are representative of three independent experiments. (B) U0126 inhibits HMGB1-induced cell migration. 3T3 fibroblasts were pretreated with or without 10 μM of the MEK1/2 inhibitor U0126 for 30 min and were allowed to migrate in Boyden chambers for 3 h toward 30 ng/ml HMGB1 or no stimulus. Each error bar represents the mean ± SD of triplicate samples. This experiment was repeated three times. (C) HMGB1 induces NF-κB nuclear translocation. 3T3 fibroblasts grown on coverslips were serum starved for 16 h and stimulated with 100 ng/ml HMGB1 or 20 ng/ml TNF-α for 20 and 10 min, respectively. Cells were fixed and immunostained indirectly for p65/RelA. (D) IκBα is phosphorylated in 3T3 fibroblasts exposed to HMGB1 or TNF-α. Cells were lysed in SDS-PAGE loading buffer at the indicated times; proteins were Western blotted with rabbit polyclonal antibodies against mouse pIκBα or mAb against β-actin. (E) MEK inhibitor U0126 prevents NF-κB nuclear translocation. 3T3 fibroblasts grown on coverslips were serum starved for 16 h, exposed where indicated to U0126, and stimulated 30 min later with 100 ng/ml HMGB1. After 20 min, cells were fixed and immunostained indirectly for p65/RelA. (F) The MEK inhibitor U0126 impairs IκBα phosphorylation. 3T3 fibroblasts were exposed to 30 μM U0126 for 30 min and stimulated with 100 ng/ml HMGB1 for 20 min. Total cellular proteins were subjected to Western blotting with rabbit polyclonal antibodies against mouse pIκBα or mAb against β-actin.
NF-κB is a family of transcription factors consisting of dimers of five different proteins—p65 (RelA), RelB, c-Rel, NF-κB1 (p105/p50), and NF-κB2 (p100/p52)—and is essential for most innate and adaptive immune responses (Pomerantz and Baltimore, 2002; Bonizzi and Karin, 2004). Different types of inactive NF-κB cytoplasmic complexes are activated by a host of stress stimuli by two routes. The classical or canonical NF-κB pathway begins with the activation of NEMO/IκB kinase γ (IKK), IKKβ, and IKKα in a cytoplasmic IKK signalsome complex. IKKβ phosphorylates the NF-κB inhibitor IκBα at two amino-terminal serines, targeting it for polyubiquitination and proteasomal destruction. This leads to the nuclear translocation of NF-κB p50/p65 and p50/c-Rel heterodimers and transcription of their target genes. The alternative or noncanonical pathway is IκBα independent and depends solely on IKKα, which phosphorylates p100 to promote its proteasomal processing to mature p52, thereby causing nuclear translocation of RelB/p52 heterodimers and a subset of p50/p65 heterodimers residing in cytoplasmic p100 complexes (Basak et al., 2007).
Indirect immunofluorescence showed that 3T3 fibroblasts accumulate p65 in their nuclei in response to extracellular HMGB1, but to a lesser extent than in response to TNF-α (Fig. 2 C). IκBα phosphorylation on Ser-32 and Ser-36 peaked between 15 and 30 min after exposure to extracellular HMGB1 (Fig. 2 D). These data indicate that HMGB1 activates the canonical NF-κB pathway.
We next tested whether the activation of ERK and NF-κB in response to HMGB1 are parallel or consecutive events. In the presence of HMGB1 and U0126, NF-κB is not translocated to the nucleus of 3T3 cells (Fig. 2 E) and IκBα phosphorylation is impaired (Fig. 2 F). Thus, HMGB1 activates the canonical NF-κB pathway via ERK.
NF-κB activation is necessary for cell migration in response to HMGB1
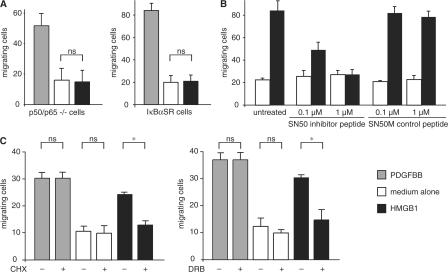
ERK participates in cytoskeleton remodeling, but NF-κB is not expected to be involved (Ridley et al., 2003). To formally rule out a role for canonical NF-κB in HMGB1-elicited cell migration, we tested immortalized MEFs, genetically deficient in both p50 and p65. To our surprise, p50/p65 knockout fibroblasts failed to migrate toward HMGB1, although they migrated as expected toward PDGF (Fig. 3 A). In further support of this result, wt MEFs stably expressing an IκBα super-repressor (IκBαSR), which cannot be phosphorylated and degraded (Brockman et al., 1995), did migrate as expected toward PDGF but not toward HMGB1 (Fig. 3 A). These experiments indicate that the classical NF-κB pathway controls HMGB1-elicited cell migration, but the possibility remains that NF-κB dimers might interact with the cytoskeleton instead of controlling transcription.
Figure 3.
NF-κB controls HMGB1-induced cell migration. Cells migrated in Boyden chambers for 3 h toward 10 ng/ml PDGFBB, 30 ng/ml HMGB1, or no chemoattractant (medium alone). (A) Immortalized p50/p65−/− MEFs and immortalized wt MEFs stably transfected with IκBαSR do not migrate toward HMGB1. Each error bar represents the mean ± SD of triplicate samples; results were replicated three times. (B) Fibroblasts were pretreated for 30 min with the indicated concentrations of SN50 or the scrambled peptide SN50M, and cell migration was assayed toward HMGB1. Error bars represent the mean of duplicate samples ± SD; results were replicated three times. The effect of treatment (SN50 vs. untreated and SN50M) and dose are significant (P < 0.05, analysis of variance). (C) Fibroblasts were pretreated for 30 min with 50 μM cycloheximide or 25 μM DRB (+) or no addition (−) and subjected to migration assays. Bars represent the mean of duplicate samples ± SD; results were replicated three times. *, P < 0.05.
We then showed that SN50, a cell-permeable peptide that competes with the nuclear transport of p50 (Lin et al., 1995), interfered in HMGB1-elicited cell migration, whereas the scrambled control peptide SN50M had no effect (Fig. 3 B). Significantly, neither SN50 nor SN50M affected PDGF-elicited cell migration (unpublished data). These results suggest that the nuclear function of NF-κB is required for cell migration toward HMGB1. Indeed, fibroblasts pretreated for 30 min with cycloheximide, an inhibitor of protein synthesis, or 5,6-dichloro-1-β-D- ribobenzimidazole (DRB), an inhibitor of transcription, failed to migrate toward HMGB1 but migrated toward 1% serum (Fig. 3 C). Similar results were obtained with endothelial and smooth muscle cells (unpublished data). These data show unequivocally that the transcriptional activity of NF-κB is necessary for cell migration toward HMGB1.
NF-κB activation is necessary for mesoangioblast migration toward HMGB1 and SDF-1/CXCL12
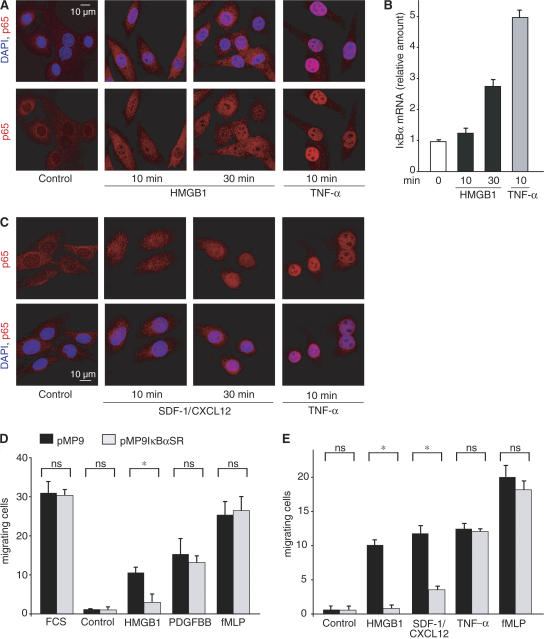
We next investigated whether NF-κB activation is also required for the HMGB1 migration response of mesoangioblasts. HMGB1 induces p65 accumulation in mesoangioblast nuclei, although to a lesser extent than TNF-α (Fig. 4 A). In addition, HMGB1 induced the transcriptional activation of the endogenous IκBα gene, a direct target of NF-κB (Fig. 4 B).
Figure 4.
NF-κB activation is required for mesoangioblast migration toward HMGB1 and CXCL12, but not other chemoattractants. (A) 100 ng/ml HMGB1 promotes nuclear translocation of p65 in mesoangioblasts. Paired images show DAPI (blue) and p65/RelA (detected via indirect immunofluorescence; red). Stimulation with 20 ng/ml TNF-α is shown as positive control. (B) 100 ng/ml HMGB1 and 20 ng/ml TNF-α induce NF-κB–dependent transcription of the endogenous IκBα gene in mesoangioblasts. Absolute quantities are normalized to β-actin. Results are shown as the mean of duplicate samples ± SD and were replicated three times. (C) 100 ng/ml SDF-1/CXCL12 promote nuclear translocation of p65 in mesoangioblasts (shown as in A). (D) Mesoangioblast migration toward HMGB1 depends on NF-κB activation. Mesoangioblasts were transfected with vectors encoding IκBαSR and GFP (pMP9IκBαSR) or GFP alone (pMP9), grown for 48 h, and induced to migrate for 6 h in Boyden chambers toward 1% serum (FCS), 100 ng/ml HMGB1, 10 ng/ml PDGFBB, 0.1 nM fMLP, or no chemoattractant (control). Only fluorescent cells were counted. Results are the mean of triplicate samples ± SD, and were replicated three times. *, P < 0.05. (E) Mesoangioblast migration toward 100 ng/ml CXCL12, but not 20 ng/ml TNF-α, depends on NF-κB activation. Results are the mean of triplicate samples ± SD; experiments were performed as described in D.
It was previously shown that mesoangioblasts respond chemotactically to several cytokines present in dystrophic muscle, including SDF-1/CXCL12 and TNF-α (Galvez et al., 2006). TNF-α is a well-known activator of the NF-κB pathway (Fig. 4, A–C); CXCL12 has been reported to activate a variety of pathways, including NF-κB, in pre-B cell lines (Ganju et al., 1998). Indeed, CXCL12 induces p65 nuclear translocation in mesoangioblasts (Fig. 4 C).
NF-κB activation and mesoangioblast migration are causally related, but only in response to a subset of chemoattractants. In fact, mesoangioblasts transiently expressing IκBαSR showed an impaired chemotactic response to HMGB1 and CXCL12, whereas their response to TNF-α was unaffected (Fig. 4, D and E).
Mesoangioblasts need NF-κB activation to ingress into damaged muscles
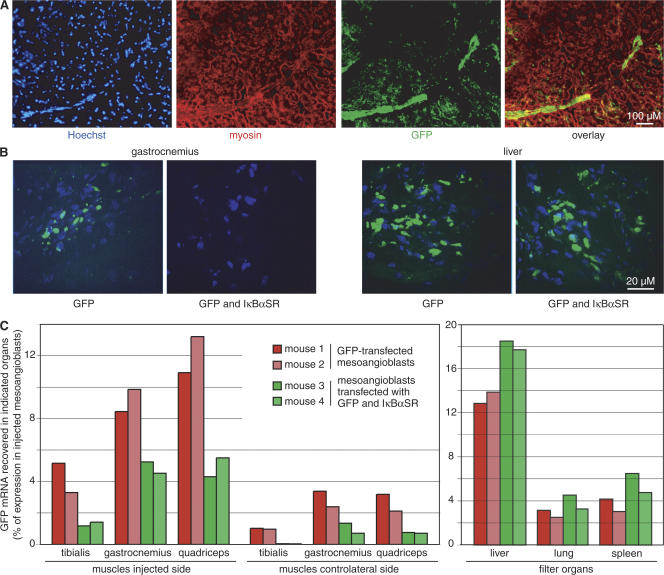
We next tested whether NF-κB activity is required for mesoangioblast ingression into the diseased muscles of α-sarcoglycan (α-SG) null dystrophic mice. Mesoangioblasts were transfected with plasmids expressing GFP and IκBαSR, or GFP alone. 48 h after transfection ∼25% of cells in each population were fluorescent. We injected 450,000 cells into the femoral artery of dystrophic mice (two per group). 6 h later we recovered filter organs (liver, spleen, and lung) and muscles (gastrocnemius, quadriceps, and tibialis) from the side of injection and the contralateral leg. Mesoangioblasts had migrated within the tissues and had not simply positioned themselves within or just outside microvessels (Fig. 5, A and B).
Figure 5.
IκBαSR impairs mesoangioblast ingression into the muscles of dystrophic mice. Mesoangioblasts were transfected with vectors encoding IκBαSR and GFP (pMP9IκBαSR) or GFP alone (pMP9), grown for 48 h, and injected intraarterially into two α-SG −/− dystrophic mice per group. After 6 h, mice were killed, and muscles and filter organs were collected. (A) The quadriceps muscle on the side injected with mesoangioblasts expressing GFP alone was excised, fixed, immunostained for myosin and GFP, and imaged. Mesoangioblasts expressing GFP (green) are seen clustering at the margin of myofiber fascicles and dispersed among the myofibers within the fascicle. (B) Representative images from sections of gastrocnemius muscles and livers from mice injected with mesoangioblasts expressing GFP alone or GFP and IκBαSR. Nuclei are stained with Hoechst (blue); GFP is stained green. (C) The number of mesoangioblasts migrating into indicated organs was inferred by RT-PCR quantification of GFP expression. Results are expressed as a percentage of GFP signal in the specific tissue; GFP expression of the entire mesoangioblast cells population before injection is set equal to 100%. This experiment was replicated three times.
To estimate the fraction of transfected mesoangioblasts arriving into the tissues, we quantified GFP mRNA in each organ and in the mesoangioblast populations before injection. In each of the muscles on the injected and contralateral sides, mesoangioblasts expressing IκBαSR and GFP were substantially fewer than those expressing GFP alone (13 ± 0.1 vs. 32.0 ± 0.2%, P < 0.001, two-tailed t test; Fig. 5 C). In contrast, filter organs contained more IκBαSR-expressing than control mesoangioblasts (20.0 ± 0.1 vs. 27.3 ± 1.9%, P < 0.05), which is consistent with the notion that mesoangioblasts not homing to muscle are mostly trapped in filter organs. This experiment was replicated three times, with similar results. Interestingly, IκBαSR reduces mesoangioblast ingression into dystrophic muscle but does not abrogate it completely, which is consistent with our in vitro results showing that not all chemotactic responses require NF-κB activation.
The role of NF-κB in cell migration and tissue repair
Collectively, our results indicate that the canonical NF-κB activation is required for migration of fibroblasts and mesoangioblasts toward specific chemoattractants associated with tissue damage. Although NF-κB is well known to direct the synthesis of cytokines and chemokines that induce the migration of immune effector cells, reports on its role in the migrating cells themselves are scarce. IκBαSR abrogates chemotaxis toward Fgf-7 in immortalized human pancreatic ductal epithelial cells and in fibroblasts forcedly expressing the Fgf-7 receptor FGF123R/IIIb (Niu et al., 2007). In a clone of the human osteogenic sarcoma cell line forcedly expressing the leukotactin-1 receptor CCR1, leukotactin-1 activates NF-κB to transcribe LZIP, and LZIP protein enhances cell migration by binding to CCR1 (Jang et al., 2007).
Our results also indicate that NF-κB activation is required for mesoangioblast ingression into damaged tissue, under conditions corresponding to the procedures being developed for cell-based therapies of muscular dystrophy (Sampaolesi et al., 2006). An important consequence of the requirement for NF-κB activation for tissue regeneration is that pharmacological regimens that suppress inflammation by interfering with NF-κB (including corticosteroids, which are commonly prescribed to dystrophic patients) might also suppress tissue regeneration and interfere with stem-cell therapies.
NF-κB activation is not required for the mechanical actions involved in cell migration because it is not needed for migration toward PDGF or formyl-met-leu-phe (fMLP). Moreover, NF-κB activation is not required for migration toward TNF-α, the archetypal canonical NF-κB activating signal. HMGB1 and CXCL12 both contribute to tissue repair (Kollet et al., 2003; Limana et al., 2005; Galvez et al., 2006) and both belong to a small group of chemoattractants that direct the navigation of stem cells (Knaut et al., 2003; Palumbo et al., 2004; Guo et al., 2005; Galvez et al., 2006). We speculate that, mechanistically, the difference between HMGB1/CXCL12 and TNF-α may be caused by differences in the timing and intensity of NF-κB activation or the concomitant activation of other signaling pathways. Physiologically, the difference may reflect specific requirements for the initiation and maintenance of migration of differentiated and stem cells in response to tissue damage.
Materials and methods
HMGB1 and reagents
Full-length, LPS-free recombinant HMGB1 protein was provided by HMGBiotech, human recombinant PDGFBB and TNF-α by R&D Systems, CXCL12 by Preprotec, and fibronectin by Roche. Anti–β-actin mAbs, fMLP, DRB, and cycloheximide were obtained from Sigma-Aldrich. U0126 and antibodies against IκBα, pIκBα, ERK, and phospho-ERK were obtained from Cell Signaling Technology. Rabbit polyclonal antibodies against p65/RelA were obtained from Calbiochem and Santa Cruz Biotechnology, Inc. SN50 and SN50M were obtained from Calbiochem.
Cell culture and transfection
3T3 mouse fibroblasts were grown in DME supplemented with 10% FCS. Mesoangioblasts (D16 clone) were grown in DME, supplemented with 20% FCS. Immortalized MEFs, wt or deficient for p50 and p65 (p50/p65 −/−; provided by A. Beg, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL), were cultured in DME supplemented with 10% FCS, 10 mM Hepes, 50 μM β-mercaptoethanol, 10 mM nonessential amino acids, and 10 mM sodium pyruvate. Primary and immortalized MEFs were obtained as described previously (Calogero et al., 1999). Constitutive expression of a transdominant IκBαSR in wt MEFs was obtained by retroviral transduction as previously described (Li et al., 2001; Li et al., 2002). A Moloney retroviral vector, which coexpresses an SV40 promoter-driven GFP gene and a long terminal repeat-driven IκBαSR Flag tagged at the amino terminus, was generated by subcloning the Flag-tagged IκBαSR ORF isolated from the pcDNA4-Flag IκBαSR vector (Brockman et al., 1995; provided by D.W. Ballard, Vanderbilt University Medical Center, Nashville, TN) into the unique BamHI site of the MP9 Moloney retroviral vector, a derivative of the PINCO vector (Grignani et al., 1998; provided by L. Lanfrancone, Istituto FIRC di Oncologia Molecolare, Milan, Italy) in which the cytomegalovirus enhancer was replaced with the SV40 promoter/enhancer. Cells were transiently transfected with each of the aforementioned vectors using FuGENE 6 reagent, according to the manufacturer's instructions (Roche).
Chemotaxis assays
Boyden chamber assays were performed as described previously (Palumbo et al., 2004). In the Dunn chamber, chemoattractants added to the outer well of the device diffuse across the bridge to the inner blind well and form a gradient within ∼30 min (Wells and Ridley, 2005). This apparatus allows for determination of the direction of migration in relation to the direction of the gradient. In our experiments, the outer well of the Dunn chamber was filled with 10 ng/ml PDGFBB and 100 ng/ml HMGB1 or serum-free medium, and the concentric inner well contained only serum-free medium. Fibroblasts were seeded on coverslips coated with 50 ng/ml fibronectin. Coverslips were inverted onto the chamber, and cell migration through the annular bridge between the concentric inner and outer wells was recorded with a microscope (Axiovert S100TV; Carl Zeiss MicroImaging, Inc.), with a still frame every 3 min for 6 h.
Western blotting and immunofluorescence for signal transduction studies
Cells were serum starved for 16 h in DME and stimulated with 100 ng/ml HMGB1, 100 ng/ml CXCL12, or 20 ng/ml TNF-α for the indicated times. Western blotting was done as described previously (Palumbo et al., 2004). Indirect immunofluorescence was done as described previously (Palumbo et al., 2004), using rabbit polyclonal anti-p65 antibody at 4°C and AlexaFluor 594–conjugated goat anti–rabbit IgG (Invitrogen). Slides were mounted in 90% glycerol, 20 mM Tris, pH 8.8, and 0.5% p-phenylenediamine. Images were taken at 37°C on a DeltaVision system consisting of a microscope (Olympus IX70), PlanApo 40×/1.35 and 60×/1.4 oil immersion objectives (Olympus), and a camera (HQ CoolSnap; Roper Scientific). Image acquisition and deconvolution (10 iterations) were done with Softworx 3.5.0 (Applied Precision).
Real-time PCR
Mesoangioblasts (100,000 per well) were stimulated with 100 ng/ml HMGB1 or 20 ng/ml TNF-α for the indicated times. cDNA was obtained with Illustra RNAspin mini RNA isolation kit (GE Healthcare) and amplified by real-time PCR on a LC480 instrument (Roche), using the relative quantification software, with LightCycler 480 SYBR Green I Master mix and primers for mouse β-actin (TGACGGGGTCACCCACACTGTGCCCATCTA and CTAGAAGCATTGCGGTGGACGATGGAGGG) and IκBα (CTTGGCTGTGATCACCAACCAG and CGAAACCAGGTCAGGATTCTGC).
Mesoangioblast ingression into dystrophic muscle
D16 mesoangioblasts were transfected with FuGene 6 (according to the manufacturer; Roche) with expression vectors encoding IκBαSR and GFP (pMP9IκBαSR) or GFP alone (pMP9), and grown for 48 h. The experiment was then conducted as previously described (Galvez et al., 2006). Cells were resuspended in PBS, and the same amount of cells (450,000 cells in 25 μl) were subjected to RT-PCR for GFP expression, or injected into the exposed femoral artery of anesthetized α-SG −/− dystrophic female mice (two per group). After 6 h, mice were killed, and the amount of GFP expression was measured by RT-PCR in filter organs (liver, lung, and spleen) and muscles (tibialis, gastrocnemius, and quadriceps) on the injected and contralateral sides. Results are expressed as the percentage of GFP signal in the specific tissue, setting GFP expression before injection as 100%. Muscles and filter organs were also processed for immunostaining as described previously (Galvez et al., 2006), using rabbit anti-GFP polyclonal antibody (Chemicon) and AlexaFluor 488–conjugated donkey anti–rabbit secondary antibody (Invitrogen) and mouse anti-myosin heavy chain mAb MF20 (Sampaolesi et al., 2003) and AlexaFluor 594–conjugated donkey anti–mouse secondary antibody (Invitrogen). Slides were mounted in fluorescent mounting medium (DakoCytomation). Images were taken at room temperature with a microscope (CTR 6000; Leica) equipped with a 40×/0.60 objective and a camera (DFC 350 FX; Leica); the acquisition software was LAS AF application suite 1.6.2 (Leica).
Digital images
Digital Images were elaborated using Photoshop 8.0 (Adobe); the luminosity of brightest and dimmest pixel in each channel were adjusted to obtain the best visual reproduction, taking care to maintain linearity in the brightness scale. Images were included in figures using Illustrator 11.0 (Adobe).
Statistical analysis
Pairwise comparisons between continuous data were done using unpaired two-tailed Student's t test; statistical analysis involving more than two groups was done using an analysis of variance model. Prism 4.0b software was used (GraphPad Software, Inc.).
Acknowledgments
We thank Dr. A. Beg for providing immortalized p50/p65 −/− MEFs, D. Covarello, J. Hering, M. Pariali, and ALEMBIC for excellent technical support, and C. Francavilla and M. Penzo for related work and discussions.
This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (M.E. Bianchi), the Association for International Cancer Research (M.E. Bianchi and R. Palumbo), Fondazione Cariplo and the Italian Ministry of Health (G. Cossu and M.E. Bianchi), the MAIN European Union Network of excellence (K.B. Marcu and M.E. Bianchi), and the National Institutes of Health (RO1-GM066882; K.B. Marcu).
The authors declare no direct financial interest. However, M.E. Bianchi is founder and part owner of HMGBiotech, a company that provides goods and services related to HMGB proteins.
K.B. Marcu and M.E. Bianchi contributed equally to this paper.
Abbreviations used in this paper: α-SG, α-sarcoglycan; DRB, 5,6-dichloro-1-β-D- ribobenzimidazole; ERK, extracellular signal-regulated kinase; fMLP, formyl-met-leu-phe; HMGB1, high mobility group box 1; IKK, IκB kinase; IκBαSR, IκBα super-repressor; MEF, mouse embryonic fibroblast; MEK, MAPK/ERK kinase; NF-κB, nuclear factor κB; RAGE, receptor for advanced glycation end products; SDF, stromal derived factor; wt, wild type.
References
- Basak, S., H. Kim, J.D. Kearns, V. Tergaonkar, E. O'Dea, S.L. Werner, C.A. Benedict, C.F. Ware, G. Ghosh, I.M. Verma, and A. Hoffmann. 2007. A fourth IκB protein within the NF-κB signaling module. Cell. 128:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, M.E. 2007. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 81:1–5. [DOI] [PubMed] [Google Scholar]
- Bierhaus, A., P.M. Humpert, M. Morcos, T. Wendt, T. Chavakis, B. Arnold, D.M. Stern, and P.P. Nawroth. 2005. Understanding RAGE, the receptor for advanced glycation end products. J. Mol. Med. 83:876–886. [DOI] [PubMed] [Google Scholar]
- Bonizzi, G., and M. Karin. 2004. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25:280–288. [DOI] [PubMed] [Google Scholar]
- Brockman, J.A., D.C. Scherer, T.A. McKinsey, S.M. Hall, X. Qi, W.Y. Lee, and D.W. Ballard. 1995. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol. Cell. Biol. 15:2809–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero, S., F. Grassi, A. Aguzzi, T. Voigtländer, P. Ferrier, S. Ferrari, and M.E. Bianchi. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth, but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276–280. [DOI] [PubMed] [Google Scholar]
- Degryse, B., T. Bonaldi, P. Scaffidi, S. Muller, M. Resnati, F. Sanvito, G. Arrigoni, and M.E. Bianchi. 2001. The high mobility group (HMG) boxes of the nuclear protein HMG1 induce chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. J. Cell Biol. 152:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez, B.G., M. Sampaolesi, S. Brunelli, D. Covarello, M. Gavina, B. Rossi, G. Costantin, Y. Torrente, and G. Cossu. 2006. Complete repair of dystrophic skeletal muscle by mesoangioblasts with enhanced migration ability. J. Cell Biol. 174:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganju, R.K., S.A. Brubaker, J. Meyer, P. Dutt, Y. Yang, S. Qin, W. Newman, and J.E. Groopman. 1998. The α-chemokine, stromal cell-derived factor-1α, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J. Biol. Chem. 273:23169–23175. [DOI] [PubMed] [Google Scholar]
- Grignani, F., T. Kinsella, A. Mencarelli, M. Valtieri, D. Riganelli, F. Grignani, L. Lanfrancone, C. Peschle, G.P. Nolan, and P.G. Pelicci. 1998. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 58:14–19. [PubMed] [Google Scholar]
- Guo, Y., G. Hangoc, H. Bian, L.M. Pelus, and H.E. Broxmeyer. 2005. SDF-1/CXCL12 enhances survival and chemotaxis of murine embryonic stem cells and production of primitive and definitive hematopoietic progenitor cells. Stem Cells. 23:1324–1332. [DOI] [PubMed] [Google Scholar]
- Hori, O., S.D. Yan, S. Ogawa, K. Kuwabara, M. Matsumoto, D. Stern, and A.M. Schmidt. 1995. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. J. Biol. Chem. 270:25752–25761. [DOI] [PubMed] [Google Scholar]
- Jang, S.W., Y.S. Kim, Y.R. Kim, H.J. Sung, and J. Ko. 2007. Regulation of human LZIP expression by NF-κB and its involvement in monocyte cell migration induced by Lkn-1. J. Biol. Chem. 282:11092–11100. [DOI] [PubMed] [Google Scholar]
- Knaut, H., C. Werz, R. Geisler, and C. Nusslein-Volhard. 2003. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 421:279–282. [DOI] [PubMed] [Google Scholar]
- Kollet, O., S. Shivtiel, Y.Q. Chen, J. Suriawinata, S.N. Thung, M.D. Dabeva, J. Kahn, A. Spiegel, A. Dar, S. Samira, et al. 2003. HGF, SDF-1, and MMP-9 are involved in stress-induced human CD34+ stem cell recruitment to the liver. J. Clin. Invest. 112:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., G.W. Peet, D. Balzarano, X. Li, P. Massa, R.W. Barton, and K.B. Marcu. 2001. Novel NEMO/IκB kinase and NF-κB target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276:18579–18590. [DOI] [PubMed] [Google Scholar]
- Li, X., P.E. Massa, A. Hanidu, G.W. Peet, P. Aro, A. Savitt, S. Mische, J. Li, and K.B. Marcu. 2002. IKKα, IKKβ, and NEMO/IKKγ are each required for the NF-κB-mediated inflammatory response program. J. Biol. Chem. 277:45129–45140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limana, F., A. Germani, A. Zacheo, J. Kajstura, A. Di Carlo, G. Borsellino, O. Leoni, R. Palumbo, L. Battistini, R. Rastaldo, et al. 2005. Exogenous high-mobility group box 1 protein induces myocardial regeneration after infarction via enhanced cardiac C-kit+ cell proliferation and differentiation. Circ. Res. 97:e73–e83. [DOI] [PubMed] [Google Scholar]
- Lin, Y.Z., S.Y. Yao, R.A. Veach, T.R. Torgerson, and J. Hawiger. 1995. Inhibition of nuclear translocation of transcription factor NF-κB by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J. Biol. Chem. 270:14255–14258. [DOI] [PubMed] [Google Scholar]
- Minasi, M.G., M. Riminucci, L. De Angelis, U. Borello, B. Berarducci, A. Innocenzi, A. Caprioli, D. Sirabella, M. Baiocchi, R. De Maria, et al. 2002. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development. 129:2773–2783. [DOI] [PubMed] [Google Scholar]
- Niu, J., Z. Chang, B. Peng, Q. Xia, W. Lu, P. Huang, M.S. Tsao, and P.J. Chiao. 2007. Keratinocyte growth factor/fibroblast growth factor-7-regulated cell migration and invasion through activation of NF-κB transcription factors. J. Biol. Chem. 282:6001–6011. [DOI] [PubMed] [Google Scholar]
- Palumbo, R., M. Sampaolesi, F. De Marchis, R. Tonlorenzi, S. Colombetti, A. Mondino, G. Cossu, and M.E. Bianchi. 2004. Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J. Cell Biol. 164:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.S., D. Svetkauskaite, Q. He, J.Y. Kim, D. Strassheim, A. Ishizaka, and E. Abraham. 2004. Involvement of TLR2 and TLR4 in cellular activation by high mobility group box 1 protein (HMGB1). J. Biol. Chem. 279:7370–7377. [DOI] [PubMed] [Google Scholar]
- Pomerantz, J.L., and D. Baltimore. 2002. Two pathways to NF-κB. Mol. Cell. 10:693–695. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., M.A. Schwartz, K. Burridge, R.A. Firtel, M.H. Ginsberg, G. Borisy, J.T. Parsons, and A.R. Horwitz. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. [DOI] [PubMed] [Google Scholar]
- Sampaolesi, M., Y. Torrente, A. Innocenzi, R. Tonlorenzi, G. D'Antona, M.A. Pellegrino, R. Barresi, N. Bresolin, M.G. Cusella De Angelis, K.P. Campbell, et al. 2003. Cell therapy of α-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 301:487–492. [DOI] [PubMed] [Google Scholar]
- Sampaolesi, M., S. Blot, G. D'Antona, N. Granger, R. Tonlorenzi, A. Innocenzi, P. Mognol, J.L. Thibaud, B.G. Galvez, I. Barthelemy, et al. 2006. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 444:574–579. [DOI] [PubMed] [Google Scholar]
- Scaffidi, P., T. Misteli, and M.E. Bianchi. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 418:191–195. [DOI] [PubMed] [Google Scholar]
- Wells, C.M., and A.J. Ridley. 2005. Analysis of cell migration using the Dunn chemotaxis chamber and time-lapse microscopy. Methods Mol. Biol. 294:31–41. [DOI] [PubMed] [Google Scholar]