Abstract
In this study, we describe the identification and characterization of a novel transcription factor GLI-similar 3 (GLIS3). GLIS3 is an 83.8 kDa nuclear protein containing five C2H2-type Krüppel-like zinc finger motifs that exhibit 93% identity with those of GLIS1, however, little homology exists outside their zinc finger domains. GLIS3 can function as a repressor and activator of transcription. Deletion mutant analysis determined that the N- and C-termini are required for optimal transcriptional activity. GLIS3 binds to the GLI-RE consensus sequence and is able to enhance GLI-RE-dependent transcription. GLIS3(ΔC496), a dominant-negative mutant, inhibits transcriptional activation by GLIS3 and GLI1. Whole mount in situ hybridization on mouse embryos from stage E6.5 through E14.5 demonstrated that GLIS3 is expressed in specific regions in developing kidney and testis and in a highly dynamic pattern during neurulation. From E11.5 through E12.5 GLIS3 was strongly expressed in the interdigital regions, which are fated to undergo apoptosis. The temporal and spatial pattern of GLIS3 expression observed during embryonic development suggests that it may play a critical role in the regulation of a variety of cellular processes during development. Both the repressor and activation functions of GLIS3 may be involved in this control.
INTRODUCTION
GLI and ZIC constitute two closely related subfamilies of Krüppel-like zinc finger proteins (1–8). Cubitus interruptus (Ci) and odd-paired (Opa) are the Drosophila homologs of GLI and ZIC, respectively, while odd-paired-like (Opl) is the Xenopus homolog of ZIC (3,9–13). GLI and ZIC proteins share a highly homologous zinc finger domain (ZFD) consisting of five Cys2-His2-type zinc finger motifs. These proteins are multifunctional transcription factors that can activate as well as repress transcription. Ci and GLI proteins act downstream of sonic hedgehog protein (Shh)–Patched–Smoothened (2,14). Activation of GLI/Ci involves proteolytic cleavage and translocation to the nucleus, a process that is regulated by interactions with Fused and Suppressor of Fused (15–17). There is growing evidence indicating links between GLI proteins and other signaling pathways, including Wnt and bone morphogenic protein (BMP). Wnt and BMP signaling has been implicated both in GLI regulation (18,19) and as downstream targets of GLI proteins (20,21).
GLI and ZIC proteins are important in mammalian development while the Drosophila homologs are critical in invertebrate development (1,5,22–26). GLI and ZIC proteins regulate various aspects of neural and skeletal development. GLI2 has been reported to be required for normal mammary gland development while GLI2 and GLI3 are essential for the formation of lung, trachea and esophagus (27,28). Studies of mice deficient in ZIC expression indicated a role for ZIC1 and ZIC2 in cerebellar development (23). ZIC and GLI genes have been implicated in several human diseases, including birth defects and cancer (5,14). Mutations in GLI3 are implicated in Greig cephalopolysyndactyly and Pallister–Hall syndromes (5,29,30), while deficiencies in ZIC2 and ZIC3 result in malformations of the forebrain (holoproencephaly) and in disturbance of the left to right body axis (heterotaxy), respectively (31,32). Several studies have indicated a role for GLI and ZIC in cancer (5,14). GLI1 is overexpressed in most basal carcinomas of the skin and exogenous expression of GLI1 has been reported to induce the formation of basal cell carcinoma (33–36). ZIC1 was found to be overexpressed in many medulloblastomas (37). Recent studies have shown that the Shh/GLI signaling pathway is highly activated in small cell lung carcinomas (38).
In this study, we describe the identification and characterization of a novel protein, referred to as GLI-similar 3 (GLIS3), containing five C2H2-type zinc finger motifs. Its ZFD has high homology to those of the GLI and ZIC proteins but shows highest identity (93%) to that of the recently described GLIS1 (6). The ZFDs of GLIS1 and GLIS3 exhibit high homology with that of the Drosophila protein gleeful (gfl), also named lame duck (lmd) (11,12), suggesting that it might be the Drosophila homolog. We demonstrate that GLIS3 can function both as an activator and repressor of transcription and provide evidence for cross-talk between the GLIS and GLI signaling pathways. Whole mount in situ hybridization demonstrated that during embryonic development GLIS3 is expressed in a temporal and spatial pattern, suggesting that it may play a critical role in the regulation of a variety of cellular processes during development. Both the repressor and activation functions of GLIS3 may be important in the regulation of these physiological processes.
MATERIALS AND METHODS
Cloning of full-length mGLIS3 cDNA
A search in GenBank for additional members of the GLIS subfamily identified a partial sequence (GenBank accession no. XM_140589) containing two zinc finger motifs that exhibit a high homology to the first two zinc fingers of GLIS1. To obtain the full-length sequence of mGLIS3, a mouse kidney cDNA library was screened by PCR using mGLIS3-specific primers. This screening was performed by OriGene (Rockville, MD) and yielded one clone containing a 5 kb insert encoding the majority of the GLIS3 coding region. To obtain the remaining 5′-end, 5′-SMART RACE was performed following the manufacturer’s protocol (Clontech, Palo Alto, CA). Subsequent screening of the GenBank database identified an unpublished sequence (IMAGE clone 4828458) encoding the human homolog of mGLIS3.
DNA sequencing
Plasmids were purified using miniprep or midiprep kits from Qiagen (Valencia, CA). Automatic sequencing was carried out using a Dynamic ET Terminator Cycle Sequencing Ready reaction kit (Perkin-Elmer) and an ABI Prism 377 automatic sequencer. DNA and deduced protein sequences were analyzed by the seqWEB sequence analysis software packages.
Northern blot analysis
Multi-tissue blots containing total RNA (25 µg) from 14 different mouse tissues or poly(A)+ RNA from 12 human tissues were purchased from Seegene (Seoul, Korea) and Ambion (Austin, TX), respectively. The blots were hybridized to a 32P-labeled probe for GLIS3. Hybridizations were performed at 68°C for 1 h, the membranes were then washed twice with 2× SSC, 0.1% SDS at room temperature for 30 min and 0.1% SSC, 0.1% SDS at 58°C for 30 min. Auto radiography was carried out with Hyperfilm-MP (Amersham) at –70°C.
Whole mount in situ hybridization
Whole mount in situ hybridization studies were performed according to Haramis and Carrasco (39). Mouse embryos from 7.75 to 14.5 d.p.c. (E7.75–E14.5) were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS), washed twice for 10 min in PBS + 0.1% Tween 20 (PBST) and then dehydrated with a series of 10 min methanol/PBST washes (25, 50, 75 and 2 × 100%). Embryos were stored at –20°C until probed. mGLIS3 probes, encoding the region from nucleotide 233 to 1147 and from nucleotide 1803 to 2375, were generated by PCR using primers containing either an EcoRI or a HindIII site. The PCR products were then cloned into pGEM3Zf(+). For labeling, plasmid DNA was linearized either with HindIII for T7-generated sense transcripts or with EcoRI for Sp6-generated antisense transcripts, and riboprobes were produced using digoxigenin-substituted UTP (Lofstrand Labs Ltd).
Plasmids
For mammalian mono-hybrid analysis the following plasmids were used. The reporter plasmid pFR-LUC, containing five copies of the Gal4 upstream activating sequence (UAS) and referred to as (UAS)5-LUC, was obtained from Stratagene. The vectors pM and VP16, and the β-galactosidase reporter plasmid pCMVβ were purchased from Clontech. pM-GLIS3 deletion mutants were created by placing different GLIS3 cDNA fragments at the 5′-end of Gal4(DBD). These fragments were generated by PCR using GLIS3-specific 5′- and 3′-primers that included either an EcoRI or BamHI restriction site, respectively, to allow the PCR fragments to be subcloned into the EcoRI or BamHI sites of the pM vector. Details on the length of each deletion are described in the text and figures. The reporter plasmid (GLI-RE)12-LUC was kindly provided by Dr R. Toftgard (Karolinska Institute, Huddinge, Sweden) (40).
To determine the subcellular localization of GLIS3 the following constructs were generated. Full-length GLIS3 was cloned into EcoRI and BamHI sites of the pEGFP-N1 vector (Clontech) creating pEGFP-GLIS3. pEGFP-GLIS3(ΔN307) and pEGFP-GLIS3(ΔN551), encoding GLIS3 from, respectively, His307 and Leu551 to the C-terminus, and pEGFP-GLIS3(ΔC528) and pEGFP-GLIS3(ΔC458), encoding GLIS3 from the N-terminus to Pro528 and Arg458, respectively, were generated by PCR using GLIS3-specific primers containing either a 5′ EcoRI or 3′ BamHI site. To generate pQE32-GLIS3(ZFD) encoding (His)6-GLIS3(ZFD) (from His307 to Ser525), the GLIS3(ZFD) region was amplified by PCR and inserted into the BamHI or SalI sites of pQE32 (Qiagen). To generate pQE32-GLI1(ZFD) encoding (His)6-GLI1(ZFD) (from Lys215 to Ser410), the GLI1(ZFD) region was amplified by PCR and inserted into the BamHI or HindIII sites of pQE32. All constructs were verified by restriction analysis and DNA sequencing.
Electrophoretic mobility shift assay (EMSA)
Escherichia coli BL21(DE3) transformed with pQE32-GLIS3(ZFD) or pQE32-GLI1(ZFD) were grown at 37°C to mid-log phase and then treated with isopropyl-β-d-thiogalactopyranoside (0.5 mM final concentration) for 3 h. GST-GLIS3(ZFD) was purified over glutathione–Sepharose 4B beads. (His)6-GLIS3(ZFD) and (His)6-GLI1(ZFD) were purified using Ni–NTA+ resin (Qiagen). Double-stranded oligonucleotides containing the consensus GLI-binding site TCTAAGAGCTCCCGAAGACCACCCACAATGATGGTTGTA were end-labeled with [32P]ATP by T4 polynucleotide kinase (Promega). (His)6-GLIS3(ZFD) or (His)6-GLI1(ZFD) recombinant protein (0.5 µg) was incubated in binding buffer (25 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 10 µM ZnSO4, 1 mM DTT, 0.1% NP40, 12% glycerol) with 32P-end-labeled, double-stranded oligonucleotides for 1 h at room temperature. The protein–DNA complexes were then separated on a 6% native polyacrylamide gel and visualized by autoradiography as described (41).
Cell culture
Green monkey kidney fibroblast CV-1 and COS-7, Chinese hamster ovary CHO, NIH 3T3 and human embryonic kidney 293 cells were obtained from ATCC. Cells were routinely maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) except for CHO, which were cultured in Ham’s F12 plus 10% FBS.
Cellular localization
pEGFP-GLIS3, pEGFP-GLIS3(ΔN307), pEGFP-GLIS3 (ΔN551), pEGFP-GLIS3(ΔC528), pEGFP-GLIS3(ΔC458) or pEGFP-N1 plasmid DNA was transfected into CV-1 or CHO cells using Fugene 6. After 30 h, enhanced green fluorescent protein (EGFP) expression was examined in a Zeiss confocal microscope LSM 510 NLO (Zeiss, Thornwood, NY) at excitation and emission frequencies of 488 and 505 nm, respectively.
The localization of 3×Flag-GLIS3 was examined after transfected cells were fixed in 4% paraformaldehyde/PBS for 15 min and then treated with 0.3% Triton X-100/PBS for 15 min. Cells were subsequently incubated with anti-Flag M2 monoclonal antibody (Sigma) for 1 h. After washing, cells were incubated with Alexa Fluor 594-conjugated anti-mouse antibody (Molecular Probes, OR) for 1 h. After several washings, Alexa Fluor 594 fluorescence was examined at excitation and emission frequencies of 590 and 617 nm, respectively.
Reporter gene assays
Cells were plated in 6-well dishes at 2 × 105 cells/well and 20 h later co-transfected (1–2 µg total DNA in 1 ml) with 0.25 µg (UAS)5-LUC, 1.0–0 µg pBSK, 0–1.0 µg of the indicated pM-GLIS3 plasmid and 0.25 µg pCMVβ, which served as an internal control to monitor transfection efficiency. Cells were transfected in Opti-MEM (Life Technologies) and 3–6 µl Fugene 6 transfection reagent (Roche). Cells were incubated for 30 h and then assayed for β-galactosidase and luciferase activity. Luciferase activity was assayed with a luciferase kit (Promega). The level of β-galactosidase activity was determined using a luminescent β-galactosidase detection kit (Clontech) according to the manufacturer’s instructions. Transfections were performed in triplicate and each experiment was repeated at least twice. The level of expression of GAL4(DBD)–GLIS3 fusion proteins was examined by western blot analysis using an anti-GAL4(DBD) antibody (Clontech).
Pulldown assays
p3×FlagCMV-GLIS1, p3×FlagCMV-GLIS2 or p3× FlagCMV-GLI1 was co-transfected with pCMV-myc-GLIS3 in CHO cells. After 48 h, cells were harvested and lysed in RIPA lysis buffer (PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate and 0.1% SDS). Subsequently, half of the cell lysates were then incubated with anti-Flag agarose resin (Sigma) for 1 h at 4°C with agitation. The resin was then washed five times with PBS. The pulled-down protein complexes were then examined by western blot analysis using an anti-myc antibody (Invitrogen). Proteins in the other half of the cell lysates were mixed with sample buffer and examined by western blot analysis using either an anti-c-myc or anti-Flag M2 antibody (Sigma).
RESULTS
Identification of GLIS3
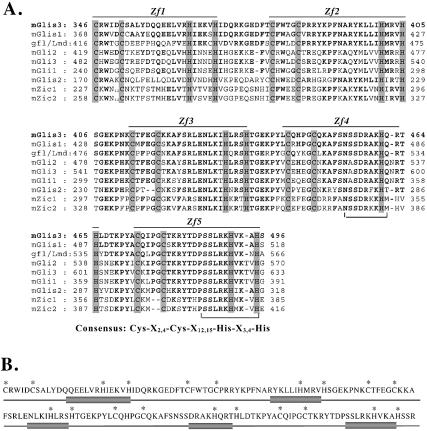
A search in GenBank for sequences encoding additional members of the GLIS subfamily identified a mouse sequence (XM_140589) encoding a partial protein sequence containing two zinc finger motifs that exhibited high homology to the first two zinc fingers of mGLIS1 (6). The upstream N-terminal sequence did not show any similarity with GLIS1. The full-length nucleotide sequence encoding this protein, referred to as GLIS3, was obtained by PCR screening of a mouse kidney cDNA library and 5′-RACE. The nucleotide and deduced amino acid sequences of mGLIS3 are shown in Figure 1. The putative translation initiation sequence CCCATATGG differs by three nucleotides from the Kozak consensus sequence CCA/GCCATGG (42). The open reading frame encodes a protein of 779 amino acids with a calculated mass of 83.8 kDa and a pI of 7.87. Human GLIS3 is five amino acids shorter than mGLIS3 and exhibits 86.2% sequence homology with mGLIS3 (Fig. 1). Analysis of the amino acid sequence by ScanProsite and PFSCAN identified, next to the ZFD, two proline-rich regions between Pro289 and Pro305 and Pro578 and Pro605. The ZFD extends from Cys346 to His495 and is composed of five tandem C2H2-type zinc finger motifs with the consensus Cys-X4-Cys-X12,15-His-X3,4-His. The interfinger sequences resemble the consensus sequence T/SGEKPY/FX typically found in Krüppel-like zinc finger proteins (43). Analysis of its secondary structure using the GOR3 secondary structure prediction method indicated that the second half of each zinc finger motif of GLIS3 consists of an α-helix (Fig. 2B).
Figure 1.
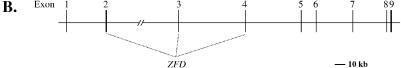
(A) The nucleotide and amino acid sequences of mouse GLIS3. The nucleotide and deduced amino acid sequences of mouse GLIS3 are shown in the first and second lines, respectively. The third line shows the amino acid sequence of human GLIS3. . indicates a gap; – indicates an amino acid conserved with mGLIS3. The start and stop codons are indicated in bold. The two proline-rich regions are underlined. The ZFD is shaded. The Cys and His residues involved in the tetrahedral configuration in the zinc finger motifs are underlined and in bold. The putative bipartite NLS is indicated by a dotted line. The GLIS3 sequence was submitted to GenBank under accession no. AY220846. (B) Schematic presentation of the genomic structure of GLIS3. The locations of the intron–exon junctions are indicated by arrowheads in (A). Bar indicates 10 kb.
Figure 2.
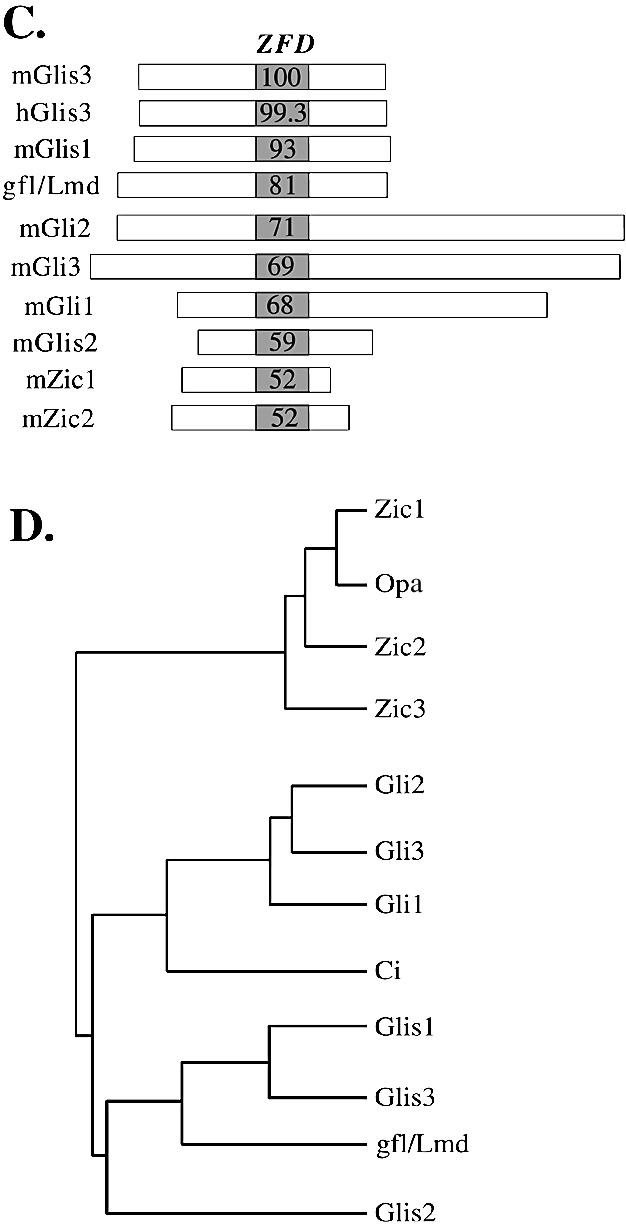
(A) Amino acid sequence alignment of the ZFD of mouse GLIS3 with those of GLIS1, gfl/lmd, GLI2, GLI3, GLI1, GLIS2, ZIC1 and ZIC2. Zf1–Zf5 indicate the five zinc finger motifs. Bold residues indicate amino acids conserved with mGLIS3. The Cys and His residues conserved in the zinc finger motifs are shaded. The consensus sequence of the zinc finger motifs is shown at the bottom. The amino acids between brackets indicate regions in the fourth and fifth zinc fingers involved in making DNA contacts. (B) Localization of α-helices in the ZFD of mGLIS3 as determined by GOR3 secondary structure prediction analysis. Bars indicate α-helix. The asterisks indicate the Cys and His residues involved in the tetrahedral configuration in the zinc finger motifs. (C) Schematic comparison of murine GLIS3, GLIS1, GLI1-3, GLIS2, ZIC1 and ZIC2 and Drosophila gfl/lmd. The percent identity of the ZFDs with mGLIS3 ZFD is indicated. Little homology was observed in the regions outside the ZFD. (D) Evolutionary relationship between the ZFDs of GLIS, GLI and ZIC proteins and their Drosophila homologs gfl/lmd, Ci and Opa. The phylogenetic tree was derived using the neighbor joining tree construction and Jukes–Cantor distance correction methods.
Exploring GenBank for genomic sequences encoding GLIS3 identified two supercontigs (NW_000143.1 and NT_ 008413.13) that encompass the whole coding region of mouse and human GLIS3, respectively. From the chromosomal locations of these supercontigs it could be concluded that the human and mouse GLIS3 genes map to chromosomes 9p23–24.3 and 19C1, respectively. Based on this comparison the genomic structure of the mouse and human GLIS3 gene could be determined. As shown in Figure 1B, the GLIS3 gene spans >300 kb and consists of nine exons and eight introns. The sites of the intron–exon junctions are indicated in Figure 1A. The ZFD is encoded by exons 2–4. Interestingly, the locations of the exon–intron junctions within the region encoding the ZFD of GLIS3 are conserved with those of GLIS1 and distinct from the locations in GLIS2, GLI and ZIC.
The ZFD of GLIS3 shows high homology to those of other GLIS proteins and members of the GLI and ZIC subfamily of Krüppel-like zinc finger proteins (1,3,6,7,44–46) (Fig. 2A). The ZFD of GLIS3 exhibits 93% identity with that of GLIS1 and 81% identity with the ZFD of the Drosophila Krüppel-like zinc finger protein gfl/lmd (11,12) (Fig. 2C). GLIS3 exhibited 68–71% identity with GLI1–GLI3, 59% with GLIS2 and 52% with ZIC. Outside the ZFD, GLIS3 showed little homology with GLIS1 or any of the other Krüppel-like zinc finger proteins.
The high sequence homology observed within their ZFDs (Fig. 2A and C) and the conserved intron–exon junctions (Fig. 1) (6) indicate that GLIS1 and GLIS3 are closely related and suggest that the ZFD of GLIS1 and GLIS3 are derived from a common ancestral gene. This evolutionary relationship between GLIS1 and GLIS3 was supported further by phylogenetic analysis (Fig. 2D). The phylogenetic tree suggests that GLIS1 and GLIS3 belong to the same subfamily and that gfl/lmd is the apparent Drosophila homolog of GLIS1 and GLIS3. Although GLIS2 is closely related, it appears not to belong to the GLIS1 and GLIS3 subfamily.
Tissue-specific expression
To examine the pattern of tissue-specific expression of GLIS3, northern blot analysis was performed using RNA from placenta and several adult human and mouse tissues. The radiolabeled GLIS3 probe hybridized to a single 8 or 9 kb transcript in mouse and human, respectively (Fig. 3). GLIS3 mRNA was most abundant in kidney and uterus and expressed at moderate levels in brain, thymus, lung, skeletal muscle, pancreas, liver and ovary. The pattern of GLIS3 expression between mouse and human was very similar, although a few differences (e.g. liver) were noted.
Figure 3.
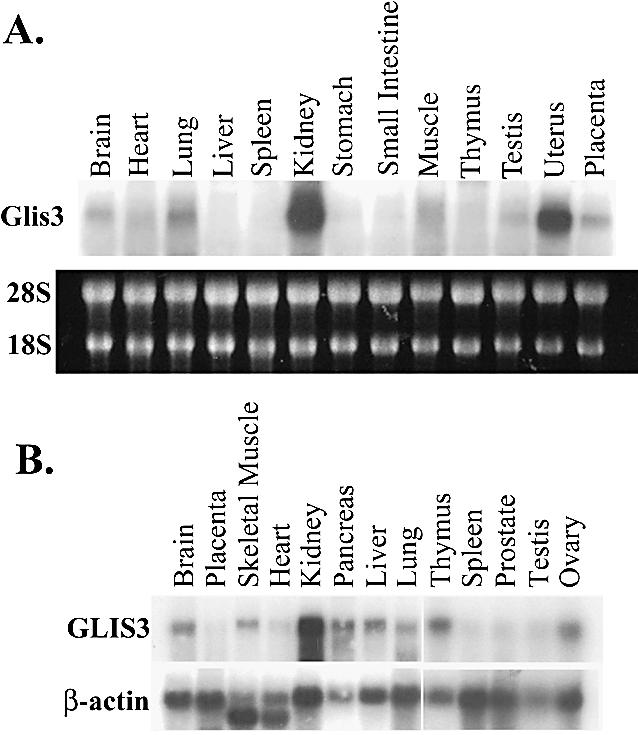
Tissue-specific expression of GLIS3. Total RNA isolated from placenta and various adult tissues was examined by northern blot analysis. (A) Mouse; (B) human. Blots were hybridized to a 32P-labeled probe for GLIS3 or β-actin as described in Materials and Methods. In (A) the pattern of 18–28S rRNA is shown to demonstrate equal loading of RNA samples.
Whole mount in situ hybridization
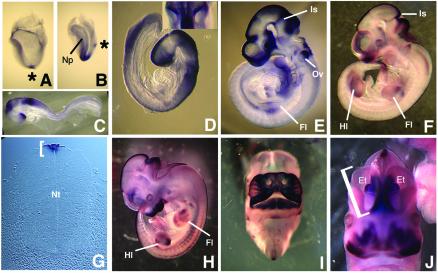
The temporal and spatial patterns of GLIS3 expression during development was determined by whole mount in situ hybridization on mouse embryos from stage E6.5 through E14.5. The earliest stage at which expression could be detected was at the early headfold stage (∼E8.0) when GLIS3 transcripts were detected specifically in the node, a structure located at the anterior end of the primitive streak and which will give rise to the embryonic notochord (Fig. 4A). At late headfold stages GLIS3 expression was maintained in the node and was initiated in the neural plate in the presumptive hindbrain (Fig. 4B). At E8.5, prior to embryonic turning, GLIS3 expression was restricted to the neural epithelium with prominent expression in three domains: the hindbrain region, the presumptive mid/hindbrain (the cephalic flexure) and the anterior forebrain (Fig. 4C). At E8.75, after embryonic turning, diffuse GLIS3 expression remained prominent in these three regions of the neural tube and began to be up-regulated in the dorsal neural tube (Fig. 4D). Also at this stage, GLIS3 transcripts were evident in the anterior/dorsal region of the otic vesicles (insert in Fig. 4D). By E9.5 expression in the mid/hindbrain junction (isthmus) and dorsal neural tube sharpened and continued through the basal plate of the forebrain (Fig. 4E). At E10.5 neural expression was mostly restricted to the roof plate of the neural tube (Fig. 4F and G). At this stage GLIS3 transcripts were also evident for the first time around the nasal placodes. This facial expression of GLIS3 was greatly increased at E11.5 and 12.5 (Fig. 4H and I) and dissection of stained tissue revealed that expression was mesenchymal. At E12.5, GLIS3 was strongly expressed along the medial walls of the two telencephalic vesicles as well as in the forebrain region known as the eminentia thalami, which connects the dorsal telencephalon and diencephalon (Fig. 5J).
Figure 4.
Whole mount preparations demonstrating GLIS3 RNA localization during mouse development. (A) Expression in the node [asterisk in (A) and (B)] of ∼E8.0 embryo. (B) Expression in node and neural plate of ∼E8.25 embryo. (C) Lateral view of E8.5 embryo. (D) Lateral view of E8.75 embryo. (Inset) Dorsal view of embryo showing otic vesicles. (E) Lateral view of E9.5 embryo. Note expression in forelimb. (F) Lateral view of E10.5 embryo. (G) Transverse section through E11.5 neural tube illustrating GLIS3 expression in roof plate (bracket). (H) Lateral view of E11.5 embryo. (I) Frontal view of E12.5 day embryo illustrating facial GLIS3 expression. (J) Frontal view of E12.5 embryo illustrating GLIS3 expression in telencephalic vesicles (one of which is bracketed) and eminentia thalami. Abbreviations: Et, eminentia thalami; Hl, hindlimb; Fl, forelimb; Is, isthmus; Np, neural plate; Nt, neural tube; Ov, otic vesicle.
Figure 5.
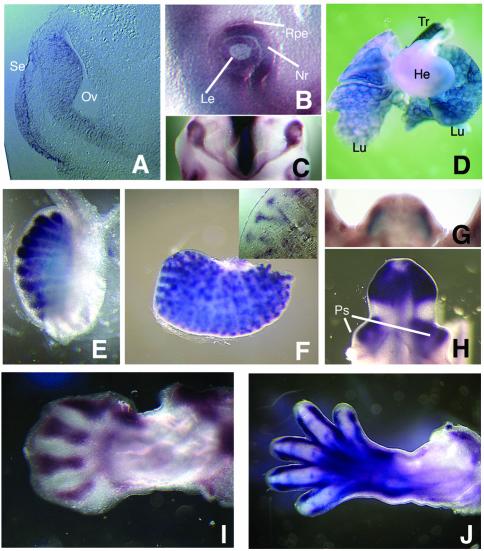
GLIS3 expression in specific embryonic organs and outgrowths. (A) Frontal section through E9.5 head demonstrating GLIS3 expression in optic vesicle and surface ectoderm. (B) Lateral view of E0.5 eye. (C) View of optic structures from the bottom of a E10.5 head demonstrating GLIS3 expression in the optic cups and nasal portion of the optic stalk. Anterior is down. (D) Lungs, heart and trachea of E4.5 embryo. (E) Testis of E14.5 embryo demonstrating GLIS3 expression in the seminiferous tubules. (F) Metanephros of E14.5 embryo. (Inset) Section of E14.5 metanephros demonstrating GLIS3 expression in the branches of the ureteric bud. (G) Genital tubercle of an E12.5 embryo viewed from the anterior perspective. (H) Genital tubercle of an E14.5 embryo viewed from the anterior perspective. (I) E12.5 forelimb, dorsal view with anterior aspect up. (J) E14.5 forelimb, dorsal view with anterior aspect up. Abbreviations: He, heart; Le, lens; Lu, lung; Nr, neural retina; Oc, optic cup; Os, optic stalk; Ov, optic vesicle; Ps, preputial swelling; Rpe, retinal pigmented epithelium; Se, surface ectoderm; Tr, trachea.
GLIS3 was expressed in a dynamic pattern during eye development. By E9.5, lateral protrusions of the ventral telencephalon develop into the optic vesicles and the distal portion of these structures contacts the head surface ectoderm. At this stage, GLIS3 expression was detected in the ventral optic vesicle and surface ectoderm (Fig. 5A). By E10.5, the surface ectoderm has differentiated into lens, the distal optic vesicle has invaginated to form the neural retina and retinal pigmented epithelium and the proximal optic vesicle has formed the optic stalk. GLIS3 expression was evident in the lens and neural retina as well as the nasal portion of the optic stalk (Fig. 5B and C). By E11.5, GLIS3 expression was not evident in the optic stalk although expression remained in both neural retina and lens, and by E12.5, expression was not evident in any optic structures (data not shown).
To assay expression during organogenesis, we performed whole mount in situ hybridizations on dissected viscera from E14.5 embryos. Expression was evident in lungs and trachea (Fig. 5D), the seminiferous tubules of the testes (Fig. 5E) and branches of the ureteric bud in the metanephros (Fig. 5F). Starting at E12.5, GLIS3 expression was also evident in lateral mesenchymal stripes in the genital tubercle (Fig. 5G). At E14.5, GLIS3 expression was also detected in the preputial swellings (Fig. 5H), which are outgrowths lateral to the genital tubercle that will form the prepuce of the external genitalia. During limb development, GLIS3 expression was restricted to specific mesenchymal patterns. The pattern was identical between the fore- and hindlimb, if we took into account the fact that the forelimb is about half a day more advanced developmentally than the hindlimb. At E9.5, GLIS3 expression was detected in the proximal central limb bud mesenchyme (Fig. 4E) and by E10.5, became restricted to the anterior proximal mesenchyme (Fig. 4F). Starting at E11.5 (Fig. 4H) through E12.5 (Fig. 5I), GLIS3 was strongly expressed in the interdigital region, which is fated to undergo cell death as digits form. At E14.5, GLIS3 expression was detected along the digits, in the interphalangeal joints of each digit and in muscles of the hand plate and proximal digit (Fig. 5J).
Subcellular localization
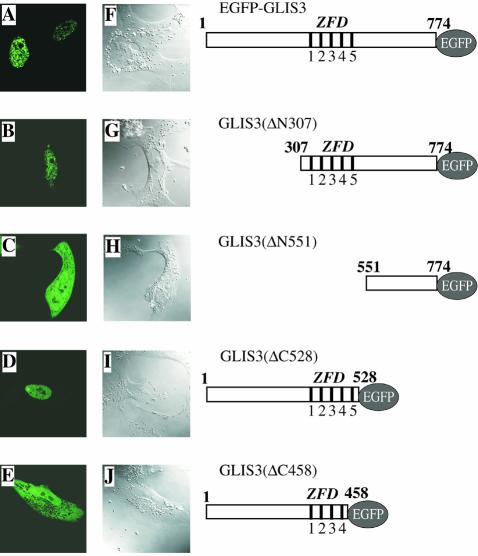
ScanProsite and PFSCAN analysis indicated the presence of a putative bipartite nuclear localization signal (NLS) at Arg489–Lys505. To examine the subcellular localization of GLIS3, CV-1 cells were transfected with an expression vector encoding an EGFP–GLIS3 fusion protein. Thirty hours later the subcellular distribution of EGFP–GLIS3 was analyzed by confocal microscopy. As shown in Figure 6A, full-length GLIS3 localized to the nucleus where it was distributed in a punctate manner. GLIS3 was detected predominantly in the nucleus in >90% of the cells. Analysis of the localization of EGFP–GLIS3 in NIH 3T3 and 293 cells or of 3× Flag-GLIS3 in CV-1 cells gave similar results. To determine what region of GLIS3 is important in its nuclear localization, the effect of four different deletions on the nuclear localization of GLIS3 was examined. GLIS3(ΔN307), which lacks the N-terminus but still contains the ZFD, still localized to the nucleus (Fig. 6B). However, GLIS3(ΔN551) lacking the N-terminus but including the entire ZFD and NLS, predominantly localized to the cytoplasm (Fig. 6C). These results suggest that the ZFD/NLS region is an important determinant in the nuclear localization of GLIS3. This was supported by the predominant nuclear localization of GLIS3(ΔC528), in which the C-terminus up to the ZFD/NLS region was deleted (Fig. 6D). The deletion mutant GLIS3(ΔC458) lacking the C-terminal region, including the fifth zinc finger motif and NLS, was present both in the nucleus and cytoplasm (Fig. 6E). These observations suggest that the region from Arg458 to Pro528, containing the NLS and the fifth zinc finger motif, is required for the nuclear localization of GLIS3.
Figure 6.
GLIS3 localizes primarily to the nucleus. Plasmids pEGFP-GLIS3 (A and F), pEGFP-GLIS3(ΔN307) (B and G), pEGFP-GLIS3(ΔN551) (C and H), pEGFP-GLIS3(ΔC528) (D and I) or pEGFP-GLIS3(ΔC458) (E and J) were transfected into CV-1 cells and after 30 h the cellular localization of EGFP–GLIS3 fusion proteins examined by fluorescence confocal microscopy as described in Materials and Methods (A–E). EGFP was equally divided between cytoplasm and nucleus (not shown). 1–5 indicate the five zinc finger motifs. (F)–(J) Confocal images.
Transcriptional activity of GLIS3 is cell type dependent
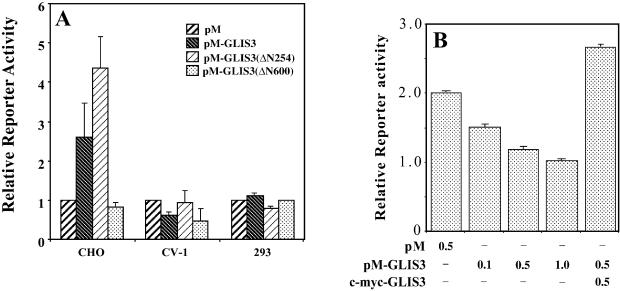
The transcriptional activity of GLIS3 was assessed in CV-1, CHO and 293 cells by mono-hybrid analysis. Cells were co-transfected with (UAS)5-LUC reporter and either pM-GLIS3, pM-GLIS3(ΔN254) or pM-GLIS3(ΔN600) DNA. In 293 cells, none of the three proteins had any effect on basal transcriptional activity, however, in CV-1 cells they suppressed basal transcription (Fig. 7). The suppression of basal transcription in CV-1 cells by GAL4(DBD)–GLIS3 was dose dependent and could be reversed by the expression of GLIS3. This reversal may be due to competition between GLIS3 and GAL4(DBD)–GLIS3 for the binding of endogenous co-repressors (squelching).
Figure 7.
GLIS3 is able to function as a transcriptional repressor and activator. (A) (UAS)5-LUC, pCMVβ and pM-GLIS3 plasmid DNAs were co-transfected into CV-1, CHO and 293 cells. After 30 h, cells were assayed for luciferase (LUC) and β-galactosidase activity as described in Materials and Methods. The relative LUC reporter activity was calculated and plotted. (B) Repression of basal transcription by Gal4(DBD)–GLIS3 and its reversal by GLIS3. CV-1 cells were co-transfected with (UAS)5-LUC, pCMVβ and pM or different amounts of pM-GLIS3 in the presence or absence of pCMV-mycGLIS3 expression plasmid as indicated. Reporter activity was measured 30 h later.
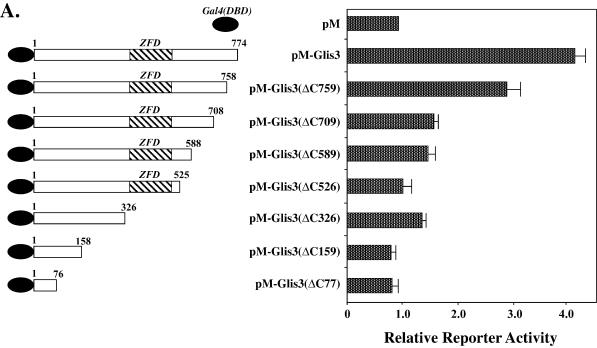
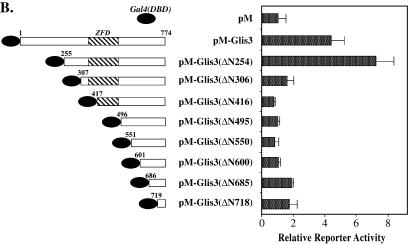
In contrast to 293 and CV-1 cells, GAL4(DBD)–GLIS3 and GAL4(DBD)–GLIS3(ΔN254) induced transcription several-fold in CHO cells. The transactivation function of GLIS3 was further characterized by examining the effect of several N- and C-terminal deletions on the activity of GLIS3. Deletion of the C-terminus up to Ser758 reduced GLIS3-mediated transactivation while deletion up to Thr708 almost totally abolished GLIS3 activity (Fig. 8A), suggesting that the C-terminus from Thr708 is critical for GLIS3 activity. Deletion of the N-terminus up to Met254 further enhanced transcriptional activity while this activity was greatly diminished in GLIS3(ΔN307). Further N-terminal deletions did not have any additional effects (Fig. 8B). These deletion analyses suggest that both the C- and N-termini are important for the transactivation function of GLIS3.
Figure 8.
Effect of various N- and C-terminal deletions on the transcriptional activity of GLIS3. CHO cells were co-transfected with (UAS)5-LUC, pCMVβ and pM or pM-GLIS3 containing various C- (A) or N-terminal (B) deletions in GLIS3 as indicated. Forty-eight hours after transfection, cells were assayed for LUC and β-galactosidase activity as described in Materials and Methods. The relative LUC activity was calculated and plotted.
Binding of GLIS3 to the GLI response element (GLI-RE)
Previous studies have provided evidence indicating that the region containing the third to fifth zinc finger motifs are important in the interaction of GLI with GLI-RE (47,48). Since this region is highly conserved among GLIS3 and GLI proteins, one might predict that GLIS3 is able to bind the consensus GLI-RE sequence GACCACCCAC. As demonstrated in Figure 9, GLIS3(ZFD) was able to interact with the consensus GLI-RE. Addition of unlabeled GLI-RE competed effectively for GLIS3 binding.
Figure 9.
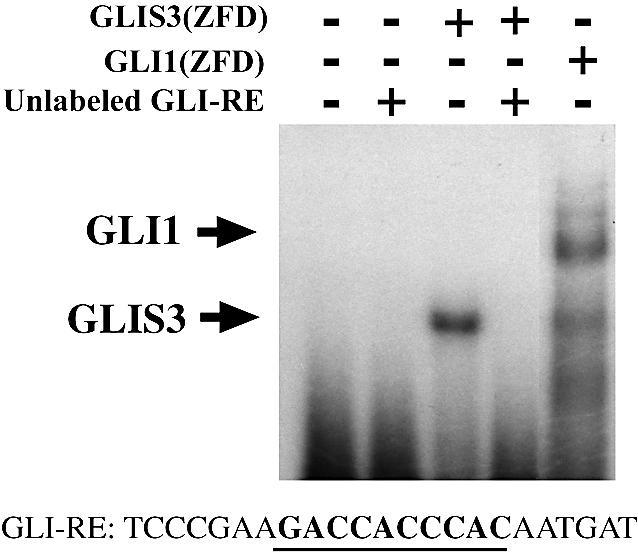
GLIS3 is able to bind the consensus GLI response element (GLI-RE). Electrophoretic mobility shift assay was performed with (His)6-GLIS3(ZFD) or (His)6-GLI1(ZFD) fusion proteins using a 32P-labeled oligonucleotide containing the consensus GLI-RE. The sequence of the GLI-RE is shown at the bottom; the consensus core motif is indicated in bold. The positions of the shifted protein–DNA complexes are indicated by arrows. Unlabeled GLI-RE (100× excess) competed with 32P-labeled GLI-RE for His6-GLIS3 binding (lane 4).
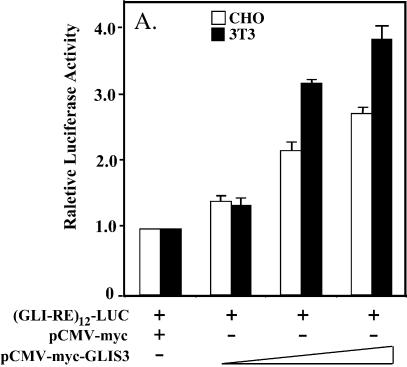
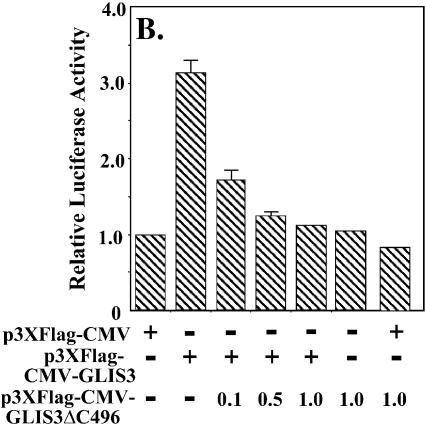
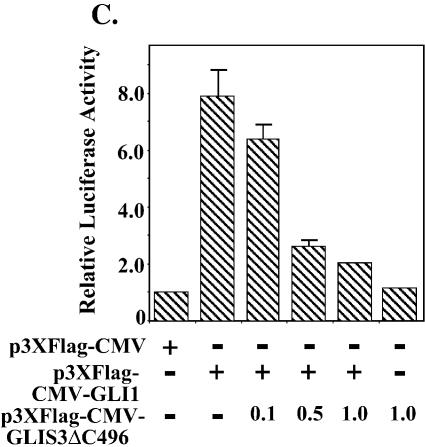
Since GLIS3 is able to bind GLI-RE in EMSA, we examined whether GLIS3 was able to induce transcription through GLI-RE. As shown in Figure 10A, transfection of CHO and NIH 3T3 cells with a (GLI-RE)12-LUC reporter and GLIS3 expression vector caused a 4-fold increase in reporter activity. Co-expression of GLIS(ΔC496) inhibited the induction of transcription by GLIS3 (Fig. 10B). Similarly, the induction of reporter activity by GLI1 was repressed by GLIS(ΔC496) (Fig. 10C). These results demonstrate that mutant GLIS(ΔC496) functions as a dominant-negative GLIS3, consistent with the results obtained in Figure 8A. This inhibition likely involves competition between GLI1 and GLIS(ΔC496) for binding to GLI-RE.
Figure 10.
(A) Induction of GLI-RE-mediated transcriptional activation by GLIS3. CHO or NIH 3T3 cells were transfected with (GLI-RE)12-LUC and increasing amounts (0.1–1.0 µg) of p3×FlagCMV-GLIS3 expression vector as indicated. (B) Inhibition of GLIS3-induced transcription by dominant-negative GLIS3(ΔC496). CHO cells were transfected with (GLI-RE)12-LUC, p3×FlagCMV-GLIS3 and increasing amounts of the p3×FlagCMV-GLIS3(ΔC496) expression vector as indicated. (C) Inhibition of GLI1- induced transcription by GLIS3(ΔC496). CHO cells were transfected with (GLI-RE)12-LUC, p3×FlagCMV-GLI1 and increasing amounts of the p3×FlagCMV-GLIS3(ΔC496) expression vector as indicated. After 30 h, cells were analyzed for reporter activities. The relative LUC activity was calculated and plotted.
DISCUSSION
In this study, we describe the cloning and sequence of a cDNA encoding a Krüppel-like zinc finger protein not reported previously, that was named GLI-similar 3 (GLIS3), based on its close relationship to GLIS1 (6). GLIS3 contains a ZFD that exhibits highest identity, 93 and 81%, respectively, with the ZFD of GLIS1 (6) and the apparent Drosophila homolog gfl/lmd (11,12). Interestingly, the exon–intron junctions within the region encoding the ZFD of GLIS3 are conserved with those of GLIS1 but distinct from the splice junctions in GLIS2, GLI and ZIC. This conservation in splice junctions indicates that GLIS1 and GLIS3 form a novel subfamily. Phylogenetic analysis further supports the conclusion that GLIS1, GLIS3 and gfl/lmd belong to a distinct subfamily of Krüppel-like zinc finger proteins different from, but closely related to, GLI, ZIC and GLIS2. GLIS1 and GLIS3 have little homology outside their ZFDs.
Analysis of its subcellular distribution showed that in several cell types GLIS3 localized predominantly to the nucleus. Deletion of a region containing the fifth zinc finger motif as well as a putative nuclear localization signal targeted the protein to the cytoplasm, suggesting that this region is required for the nuclear localization of GLIS3. In a number of zinc finger proteins, the zinc finger motifs have been implicated in nuclear localization (49). Although the precise mechanism by which this region mediates nuclear transport is not yet quite understood, it likely involves protein–protein interaction. Whether the subcellular localization of GLIS proteins is regulated by protein–protein interactions, kinases or proteolytic processing, as has been reported for GLI proteins, has yet to be established (15–17).
GLI and ZIC proteins regulate transcription by interacting with specific DNA response elements (GLI-REs) containing the consensus GACCACCCA in the promoter region of target genes (47,50–52). GLIS3 is also able to interact with GLI-REs (Fig. 8), as is GLIS1 (6,53). However, GLI, ZIC and GLIS proteins can exhibit different affinities for distinct GLI-RE-like DNA elements. It has been shown that the sequences adjacent to the GACCACCCA consensus is important in determining the binding affinity of these zinc finger proteins to GLI-REs (52). The third to fifth zinc finger motifs, which constitute the most highly conserved region within the ZFD of members of the GLI, ZIC and GLIS subfamilies (Fig. 2A), appear critical in the recognition of this specific sequence (47,50). Differences in this region between these proteins may be responsible for the observed differences in their affinity for different GLI-REs.
Experiments examining the transcriptional activity of GLIS3 showed that it can function as a repressor and activator of transcription depending on the cell type analyzed. In CV-1 cells, GLIS3 repressed transcription in a dose-dependent manner, suggesting that in these cells GLIS3 acts as a repressor. However, in CHO cells GLIS3 functions as a weak transcriptional activator. The weak transactivation may be due to suboptimal interaction with the GLI-RE used, post-translational modifications or lack of appropriate co-activators. Deletion analysis demonstrated that deletion of the N-terminus up to His254 enhanced transactivation, while further deletion up to Tyr306 almost totally abolished GLIS3 activity. Similarly, deletion of its C-terminus up to Ser709 greatly diminished transcriptional activity. These results suggest that the N- and C-termini contain domains required for optimal transcriptional activation. In CHO cells, GLIS3 also induces GLI-RE-dependent transcription. The mutant GLIS3(ΔC496), lacking the activation function at the C-terminus, repressed the induction of transcription by either GLIS3 or GLI1, suggesting that this mutant acts as a dominant-negative factor. This inhibition may at least in part be due to competition for GLI-RE binding.
Repression and activation of transcription by GLI/ZIC/GLIS proteins is likely mediated through recruitment of intermediary proteins that function either as co-repressors or co-activators, respectively. These interactions involve specific motifs/domains in GLI/ZIC/GLIS. For example, the repression by a large number of Krüppel-like zinc finger proteins involves the specific repressor domain Krüppel-associated box (KRAB) (54,55) or SCAN box (56). The repressor function of the KRAB is mediated through its interaction with the co-repressor TIF1 (57–59). However, the repressor activity of GLIS3 does not involve either a KRAB or SCAN box since the N-terminus of GLIS3 does not have any resemblance to these motifs. Studies are in progress to identify co-repressors and co-activators that mediate the activity of GLIS proteins.
We demonstrate that GLIS3 expression is restricted to specific tissues and structures in the mouse embryo. GLIS3 expression was first detected in the node, which is the murine equivalent of Spemann’s organizer in the frog and Hensen’s node in the chick (60). However, GLIS3 is quickly down-regulated, as it is not expressed in the notochord, the midline structure derived from cells in the node. During neurulation, GLIS3 is first expressed in the early neural plate in the prospective hindbrain region and is then expressed in the mid/hindbrain junction and anterior forebrain by E8.5 and in the roof plate (the dorsal-most region of the neural tube) by E9.0. These expression domains sharpen over the next 3 days of development, but by E12.5 GLIS3 is mostly found in specific structures of the telencephalon. This highly dynamic pattern of GLIS3 expression during neurulation includes eye development. GLIS3 is first expressed in the dorsal optic vesicle and lens placode in the surface ectoderm. As eye development progresses, expression is evident in the lens, neural retina and nasal optic stalk. GLIS3 is prominently expressed in the mesenchyme of two outgrowths: the genital tubercle and limbs. Finally, during organogenesis GLIS3 is expressed in cell-specific patterns in lungs, trachea, testes and kidneys.
Expression of GLIS3 in multiple embryonic domains implicates this factor in a variety of cellular processes during development. Thus, GLIS3 may play a role in cell migration as it is expressed in structures that are sources of migratory cells: the node, the dorsal neural tube and anterior rhombic lip of the isthmus. However, GLIS3 is not expressed in the cells that migrate from these tissues (notochord, neural crest and granule cell precursors, respectively). In addition to the neural crest, the neural roof plate is also an organizing center that patterns the neural tube and functions as a source of progenitor cells that will give rise to the dorsal sensory interneurons. GLIS3 may play a role in either of these processes. Another role for GLIS3 is suggested by its strong expression in the interdigital regions before (E11.5) and during the onset (E12.5) of apoptosis. Thus, GLIS3 may play a role in the programmed cell death that removes this tissue and sculpts the digits.
One commonality in nearly all of the GLIS3 expression domains is that these are regions where signaling through BMPs or other members of the transforming growth factor (TGF)-β superfamily play a central role in embryogenesis. For example, regulation of BMP4 signaling is required for normal node function (61). BMP signaling from the roof plate opposes SHH signaling from the floor plate and together these inductive signals generate dorsal/ventral positional patterning of the neural tube (62). BMP4 is expressed in both the optic vesicle and surface ectoderm and is required for the inductive interactions between these tissues for normal eye development (63). During limb development, BMPs and TGF-β2/3 are thought to be major determinants of interdigit cell death (64,65), and Gdf5/6/7 are required in interphalangeal joints of the digits zone for proper joint development (66). Gene inactivation studies in mouse embryos have shown that BMP7, which is expressed initially in the metanephric ureteric bud, is required for normal kidney development (67,68), and TGF-β2/3 are required in lung development (69,70). Finally, conditional Cre-mediated inactivation of BMP receptor 1A in facial mesenchyme has demonstrated that BMP signals are required for normal facial development (M. Lewandoski and T. Williams, unpublished observations). Therefore, we speculate that ligands of the TGF-β superfamily may be GLIS3 targets, however, we have not ruled out the possibility that GLIS3 may be downstream of BMP signaling.
As discussed above, gfl/lmd is the putative Drosophila homolog of GLIS1 and GLIS3. gfl/lmd has been reported to play an important role in muscle differentiation. Embryos lacking lmd function show a loss of expression of Mef2 and stick-and-stones, two key differentiation and fusion genes, and are completely devoid of multinucleated muscle fibers (11,12). Evidence has been provided that lmd may directly control transcription of the Mef2 gene. Although GLIS3 has been found in adult skeletal muscle, future studies have to identify its precise role in this tissue.
In the present study, we describe the identification of GLIS3 and provide evidence indicating that GLIS1 and GLIS3 form a distinct subfamily of Krüppel-like zinc finger proteins that is different from but closely related to GLI and ZIC. Whole mount in situ hybridization has demonstrated that during embryonic development GLIS3 is expressed in a temporal and spatial pattern suggesting that it may play a critical role in the regulation of a variety of cellular processes during development.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Catherine P. Wilson for expert technical assistance and Jeff Reece for his advice and assistance with confocal microscopy. We also thank Alan Perantoni, Sergei Kozlov, Jean Herbert and John Rubenstein for discussion and advice.
DDBJ/EMBL/GenBank accession no. AY220846
REFERENCES
- 1.Nagai T., Aruga,J., Takada,S., Gunther,T., Sporle,R., Schughart,K. and Mikoshiba,K. (1997) The expression of the mouse Zic1, Zic2 and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol., 182, 299–313. [DOI] [PubMed] [Google Scholar]
- 2.Walterhouse D.O., Yoon,J.W. and Iannaccone,P.M. (1999) Developmental pathways: Sonic hedgehog-Patched-GLI. Environ. Health Perspect., 107, 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aruga J., Nagai,T., Tokuyama,T., Hayashizaki,Y., Okazaki,Y., Chapman,V.M. and Mikoshiba,K. (1996) The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J. Biol. Chem., 271, 1043–1047. [DOI] [PubMed] [Google Scholar]
- 4.Kinzler K.W., Ruppert,J.M., Bigner,S.H. and Vogelstein,B. (1988) The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature, 332, 371–374. [DOI] [PubMed] [Google Scholar]
- 5.Villavicencio E.H., Walterhouse,D.O. and Iannaccone,P.M. (2000) The Sonic Hedgehog-Patched-Gli pathway in human development and disease. Am. J. Hum. Genet., 67, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y.S., Lewandoski,M., Perantoni,A.O., Kurebayashi,S., Nakanishi,G. and Jetten,A.M. (2002) Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem., 277, 30901–30913. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F., Nakanishi,G., Kurebayashi,S., Yoshino,K., Perantoni,A., Kim,Y.S. and Jetten,A.M. (2001) Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcriptional factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J. Biol. Chem., 12, 10139–10149. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F. and Jetten,A.M. (2001) Genomic structure of the gene encoding the human GLI-related, Kruppel-like zinc finger protein GLIS2. Gene, 280, 49–57. [DOI] [PubMed] [Google Scholar]
- 9.Benedyk M.J., Mullen,J.R. and DiNardo,S. (1994) odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev., 8, 105–117. [DOI] [PubMed] [Google Scholar]
- 10.Lessing D. and Nusse,R. (1998) Expression of wingless in the Drosophila embryo: a conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development, 125, 1469–1476. [DOI] [PubMed] [Google Scholar]
- 11.Duan H., Skeath,J.B. and Nguyen,H.T. (2001) Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development, 128, 4489–4500. [DOI] [PubMed] [Google Scholar]
- 12.Furlong E.E. andersen,E.C., Null,B., White,K.P. and Scott,M.P. (2001) Patterns of gene expression during Drosophila mesoderm development. Science, 293, 1629–1633. [DOI] [PubMed] [Google Scholar]
- 13.Kuo J.S., Patel,M., Gamse,J., Merzdorf,C., Liu,X., Apekin,V. and Sive,H. (1998) Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development, 125, 2867–2882. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz i Altaba A., Sanchez,P. and Dahmane,N. (2002) GLI and hedgehog in cancer: tumours, embryos and stem cells. Nature Rev., 2, 361–372. [DOI] [PubMed] [Google Scholar]
- 15.Ding Q., Fukami,S., Meng,X., Nishizaki,Y., Zhang,X., Sasaki,H., Dlugosz,A., Nakafuku,M. and Hui,C. (1999) Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol., 9, 1119–1122. [DOI] [PubMed] [Google Scholar]
- 16.Murone M., Luoh,S.M., Stone,D., Li,W., Gurney,A., Armanini,M., Grey,C., Rosenthal,A. and de Sauvage,F.J. (2000) Gli regulation by the opposing activities of fused and suppressor of fused. Nature Cell. Biol., 2, 310–312. [DOI] [PubMed] [Google Scholar]
- 17.Kogerman P., Grimm,T., Kogerman,L., Krause,D., Unden,A.B., Sandstedt,B., Toftgard,R. and Zaphiropoulos,P.G. (1999) Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nature Cell. Biol., 1, 312–319. [DOI] [PubMed] [Google Scholar]
- 18.Borycki A., Brown,A.M. and Emerson,C.P.,Jr (2000) Shh and Wnt signaling pathways converge to control Gli gene activation in avian somites. Development, 127, 2075–2087. [DOI] [PubMed] [Google Scholar]
- 19.Meyer N.N. and Roelink,H. (2003) The amino-terminal region of Gli3 antagonizes the Shh response and acts in dorsoventral fate specification in the developing spinal cord. Dev. Biol., 257, 343–355. [DOI] [PubMed] [Google Scholar]
- 20.Kawai S. and Sugiura,T. (2001) Characterization of human bone morphogenetic protein (BMP)-4 and -7 gene promoters: activation of BMP promoters by Gli, a sonic hedgehog mediator. Bone, 29, 54–61. [DOI] [PubMed] [Google Scholar]
- 21.Mullor J.L., Dahmane,N., Sun,T. and Ruiz i Altaba,A. (2001) Wnt signals are targets and mediators of Gli function. Curr. Biol., 11, 769–773. [DOI] [PubMed] [Google Scholar]
- 22.Walterhouse D., Ahmed,M., Slusarski,D., Kalamaras,J., Boucher,D., Holmgren,R. and Iannaccone,P. (1993) gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev. Dyn., 196, 91–102. [DOI] [PubMed] [Google Scholar]
- 23.Aruga J., Inoue,T., Hoshino,J. and Mikoshiba,K. (2002) Zic2 controls cerebellar development in cooperation with Zic1. J. Neurosci., 22, 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz i Altaba A. (1999) Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development, 126, 3205–3216. [DOI] [PubMed] [Google Scholar]
- 25.Mo R., Freer,A.M., Zinyk,D.L., Crackower,M.A., Michaud,J., Heng,H.H., Chik,K.W., Shi,X.M., Tsui,L.C., Cheng,S.H. et al. (1997) Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development, 124, 113–123. [DOI] [PubMed] [Google Scholar]
- 26.Mikoshiba K. (1999) Mammalian neural induction and brain pattern formation. Keio J. Med., 48, 111–123. [DOI] [PubMed] [Google Scholar]
- 27.Grindley J.C., Bellusci,S., Perkins,D. and Hogan,B.L. (1997) Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Dev. Biol., 188, 337–348. [DOI] [PubMed] [Google Scholar]
- 28.Motoyama J., Liu,J., Mo,R., Ding,Q., Post,M. and Hui,C.C. (1998) Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nature Genet., 20, 54–57. [DOI] [PubMed] [Google Scholar]
- 29.Kalff-Suske M., Wild,A., Topp,J., Wessling,M., Jacobsen,E.M., Bornholdt,D., Engel,H., Heuer,H., Aalfs,C.M., Ausems,M.G. et al. (1999) Point mutations throughout the GLI3 gene cause Greig cephalopolysyndactyly syndrome. Hum. Mol. Genet., 8, 1769–1777. [DOI] [PubMed] [Google Scholar]
- 30.Kang S., Graham,J.M.,Jr, Olney,A.H. and Biesecker,L.G. (1997) GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nature Genet., 15, 266–268. [DOI] [PubMed] [Google Scholar]
- 31.Gebbia M., Ferrero,G.B., Pilia,G., Bassi,M.T., Aylsworth,A.S., Penman-Splitt,M., Bird,L.M., Bamforth,J.S., Burn,J., Schlessinger,D. et al. (1997) X-linked situs abnormalities result from mutations in Zic3. Nature Genet., 17, 305–308. [DOI] [PubMed] [Google Scholar]
- 32.Brown L.Y., Hodge,S.E., Johnson,W.G., Guy,S.G., Nye,J.S. and Brown,S. (2002) Possible association of NTDs with a polyhistidine tract polymorphism in the Zic2 gene. Am. J. Med. Genet., 108, 128–131. [DOI] [PubMed] [Google Scholar]
- 33.Dahmane N., Lee,J., Robins,P., Heller,P. and Ruiz i Altaba,A. (1997) Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature, 389, 876–881. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson M., Unden,A.B., Krause,D., Malmqwist,U., Raza,K., Zaphiropoulos,P.G. and Toftgard,R. (2000) Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc. Natl Acad. Sci. USA, 97, 3438–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grachtchouk M., Mo,R., Yu,S., Zhang,X., Sasaki,H., Hui,C.C. and Dlugosz,A.A. (2000) Basal cell carcinomas in mice overexpressing Gli2 in skin. Nature Genet., 24, 216–217. [DOI] [PubMed] [Google Scholar]
- 36.Green J., Leigh,I.M., Poulsom,R. and Quinn,A.G. (1998) Basal cell carcinoma development is associated with induction of the expression of the transcription factor Gli-1. Br. J. Dermatol., 139, 911–915. [DOI] [PubMed] [Google Scholar]
- 37.Yokota N., Aruga,J., Takai,S., Yamada,K., Hamazaki,M., Iwase,T., Sugimura,H. and Mikoshiba,K. (1996) Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res., 56, 377–383. [PubMed] [Google Scholar]
- 38.Watkins D.N., Berman,D.M., Burkholder,S.G., Wang,B., Beachy,P.A. and Baylin,S.B. (2003) Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature, 422, 313–317. [DOI] [PubMed] [Google Scholar]
- 39.Haramis A.G. and Carrasco,A.E. (2001) In situ hybridization and immunohistochemistry. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.M., Seidman,J.G., Smith,J.A., Stuhl,K. (eds), Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY, Unit 14.9. [Google Scholar]
- 40.Dunaeva M., Michelson,P., Kogerman,P. and Toftgard,R. (2003) Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem., 278, 5116–5122. [DOI] [PubMed] [Google Scholar]
- 41.Yan Z.H., Medvedev,A., Hirose,T., Gotoh,H. and Jetten,A.M. (1997) Characterization of the response element and DNA binding properties of the nuclear orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor. J. Biol. Chem., 272, 10565–10572. [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuh R., Aicher,W., Gaul,U., Cote,S., Preiss,A., Maier,D., Seifert,E., Nauber,U., Schroder,C., Kemler,R. et al. (1986) A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell, 47, 1025–1032. [DOI] [PubMed] [Google Scholar]
- 44.Ruppert J.M., Kinzler,K.W., Wong,A.J., Bigner,S.H., Kao,F.T., Law,M.L., Seuanez,H.N., O’Brien,S.J. and Vogelstein,B. (1988) The GLI-Kruppel family of human genes. Mol. Cell. Biol., 8, 3104–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruppert J.M., Vogelstein,B., Arheden,K. and Kinzler,K.W. (1990) GLI3 encodes a 190-kilodalton protein with multiple regions of GLI similarity. Mol. Cell. Biol., 10, 5408–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes D.C., Allen,J., Morley,G., Sutherland,K., Ahmed,W., Prosser,J., Lettice,L., Allan,G., Mattei,M.G., Farrall,M. et al. (1997) Cloning and sequencing of the mouse Gli2 gene: localization to the Dominant hemimelia critical region. Genomics, 39, 205–215. [DOI] [PubMed] [Google Scholar]
- 47.Klug A. and Schwabe,J.W. (1995) Protein motifs 5. Zinc fingers. FASEB J., 9, 597–604. [PubMed] [Google Scholar]
- 48.Pavletich N.P. and Pabo,C.O. (1993) Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science, 261, 1701–1707. [DOI] [PubMed] [Google Scholar]
- 49.Koyabu Y., Nakata,K., Mizugishi,K., Aruga,J. and Mikoshiba,K. (2001) Physical and functional interactions between Zic and Gli proteins. J. Biol. Chem., 276, 6889–6892. [DOI] [PubMed] [Google Scholar]
- 50.Vortkamp A., Gessler,M. and Grzeschik,K.H. (1995) Identification of optimized target sequences for the GLI3 zinc finger protein. DNA Cell Biol., 14, 629–634. [DOI] [PubMed] [Google Scholar]
- 51.Yoon J.W., Liu,C.Z., Yang,J.T., Swart,R., Iannaccone,P. and Walterhouse,D. (1998) GLI activates transcription through a herpes simplex viral protein 16-like activation domain. J. Biol. Chem., 273, 3496–3501. [DOI] [PubMed] [Google Scholar]
- 52.Mizugishi K., Aruga,J., Nakata,K. and Mikoshiba,K. (2001) Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J. Biol. Chem., 276, 2180–2188. [DOI] [PubMed] [Google Scholar]
- 53.Nakashima M., Tanese,N., Ito,M., Auerbach,W., Bai,C., Furukawa,T., Toyono,T., Akamine,A. and Joyner,A.L. (2002) A novel gene, GliH1, with homology to the Gli zinc finger domain not required for mouse development. Mech. Dev., 119, 21. [DOI] [PubMed] [Google Scholar]
- 54.Bellefroid E.J., Poncelet,D.A., Lecocq,P.J., Revelant,O. and Martial,J.A. (1991) The evolutionarily conserved Kruppel-associated box domains defines a subfamily of eukaryotic multifingered proteins. Proc. Natl Acad. Sci. USA, 88, 3608–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margolin J.F., Friedman,J.R., Meyer,W.K., Vissing,H., Thiesen,H. and Rauscher,F.J. (1994) Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl Acad. Sci. USA, 91, 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams A.J., Khachigian,L.M., Shows,T. and Collins,T. (1995) Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J. Biol. Chem., 270, 22143–22152. [DOI] [PubMed] [Google Scholar]
- 57.Le Douarin B., You,J., Nielsen,A.L., Chambon,P. and Losson,R. (1998) TIF1alpha: a possible link between KRAB zinc finger proteins and nuclear receptors. J. Steroid Biochem. Mol. Biol., 65, 43–50. [DOI] [PubMed] [Google Scholar]
- 58.Peng H., Begg,G.E., Harper,S.L., Friedman,J.R., Speicher,D.W. and Rauscher,F.J.I. (2000) Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J. Biol. Chem., 275, 18000–18010. [DOI] [PubMed] [Google Scholar]
- 59.Poncelet D.A., Bellefroid,E.J., Bastiaens,P.V., Demoitie,M.A., Marine,J.C., Pendeville,H., Alami,Y., Devos,N., Lecocq,P., Ogawa,T. et al. (1998) Functional analysis of ZNF85 KRAB zinc finger protein, a member of the highly homologous ZNF91 family. DNA Cell Biol., 17, 931–943. [DOI] [PubMed] [Google Scholar]
- 60.Beddington R.S. (1994) Induction of a second neural axis by the mouse node. Development, 120, 613–620. [DOI] [PubMed] [Google Scholar]
- 61.Fujiwara T., Dehart,D.B., Sulik,K.K. and Hogan,B.L. (2002) Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development, 129, 4685–4696. [DOI] [PubMed] [Google Scholar]
- 62.Helms A.W. and Johnson,J.E. (2003) Specification of dorsal spinal cord interneurons. Curr. Opin. Neurobiol., 13, 42–49. [DOI] [PubMed] [Google Scholar]
- 63.Furuta Y. and Hogan,B.L. (1998) BMP4 is essential for lens induction in the mouse embryo. Genes Dev., 12, 3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuzarte-Luis V. and Hurle,J.M. (2002) Programmed cell death in the developing limb. Int. J. Dev. Biol., 46, 871–876. [PubMed] [Google Scholar]
- 65.Dunker N., Schmitt,K. and Krieglstein,K. (2002) TGF-beta is required for programmed cell death in interdigital webs of the developing mouse limb. Mech. Dev., 113, 111–120. [DOI] [PubMed] [Google Scholar]
- 66.Settle S.H. Jr, Rountree,R.B., Sinha,A., Thacker,A., Higgins,K. and Kingsley,D.M. (2003) Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol., 254, 116–130. [DOI] [PubMed] [Google Scholar]
- 67.Dudley A.T., Lyons,K.M. and Robertson,E.J. (1995) A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev., 9, 2795–2807. [DOI] [PubMed] [Google Scholar]
- 68.Luo G., Hofmann,C., Bronckers,A.L., Sohocki,M., Bradley,A. and Karsenty,G. (1995) BMP-7 is an inducer of nephrogenesis and is also required for eye development and skeletal patterning. Genes Dev., 9, 2808–2820. [DOI] [PubMed] [Google Scholar]
- 69.Kaartinen V., Voncken,J.W., Shuler,C., Warburton,D., Bu,D., Heisterkamp,N. and Groffen,J. (1995) Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nature Genet., 11, 415–421. [DOI] [PubMed] [Google Scholar]
- 70.Sanford L.P., Ormsby,I., Gittenberger-de Groot,A.C., Sariola,H., Friedman,R., Boivin,G.P., Cardell,E.L. and Doetschman,T. (1997) TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development, 124, 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]