Abstract
Chaperone protein BiP binds to Ire1 and dissociates in response to endoplasmic reticulum (ER) stress. However, it remains unclear how the signal transducer Ire1 senses ER stress and is subsequently activated. The crystal structure of the core stress-sensing region (CSSR) of yeast Ire1 luminal domain led to the controversial suggestion that the molecule can bind to unfolded proteins. We demonstrate that, upon ER stress, Ire1 clusters and actually interacts with unfolded proteins. Ire1 mutations that affect these phenomena reveal that Ire1 is activated via two steps, both of which are ER stress regulated, albeit in different ways. In the first step, BiP dissociation from Ire1 leads to its cluster formation. In the second step, direct interaction of unfolded proteins with the CSSR orients the cytosolic effector domains of clustered Ire1 molecules.
Introduction
In eukaryotic cells, most secretory and membrane proteins are folded and assembled in the ER. Impairment of this process is collectively called ER stress. Because accumulation of unfolded proteins is harmful to cells, defensive mechanisms against ER stress exist. The type I transmembrane protein Ire1 is conserved in fungi, animals, and plants and plays a central role in the unfolded protein response (UPR). Ire1 has protein kinase and RNase domains in the cytosolic region (Cox et al., 1993; Mori et al., 1993; Sidrauski and Walter, 1997). ER stress causes autophosphorylation of Ire1, which is followed by its activation as an RNase (Shamu and Walter, 1996; Papa et al., 2003). In budding yeast Saccharomyces cerevisiae, Ire1 contributes to unconventional RNA splicing that converts the precursor form of HAC1 mRNA (HAC1u) to the mature form (HAC1i; Cox and Walter, 1996). HAC1i is effectively translated into the transcription factor protein Hac1, which regulates a wide variety of genes to alleviate ER stress (Travers et al., 2000; Kimata et al., 2006). In contrast to yeast cells, which have only one known ER stress-sensing protein (Ire1), mammalian cells carry multiple ER stress sensors. There are two Ire1 paralogues, IREα and IREβ, as well as a second type I transmembrane protein, pancreatic ER kinase (PERK), which attenuates protein translation by phosphorylation of eukaryotic translation initiation factor 2α (Harding et al., 1999). A third system is the membrane-anchored transcription factor ATF6. Upon ER stress, ATF6 is solubilized by proteolysis and up-regulates ER chaperone and other genes (Haze et al., 1999).
Because the luminal domains of Ire1 and PERK show moderate amino acid sequence similarity, we believe that they sense ER stress by the same mechanism. One breakthrough finding toward understanding the ER stress-sensing mechanism was the observation that BiP binds to Ire1 and dissociates in response to ER stress (Bertolotti et al., 2000; Okamura et al., 2000). It is highly likely that BiP binding negatively regulates Ire1 because activity of Ire1 is considerably attenuated in yeast BiP mutants in which dissociation of the mutated BiP proteins from Ire1 is impaired (Kimata et al., 2003, 2004). However, as described in the next paragraph, dissociation of BiP from Ire1 is not sufficient for activation of Ire1.
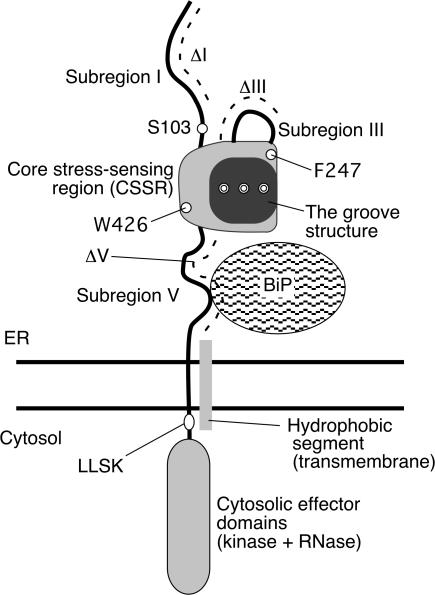
We previously predicted that the luminal domain of yeast Ire1 is composed of five subregions (I–V; Fig. 1). This is because our systematic mutagenesis analysis demonstrated that 10 aa deletions in subregions II and IV, but not in subregions I, III, or V, inactivate Ire1 (Kimata et al., 2004). Our speculation that subregions I and V are loosely folded is supported by the finding that these subregions are highly sensitive to proteolysis in the context of a recombinant Ire1 luminal domain protein (Oikawa et al., 2005). According to the crystal structure presented by Credle et al. (2005), subregions II–IV form one tightly folded domain, which we termed the core stress-sensing region (CSSR). Both this crystal structure analysis and our systematic mutational analysis suggest that subregion III exists as a flexible stretch sticking out from the CSSR. The BiP binding site is located in subregion V (Kimata et al., 2004). Importantly, an Ire1 mutant that contains a deletion of almost all of subregion V (hereafter called ΔV; see Fig. 1 for the position) is not constitutively active, but is regulated by ER stress as well as wild-type Ire1, even though BiP does not bind to this mutant (Kimata et al., 2004).
Figure 1.
Structure of yeast Ire1 and mutations used in this study. Structure of the luminal domain according to Kimata et al. (2004), Oikawa et al. (2005), and Credle et al. (2005) is illustrated. Subregions I (aa 32–111), III (aa 243–272), and V (aa 455–524) are loosely folded, whereas subregions II (aa 112–242) and IV (aa 273–454) form the tightly folded CSSR. The position of a hydrophobic segment (aa 527–570) that is deduced to be the transmembrane domain is also shown. The dashed lines indicate positions of amino acid residues deleted in ΔI (aa 32–91), ΔIII (aa 253–272), and ΔV (aa 463–524) mutants, respectively. The positions of another deletion mutation, 567LLSK570, and point mutations S103, F247, and W246 are also indicated. Double circles respectively represent M229, F285, and Y301, which are simultaneously replaced by Ala in Figs. 5 E and 7. Because we now assign the initiation methionine according to the data from the Saccharomyces genome database (http://www.yeastgenome.org/), the amino acid numbers of Ire1 in the present paper differ by 7 aa from those in two of our previous papers (Kimata et al., 2004; Oikawa et al., 2005).
The crystal structure also indicates that a CSSR dimer forms a major histocompatibility complex (MHC)–like groove (Credle et al., 2005). Analagous to the MHC, peptide fragments and, more speculatively, unfolded proteins may be captured by this groove. An idea emerging from these observations is that in addition to regulation by BiP, the CSSR, as its name denotes, directly senses unfolded proteins and regulates Ire1. However, based on the crystal structure of the mammalian IRE1α luminal domain, Zhou et al. (2006) argued that this groove is not suited to capture unfolded proteins. Moreover, no biochemical evidence has been provided for direct binding of the CSSR to unfolded proteins.
The oligomerization status of Ire1 is also enigmatic (Kohno, 2007). Epitope-tagged Ire1 coimmunoprecipitates with differently epitope-tagged Ire1 from lysate of ER-stressed cells, but only to a small extent from that of nonstressed cells (Kimata et al., 2004). We reported recently, from coimmunoprecipitation analysis, that Ire1 seems to be fully self-associated, even in the absence of ER stress, when it carries both the ΔV mutation and another deletion of the N-terminal three-quarters of subregion I (hereafter called ΔI; see Fig. 1 for the position; Oikawa et al., 2007). This finding suggests that the self-association is regulated both by subregion V (probably by binding and dissociation of BiP) and subregion I. Notably, activation of this ΔIΔV mutant Ire1 was still dependent on ER stress (although ΔIΔV Ire1, whose luminal domain consists of almost only the CSSR, was named “core mutant” in our previous paper, we do not use this name in the present paper because it can be confused with “a mutation in the CSSR”; Oikawa et al., 2007). According to density gradient fractionation of cell lysates, it seems that the oligomerization status of Ire1, upon activation by ER stress or when carrying the ΔIΔV mutation, is dimeric (Bertolotti et al., 2000; Oikawa et al., 2007). Furthermore, recombinant Ire1 luminal domain or CSSR exists in dimeric form in solution (Liu et al., 2002; Oikawa et al., 2005). However, the crystal structure suggests that the CSSR forms higher order oligomers because one CSSR molecule is in contact with two CSSR molecules via different interfaces (Credle et al., 2005). The authors argue that this higher order oligomerization is crucial for activation of Ire1 because point mutations that are deduced to deform either interface considerably weaken Ire1 activity. Nevertheless, formation of such higher order oligomers in vivo has not been demonstrated.
One scenario proposed by Credle et al. (2005) and modified by us (Oikawa et al., 2007) explains these confusing observations. In this explanation, a combination of BiP dissociation and release from a negative regulation by subregion I lead Ire1 to dimerize. Unfolded proteins bind and may tether dimerized CSSRs, which results in highly oligomerized and active Ire1. Because the higher order oligomer is unstable, it cannot be detected in cell lysates or in solutions of recombinant Ire1 fragments. In the present study, we provide some lines of evidence that in vivo Ire1 actually forms higher order oligomers, here called clusters, and interacts with unfolded proteins. Furthermore, we describe a new scenario in which these two events are positioned differently from the aforementioned scenario.
Results
Cluster formation of yeast Ire1 upon activation
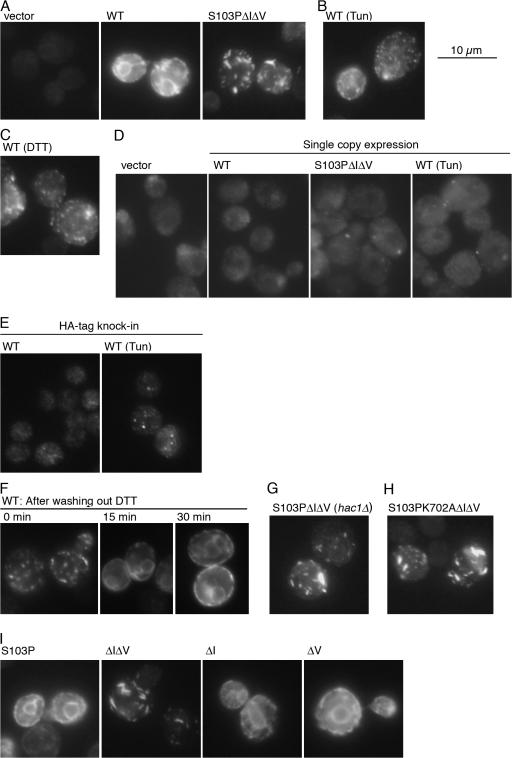
We recently reported S. cerevisiae Ire1 to be constitutively activated by a combination of ΔIΔV deletion and Ser103 to Pro (S103P) point mutations (see Fig. 1 for the positions and its legend for the amino acid numbering; Oikawa et al., 2007). Here, we checked cellular localization of Ire1 and its mutants by immunofluorescent staining of C-terminally HA-tagged molecules. As shown in Fig. 2 A, HA- tagged wild-type Ire1 (Ire1-HA) demonstrated an ER-like staining pattern in nonstressed cells, whereas the S103PΔIΔV mutation changed localization of Ire1-HA to a clumped distribution. A dotlike distribution, hereafter called clusters, was also observed in wild-type Ire1-HA cells exposed to ER stress by treatment with tunicamycin (Tun) or DTT, although there was also residual ER-like distribution (Fig. 2, B and C). To our knowledge, this paper is the first demonstration of a localization change of Ire1.
Figure 2.
Cluster formation of Ire1. Excepting E, Ire1-HA variants were expressed from wild type (WT) or mutant versions of pRS315-IRE1-HA (D; centromeric plasmid) or pRS423-IRE1-HA (A–C and E–I; 2-μm plasmid) in yeast strain KMY1015 (ire1Δ; D), YKY1004 (ire1Δ hac1Δ; G), or KMY1516 (ire1Δ; A–C, E and F, and H and I) and detected by anti-HA immunofluorescent staining. For vector control not carrying the Ire1-HA gene, cells carrying pRS423 (A) or pRS315 (D) were used. (E) An Ire1-HA gene knockin strain YKY2005 was examined. Where indicated, Tun (2 μg/ml final concentration) or DTT (10 mM final concentration) was added into cultures 1 h before harvest. (F) Cells treated with 10 mM DTT for 30 min were washed with medium and further incubated for the indicated times. Cells were observed by a conventional fluorescent microscope, and exposure times for image acquisition were 2 s for D and E and 1 s for other panels.
In this immunostaining analysis and in the immunoprecipitation analyses shown in Figs. 2, 3, 4, and 6, we mainly used cells expressing Ire1-HA from multicopy plasmids. It should be noted that, as shown in Fig. S1 (available at http://www.jcb.org/cgi/content/jcb.200704166/DC1), this multicopy expression of wild-type Ire1-HA alone did not cause activation of this molecule. As expression level of Ire1 is reported to be low, we could not detect a meaningful HA signal from nonstressed cells containing the centromeric plasmid-borne wild-type Ire1-HA gene (Fig. 2 D, compare wild type [WT] to the vector control). However, probably because of assembly of several HA epitopes, cluster formation of S103PΔIΔV Ire1-HA in nonstressed cells and of wild-type Ire1-HA in ER-stressed cells was observed even with expression from centromeric plasmids (Fig. 2 D, S103PΔIΔV and WT [Tun]), although this fluorescent signal was faint. This finding indicates that the cluster formation is not an artifact caused by high expression from multicopy plasmids. Furthermore, we constructed an Ire1-HA gene knockin strain, here named YKY2005. Western blot detection of Ire1-HA showed that the expression level of Ire1-HA in YKY2005 cells is slightly higher than that in ire1Δ cells in which Ire1-HA is expressed from a centromeric plasmid (Fig. S2). As expected, clusters of Ire1-HA were observed in YKY2005 cells when they were exposed to ER stress (Fig. 2 E).
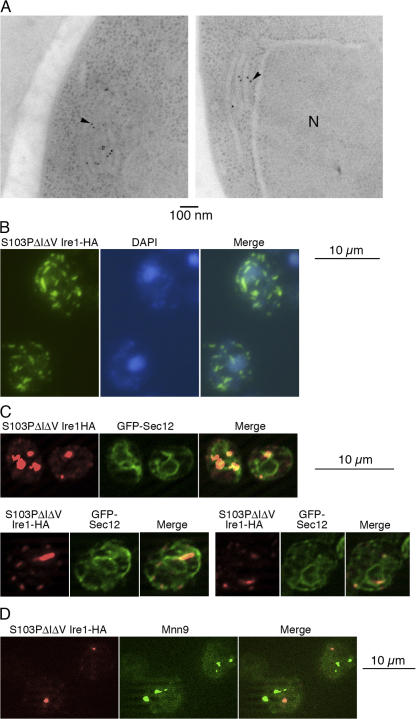
Figure 3.
Cellular localization of Ire1 clusters as examined by electron microscopy and double fluorescent staining. YKY1004 (ire1Δ hac1Δ; A and C) or KMY1516 (ire1Δ; B and D) cells carrying the S103PΔIΔV mutant version of pRS423-Ire1-HA were examined. (C) The cells also contained a GFP-Sec12 expression plasmid as described in the Materials and methods. (A) Ultrathin sections of the cells were immunogold stained with anti-HA antibody and observed with a transmission electron microscope. In each panel, the arrowhead points to one of the gold particles (N, nucleus). (B) The cells were doubly stained with anti-HA antibody (FITC) and DAPI and observed by a conventional fluorescent microscope. (C and D) Cells were doubly stained with anti-HA antibody (Cy5) and anti-GFP (C) or anti-Mnn9 (D) antibody (FITC). (C) Cy5 image in the top panels is brighter than those in the bottom panels, partly because of difference in exposure times for image acquisition.
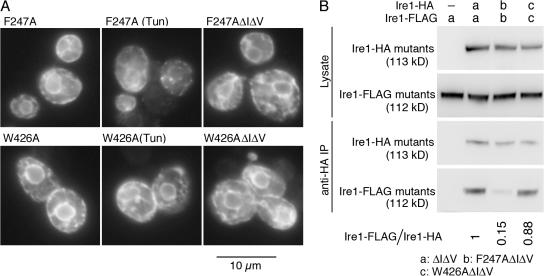
Figure 4.
Mutations that impair cluster formation. (A) KMY1516 (ire1Δ) cells carrying the indicated mutant version of pRS423-IRE1-HA were subjected to anti-HA immunofluorescent staining. Cells were observed by a conventional fluorescent microscope, and exposure times for image acquisition were 1 s for all panels. When indicated, Tun (2 μg/ml final concentration) was added into cultures 1 h before harvest. (B) Diploid cells KMY1015 × KMY1520 (ire1Δ/ire1Δ) coexpressing Ire1-HA and Ire1-FLAG carrying the indicated mutations from mutant versions of pRS315-IRE1-HA and pRS426-IRE1-FLAG, respectively, were lysed and subjected to anti-HA immunoprecipitation. The lysates and IPs were then analyzed by anti-HA or anti-FLAG Western blotting to detect the indicated proteins. For vector control (−), cells carried empty vector pRS315. The ratios of Ire1-Flag to Ire1-HA signal in anti-HA IP (Ire1-Flag/Ire1-HA) were normalized to that of the ΔIΔV mutant and are presented. Multiple independent clones were examined to obtain standard deviations < 20% of the means.
Figure 6.
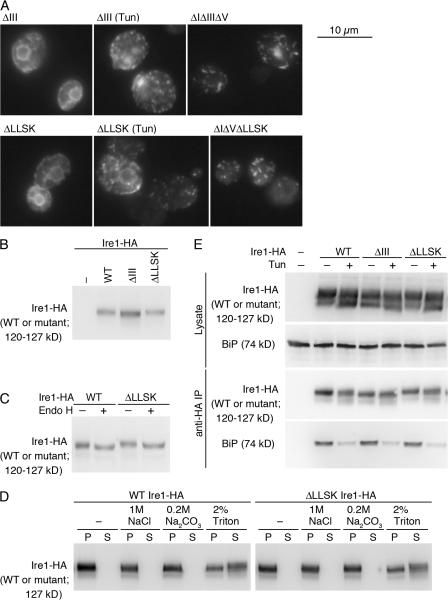
Cluster formation, localization, cellular expression, or BiP dissociation is not significantly altered by either the ΔIII or the ΔLLSK mutation. (A) KMY1516 (ire1Δ) cells carrying the indicated mutant version of pRS423-IRE1-HA were subjected to anti-HA immunofluorescent staining. Cells were observed by a conventional fluorescent microscope, and exposure times for image acquisition were 1 s for all panels. (B and C) Lysates from KMY1015 (ire1Δ) cells carrying pRS315-IRE1-HA (wild type [WT] or mutant) were analyzed by anti-HA Western blotting. (C) Cell lysate was treated with Endo H where indicated. (D) Detergent-free crude cell lysate containing 0.7 M sorbitol was prepared from KMY1516 cells carrying pRS423-IRE1-HA (WT or ΔLLSK) and incubated with the indicated reagents (final concentration) on ice for 10 min. The samples were then centrifuged at 100,000 g for 30 min, and pellet (P) and supernatant (S) fractions prepared from the same amount of samples were analyzed by anti-HA Western blotting. (E) Lysates from KMY1516 cells carrying pRS423-IRE1-HA (WT or mutant) were subjected to anti-HA immunoprecipitation, and the lysates and the IPs were analyzed by anti-HA or anti-BiP Western blotting to detect the indicated proteins. When indicated in A and E, Tun was added at 2-μg/ml final concentration into cultures 1 h before harvest. For vector control (−), cells carried an empty vector, pRS315 (B) or pRS423 (E).
Clustered Ire1-HA in DTT-treated cells quickly reverted to the normal ER-like distribution when the cells were incubated under nonstressed conditions (Fig. 2 F). This observation suggests that the cluster is not a simple aggregate of the protein.
Cluster formation of S103PΔIΔV Ire1-HA was observed even in cells carrying the hac1Δ mutation (Fig. 2 G) or when a kinase-inactive mutation, K702A, was introduced (Fig. 2 H). Thus, activation of either Ire1 or the UPR signaling pathway (Ire1-HAC1 pathway) is not a prerequisite for cluster formation. For cluster formation, the combination of the ΔI and ΔV mutations was necessary and sufficient, whereas the S103P mutation did not contribute (Fig. 2 I). Total cellular amount of Ire1-HA did not change considerably upon introduction of any of these mutations (Oikawa et al., 2007), and thus expression level is not an important determinant of the localization.
As presented in electron micrographs (Fig. 3 A), cells with multicopy expression of S103PΔIΔV Ire1-HA often showed abnormally folded cisternae that were probably derived from the ER. Importantly, immunogold labeling of anti-HA antibody demonstrated that the epitope was located on these membranous structures. These observations strongly suggest that Ire1 clusters on the membrane of the ER, which is deformed by the cluster formation. When the cluster is smaller, as in the case of single copy expression of Ire1-HA, the immunogold signals of the Ire1-HA clusters and the abnormal membranous structures are likely to be more difficult to find. Thus it is unclear whether cluster formation of endogenous Ire1 under ER-stress conditions leads to such morphological changes of the ER.
Many of the Ire1-HA clusters did not overlap with the DAPI signal (Fig. 3 B), indicating that the clustering is most likely occurring in regions that do not abut the nucleus. In Fig. 3 C, cells expressing both S103PΔIΔV Ire1-HA and ER marker protein GFP-Sec12 (Sato et al., 2003) were doubly stained with anti-HA and anti-GFP antibodies and observed using an ApoTome-based optical sectioning system. Most of the clusters (>90%) localized on or touched the anti-GFP–stained area, suggesting again that the clusters are formed in the ER. It is likely that not all of the clusters exactly localize on the anti-GFP–stained area because they may exclude other membrane proteins including GFP-Sec12. As shown in Fig. 3 D, the clusters did not colocalize with Golgi marker protein Mnn9.
Cluster formation of Ire1 is caused by two different types of homomeric interaction of the CSSR
According to the crystal structure (Credle et al., 2005), two CSSR molecules contact via two different interfaces (I and II; see Fig. 8), which suggests a possibility for formation of higher order oligomers. The crystal structure also predicts that the point mutations of Ire1, F247A and W426A (see Fig. 1 for the positions), respectively deform interfaces I and II. As shown in Fig. 4 A, both the F247A and the W426A mutations abolished cluster formation, although F247A Ire1-HA clustered in ∼20% of total cells treated with Tun. This finding strongly suggests that the cluster formation is caused by the high order oligomerization of the CSSR.
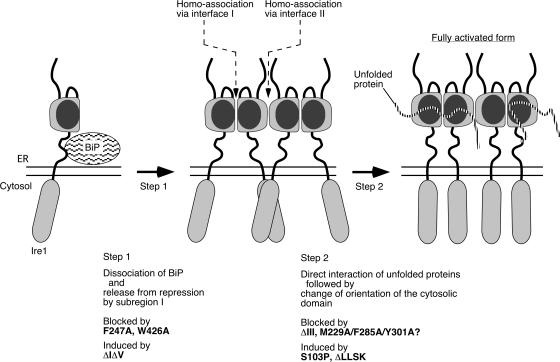
Figure 8.
Our current model for the mechanism by which ER stress activates Ire1. Our view about effects of the Ire1 mutations is also indicated. See text for details.
Notably, our present observations provide the first evidence for in vivo high order oligomerization of Ire1, which, as described in the Introduction, is not observed in analyses of cell lysates. One explanation for this discrepancy is that the homomeric association via interface I or II is too weak to maintain the cluster structure after lysis of cells. When ΔIΔV Ire1-HA is coexpressed with a C-terminally FLAG-tagged version (Ire1-FLAG) of the same mutant, the FLAG-tagged version is efficiently coimmunoprecipitated with the ΔIΔV Ire1-HA from the cell lysate even in the absence of ER stress (Fig. 4 B; Oikawa et al., 2007). The F247A mutation, but not the W426A mutation, considerably reduced the level of coimmunoprecipitation (Fig. 4 B). This finding suggests low affinity of the association via interface II.
In Fig. S3 (available at http://www.jcb.org/cgi/content/jcb.200704166/DC1), cells were treated with the noncleavable cross-linking reagent disuccinimidyl suberate, and the lysate was analyzed by anti-HA Western blotting. The amount of ΔIΔV Ire1-HA migrating to the original position was significantly decreased by treatment of cells with disuccinimidyl suberate, whereas wild-type Ire1-HA was less affected. The smear signal produced by high molecular mass protein is more intense in lane 4 than in lane 2 (Fig. S3), although highly cross-linked proteins may be harder to detect for several reasons, including extremely low mobility in the gel. Altogether, this finding supports highly efficient cross-linking and thus high order oligomerization of ΔIΔV Ire1-HA.
Cluster formation is necessary but not sufficient for activation of Ire1
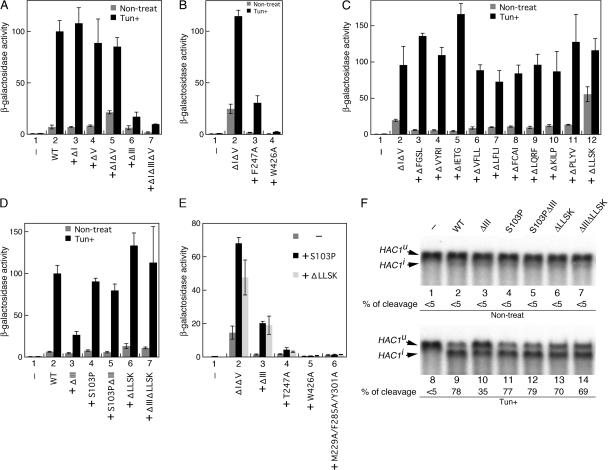
We then monitored activity of the Ire1 mutants by using a UPR element (UPRE)-reporter construct, from which expression of β-galactosidase is driven by a UPRE (Mori et al., 1992; Kohno et al., 1993). The activity of ΔI Ire1 and ΔV Ire1 was as tightly regulated as that of wild-type Ire1 (Fig. 5 A, 2–4), whereas ΔIΔV Ire1 was slightly activated even in nonstressed cells (Fig. 5 A, 5; Oikawa et al. 2007). Because extrinsic ER stress was required, even for full activation of ΔIΔV Ire1, which was constitutively clustered, we think that cluster formation is not sufficient for full activation of Ire1.
Figure 5.
Activity of Ire1 mutants. (A–E) Cellular β-galactosidase activity of KMY1015 (ire1Δ) cells carrying pRS315-IRE1-HA (wild type [WT] or mutant) and UPRE-lacZ reporter plasmid pCZY1 was assayed and is presented as the mean and standard deviation of three independent transformants, which has been normalized to the Tun + wild-type Ire1-HA control (set at 100) in all panels. Column 1 in each panel is vector control, in which the cells carried an empty vector pRS315. The value from wild-type or ΔIΔV Ire1-HA cells is presented in column 2. In column 3 and the subsequent columns, the mutations indicated on the x axis were introduced into WT (A and D) or ΔIΔV (B, C, and E) Ire1-HA. (C) The results from aa deletion scanning from aa 531–534 (ΔFGSL) to aa 567–570 (ΔLLSK) are presented in columns 2 and 3. The result from the N-terminal mutation (deletion of aa 527–530) was not reproducible (not depicted). (E) S103P, ΔLLSK, or no (−) mutation was additionally introduced as indicated. (F) Total RNA prepared from KMY1516 (ire1Δ) cells expressing untagged Ire1 variants from the WT or mutant versions of pRS313-IRE1 was analyzed by Northern blot detection of the HAC1 mRNA variants. For vector control (lanes 1 and 8), cells carried an empty vector pRS313. The percentage of HAC1u mRNA cleavage was estimated as described in Kimata et al. (2003). For Tun+ samples, Tun was added into cultures at 2 μg/ml final concentration 4 (A–D) or 1 h (F) before harvest.
When either of the clustering-impaired mutations, F247A or W426A, was introduced into ΔIΔV Ire1 (Fig. 5 B, 3 and 4) or wild-type Ire1 (Credle et al. 2005), the activity was considerably compromised. This finding strongly suggests that cluster formation is a prerequisite of Ire1 activation. Although the W426A mutation has a stronger negative effect on activity and cluster formation than the F247A mutation (Figs. 4 A and 5 B), coimmunoprecipitation of Ire1-FLAG with Ire1-HA was considerably impaired only by the F247A mutation (Fig. 4 B), suggesting again that a kind of homomeric association of Ire1 is not detected by the coimmunoprecipitation analysis.
A deletion of subregion III (hereafter called ΔIII; see Fig. 1 for position) significantly reduced activity of wild-type and ΔIΔV Ire1 (Fig. 5 A, 6 and 7). This finding was further confirmed by directly checking splicing of HAC1 mRNA (Fig. 5 F, lane 10). It should be noted that the ΔIII mutation did not impair cluster formation either by extrinsic ER stress or by the ΔIΔV mutation (Fig. 6 A), nor did it reduce cellular expression level of Ire1 (Fig. 6 B). These observations strongly suggest that in addition to cluster formation (see Step 1 in Fig. 8), the CSSR is responsible for another step, hereafter called Step 2 (see Fig. 8), which is also necessary for Ire1 activation.
Figure 7.
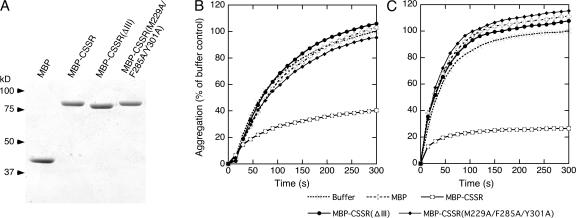
Direct interaction of CSSR with unfolded proteins as monitored by inhibition of in vitro aggregation. (A) Bacterially expressed MBP-CSSR, its mutant version, or unfused MBP were purified; run on 10% SDS-PAGE gel (1 μg of protein per lane); and stained with Coomassie blue. (B and C) At time 0, 25 μM of luciferase (B) or 50 μM of citrate synthase (C) in guanidine HCl–denaturing solution was 50- (for luciferase) or 66-fold (for citrate synthase) diluted into assay buffer containing MBP-CSSR, its mutant version, unfused MBP (2 μM each for luciferase or 0.5 μM each for citrate synthase), or buffer only. Turbidity of the sample mixtures was monitored, normalized against the maximal value of the buffer sample, and presented as aggregation.
Mutations that activate clustered Ire1
To address the involvement of the transmembrane domain in the activity of Ire1, we performed mutation scanning of this domain. ΔIΔV Ire1 was mutagenized such that it had 4 aa serial deletions, from aa 527–530 to 567–570 (see Fig. 1 for the positions), and was subjected to the UPRE-lacZ reporter assay, which showed that none of these mutations inactivate Ire1 (Fig. 5 C). On the contrary, ΔIΔV Ire1 was activated by the deletion of 4 aa residues, LLSK, located at the cytosolic end of the transmembrane domain even in the absence of ER stress (Fig. 5 C, 12; and see Fig. 1 for the position). This mutation, hereafter called ΔLLSK, did not significantly alter activity of wild-type Ire1 (Fig. 5 D [6] and F[lanes 6 and 13]). However, the impaired- activation phenotype of the ΔIII mutation was suppressed by the ΔLLSK mutation (Fig. 5 D [7] and F [compare lane 14 to 10]). Shift of mobility on SDS-PAGE by Endo H digestion indicates that ΔLLSK Ire1, as well as wild-type Ire1, is N-glycosylated (Fig. 6 C). We also checked whether wild-type and ΔLLSK Ire1 could be extracted from the yeast microsome fraction by various reagents (Fig. 6 D). Although both of the Ire1 variants were resistant to extraction with sodium chloride and sodium carbonate, they were partially extracted by Triton X-100. Together with ER localization (Fig. 6 A, bottom left) and activity of ΔLLSK Ire1, these observations show that the ΔLLSK mutation does not prevent Ire1 from being an ER-located type I transmembrane protein, although the LLSK residues may be a part of the transmembrane domain. Cluster formation of Ire1 was not affected by the ΔLLSK mutation (Fig. 6 A). Collectively, our results demonstrate that the ΔLLSK mutation abolishes the requirement of Step 2 for activation of Ire1, but not the requirement of the cluster formation.
S103P, as well as ΔLLSK, is likely to be a mutation that does not facilitate cluster formation (Fig. 2 I) but which activates clustered Ire1. The S103P mutation confers a constitutive-activation phenotype on ΔIΔV Ire1 (Fig. 5 E, 2; Oikawa et al., 2007) but not on wild-type Ire1 (Fig. 5 D, 4). Also, the S103P mutation suppressed the impaired-activation phenotype of the ΔIII mutation (Figs. 5 D [5] and F [compare lane 12 to 10]).
It is notable that the constitutive-activation phenotypes of both S103PΔIΔV Ire1 and ΔIΔVΔLLSK Ire1 were only moderately compromised by the ΔIII mutation, whereas ΔIΔIIIΔV Ire1 carrying neither the S103P nor the ΔLLSK mutation was almost completely inactive (Fig. 5 E, compare 3 to 2). In contrast, the clustering-impaired mutations F247A and W426A almost completely attenuated basal activity of ΔIΔV Ire1, even when carrying the S103P or the ΔLLSK mutation (Fig. 5 E, 4 and 5).
Fig. 6 E shows the results of a coimmunoprecipitation experiment that probed the physical interaction between Ire1 variants and BiP. As described in our previous papers (Okamura et al., 2000; Kimata et al., 2003, 2004; Oikawa et al., 2007), Ire1-HA or mutants were expressed from 2-μm plasmids, and the cells were lysed and subjected to anti-HA immunoprecipitation. Double bands of Ire1-HA in the top and third panels (Fig. 6 E), which were also shown in Figs. 6 D, S2, and S3, are caused by partial degradation in subregion I (Oikawa et al., 2007). Anti-BiP Western blotting of the anti-HA immunoprecipitates (Fig. 6 E, bottom) indicates BiP binding to Ire1-HA and its dissociation upon ER stress, neither of which were affected by either the ΔIII or the ΔLLSK mutation. S103P Ire1-HA also shows BiP binding and dissociation at similar levels to wild-type Ire1-HA (Oikawa et al., 2007).
The CSSR interacts directly with unfolded proteins
Finally, the possibility that the CSSR binds directly to unfolded proteins was explored by monitoring its ability to inhibit aggregation of denatured proteins in vitro. For this experiment, the CSSR was bacterially expressed as an N-terminally maltose binding protein (MBP)–fused and C-terminally His8-tagged recombinant protein, hereafter called MBP-CSSR. Integrity and purity of wild-type and mutant MBP-CSSRs and an unfused MBP control were verified by SDS-PAGE analysis (Fig. 7 A). Firefly luciferase and porcine citrate synthase were denatured by guanidine HCl, and then the aggregation induced by dilution of the denaturing mixtures was monitored by measuring the increase of turbidity. Fig. 7 (B and C) shows that aggregation of either denatured protein was not attenuated by the unfused MBP control, whereas MBP-CSSR showed significant effects in inhibiting the aggregation. This finding indicates that the CSSR can interact with two models of aggregation-prone substrates, which suggests that this domain binds to unfolded regions on polypeptides.
To demonstrate that activation of Ire1 is related to its direct interaction with unfolded proteins, we used the ΔIII mutation as an activation-impaired mutation. This is because, as shown in Fig. 5, the phenotype of the ΔIII mutation was suppressed either by the S103P or the ΔLLSK mutation, which indicates that the global structure of Ire1 is not perturbed by the ΔIII mutation. As shown in Fig. 7 (B and C), the ΔIII mutant version of MBP-CSSR did not attenuate aggregation of the denatured proteins.
Does the groove-like structure described in the Introduction contribute to interaction of the CSSR with unfolded proteins? We modified MBP-CSSR to carry combined substitutions of 3 aa residues (M229A/F285A/Y301A) facing into the groove, which abolish activation of Ire1 (Credle et al., 2005). This mutant MBP-CSSR did not inhibit aggregation of unfolded proteins (Fig. 7, B and C), supporting the idea that unfolded proteins are captured by the groove. Nevertheless, it is likely that the M229A/F285A/Y301A mutation confers more extensive damage to the CSSR than the ΔIII mutation, which may include perturbation of global protein structure, because the impaired-activation phenotype of M229A/F285A/Y301A Ire1 was not rescued either by the S103P or the ΔLLSK mutation (Fig. 5 E, 6).
Discussion
Here, we demonstrate that Ire1 clusters when activated. Impairment of the cluster formation, either by the F247A or the W426A mutation, strongly suggests that the molecular basis of the cluster formation is high order oligomerization of the CSSR (Credle et al. 2005). Importantly and conversely, our finding provides evidence for the high order oligomerization of Ire1, which, as detailed in the Introduction, has been unsupported by many of the previous biochemical analyses of cell lysates or solutions of recombinant Ire1 fragments. We think that the homomeric interaction via interface II is so weak that the association is undetectable in samples used in the biochemical analyses, where the concentration of Ire1 is probably much lower than its local concentration on the ER membrane. It is likely that cluster formation is not a result but a prerequisite of activation of Ire1. This is because Ire1 clustered even with the kinase mutation K702A or in hac1Δ cells and because activity of Ire1 was impaired by either the F247A or the W426A mutation. Clusters of wild-type Ire1 quickly dissociated upon removal of ER stress. This finding again implies that the cluster formation is biologically relevant. The molecular mechanism by which this cluster dissociation is promoted is currently unclear.
The cytosolic domain of yeast Ire1 carries a highly basic sequence, which, according to Goffin et al. (2006), acts as an NLS when fused with other proteins. However, our immunofluorescent analysis indicated that both unclustered and clustered Ire1 variants are distributed not only at the nuclear rim but also at other parts of the ER. We thus think that this highly basic sequence does not function as an NLS in the authentic Ire1 molecule.
Another feature of the CSSR that is predicted by the crystal structure but for which there is no supporting biochemical evidence is its direct binding to unfolded proteins (Credle et al. 2005). Here, we also demonstrate that the CSSR actually interacts with unfolded proteins by monitoring its ability to inhibit aggregation of denatured proteins in vitro. Because this property of the CSSR was abolished by the ΔIII mutation, we think that this interaction is biologically meaningful and required for activation of Ire1. The result from the M229A/F285A/Y301A mutation supports the idea that unfolded proteins are captured by the groove-like structure of the CSSR. We failed to demonstrate in vivo interaction between unfolded model proteins and Ire1 by coimmunoprecipitation, even from cells treated with a chemical cross-linker, dithiobis succinimidyl propionate. The interaction may be weak and transient, and in addition, we speculate that because of a structural reason, the cross-linking between the proteins is inefficient.
Unlike a previous scenario detailed in the last paragraph of the Introduction, we now believe that there exists a regulatory step, called here Step 2, other than the cluster formation. This is because ΔIΔV Ire1 was constitutively clustered, but extrinsic ER stress was still required for full activation of this mutant. Furthermore, the S103P and the ΔLLSK mutations are likely to abolish the requirement of Step 2 for activation of Ire1; in contrast, the ΔIII mutation impairs progression of Step 2 but not of the cluster formation. Importantly, such phenotypes of these mutations support our proposal about requirement of Step 2 for activation of Ire1.
What triggers cluster formation or Step 2? Unlike ΔIΔV Ire1, either ΔI or ΔV single mutant showed normal ER-like localization. This finding indicates that the cluster formation requires both dissociation of BiP or the BiP-nonbinding mutation ΔV, and release from repression by subregion I. The mechanism by which subregion I negatively regulates Ire1 remains unclear. Considering BiP's ability to recognize a wide variety of unfolded proteins, it is an attractive idea that BiP acts as a sensor for unfolded proteins in the cluster-formation step, although Ire1 may positively contribute to its own dissociation from BiP (Kimata et al., 2004). In contrast, BiP is not involved in Step 2 because the BiP-nonbinding mutant ΔIΔV Ire1 undergoes regulation in Step 2 and because none of the Step 2 mutants (ΔIII, S103P, or ΔLLSK) affect BiP binding and its dissociation from Ire1. Impairment of the interaction between denatured proteins and the CSSR protein carrying the ΔIII mutation strongly suggests that Step 2 is regulated by direct interaction of unfolded proteins with Ire1. Considering the constitutive cluster formation of ΔIΔV Ire1, we think that direct interaction of unfolded proteins is not required in Step 1.
Another important question is what change is produced in Step 2. Because the ΔLLSK mutation, which is located at the cytosolic end of the transmembrane domain, abolishes the requirement of Step 2 for full activation of Ire1, it is likely that orientation of the cytosolic domain is tightly related to Step 2. We propose that, as illustrated in Fig. 8, physical interaction of unfolded proteins with the CSSR causes conformational change of the luminal domain, which leads to reorientation of the cytosolic domain, without changing oligomerization status. This proposal is similar to the case for some cytokine receptors, which, upon binding of ligands, undergo not only self-association but also conformational change, causing alteration of cytosolic-domain orientation.
As a result of this work, we propose a model that is illustrated in Fig. 8. Importantly, ER stress provokes multiple events that separately contribute to activation of Ire1 at different steps. Cluster formation probably leads to considerably higher local concentration of the cytosolic effector domain of Ire1, which may be required for efficient cleavage of the HAC1u mRNA. Regulation by dual steps in different manners is likely to be important for precision of response by ensuring that Ire1 is only up-regulated by ER stress. Indeed, either ethanol or high temperature inappropriately activates ΔV Ire1 (Kimata et al., 2004).
Nevertheless, it is likely that not all events noted in Fig. 8 are required to obtain partial activation of Ire1. Indeed, the constitutively clustering mutant ΔIΔV Ire1 is slightly but significantly activated even without extrinsic ER stress. Furthermore, Liu et al. (2000) reported substantial activation of chimeric Ire1 mutants in which the luminal domain was replaced by dimer-forming fragments of transcription factor proteins. More recently, Lipson et al. (2006) reported that transient activation of mammalian IRE1α in pancreatic β cells exposed to high levels of glucose does not accompany BiP dissociation.
Is the mechanism presented here applicable to Ire1 orthologues? Because mammalian and plant orthologues of Ire1 do not carry regions corresponding to subregion I, their self-association may be regulated solely by BiP. It should be noted that Zhou et al. (2006) reported a crystal structure of the luminal domain of mammalian IRE1α, which suggests that neither high order oligomerization nor direct binding of unfolded protein is likely. Unlike cells of the unicellular organism yeast, which suffer direct environmental stress and carry only one known ER-stress sensor Ire1, mammalian cells live in sophisticatedly regulated conditions and have more complicated pathways to respond to ER stress. Thus, it is not unreasonable to postulate that the regulatory mechanisms of the mammalian ER-stress sensors are different from those of yeast Ire1. Nevertheless, we do not think that mammalian IRE1α is regulated solely by BiP because a mutant of this protein with a deletion of the entire region corresponding to subregion V was still regulated by ER stress (unpublished data).
Whereas PERK's activity is apparently regulated by binding and dissociation of BiP (Bertolotti et al., 2000; Ma et al., 2002), a recombinant fragment of its luminal domain has been shown to inhibit aggregation of unfolded proteins in vitro (Yohda, M. et al. 2006. Proceedings of the 20th International Union of Biochemistry and Molecular Biology Congress and the 11th Federation of Asian and Oceanian Biochemists and Molecular Biologists Congress). Therefore, it is likely that in a similar manner to yeast Ire1, PERK is regulated both by BiP and by direct binding of unfolded proteins. Interestingly, ATF6 may also be regulated dually in its activation upon ER stress, although it has no structural similarity to Ire1 or PERK. Activation (i.e., transport to Golgi apparatus) of ATF6 is negatively regulated by binding of BiP and by intra- and intermolecular disulfide bridge formation, both of which are lowered upon ER stress (Shen et al., 2002; Nadanaka et al., 2007). Finally, such multiplicity of regulatory mechanisms implies complexity of conditions in which these ER-stress sensors are individually activated, as suggested by DuRose et al. (2006).
Materials and methods
Yeast strains and plasmids
Yeast congenic haploid strains KMY1015, KMY1516, and KMY1520 (Kimata et al., 2004), all of which carry an ire1Δ null mutation, were used according to the difference of their mating and auxotrophic phenotypes. To generate the ire1Δ hac1Δ strain YKY1004, a EUROSCARF strain Y15650 (MATα ura3 leu2 his3 lys2 hac1∷kanMX4; provided by J.W. Goethe, University Frankfurt, Frankfurt, Germany) was modified to carry a complete deletion of the IRE1 ORF by replacement with the URA3 gene. To obtain a YKY2005 strain, the Ire1-HA gene was knocked in to replace the endogeneous IRE1 gene of wild-type strain KMY1005 (MATα ura3 leu2 his3 trp1 lys2), which is congenic to KMY1015, KMY1516, and KMY1520. Cells were exponentially cultured in SD medium (2% glucose and 0.66% yeast nitrogen base without amino acids; Difco), supplemented with appropriate nutrients, at 30°C.
In our previous studies (Okamura et al., 2000; Kimata et al., 2003, 2004), plasmids pRS313-IRE1, pRS315-IRE1-HA, pRS423-IRE1-HA, and pRS426-IRE1-FLAG were generated by insertion of the IRE1 gene (or its C-terminally epitope-tagged version) into yeast centromeric vectors pRS313 and pRS315 (Sikorski and Hieter, 1989) and 2-μm vectors pRS423 and pRS426, respectively (Christianson et al., 1992). As described in Kimata et al. (2004), we introduced IRE1 mutations into these plasmids by in vivo homologous recombination (gap repair) between the IRE1 plasmids cleaved by restriction enzymes and IRE1 mutant fragments created by overlap PCR, primers for which are listed in Table S1 (available at http://www.jcb.org/cgi/content/jcb.200704166/DC1). A UPRE-lacZ reporter plasmid pCZY1 (URA3 2 μm) was provided by K. Mori (Kyoto University, Kyoto, Japan). A GFP-Sec12 expression plasmid pSKY54-GFP-SEC12 was provided by A. Nakano (Institute of Physical and Chemical Research, Saitama, Japan). A pRS315-based derivative of pSKY54-GFP-SEC12 was generated by in vivo homologous recombination between PvuII-digested pSKY54-GFP-SEC12 and XhoI–NotI–digested pRS315.
Fluorescent microscopy
For fluorescent microscopic examination, cells were fixed in 0.1 M potassium acetate buffer, pH 6.8, containing 3.3% formaldehyde for 2 h and processed according to Oka et al. (1998). Antibodies used are listed in Table S2 (available at http://www.jcb.org/cgi/content/jcb.200704166/DC1), including anti-Mnn9 antiserum, which was provided by Y. Noda (The University of Tokyo, Tokyo, Japan). The mounting medium was 90% glycerol containing 0.1% p-phenylenediamine. For conventional fluorescent microscopy, Axiophoto (Carl Zeiss MicroImaging, Inc.) was used with an oil immersion lens (Plan-Neofluor 100/1.30), and images were captured by a digital charge-coupled device (CCD) camera system (DP70; Olympus) carrying built-in software for image acquisition. For deconvolution microscopy, an Axiovert 200M (100/1.40 oil immersion Plan Apochromat objective; Carl Zeiss MicroImaging, Inc.) with the Apotome system was used. Unless noted, FITC was used as fluorochrome. Photoshop software (Adobe) was used for conversion to grayscale images (Figs. 2, 4 A, and 6 A) and image overlapping (Fig. 3 B).
Electron microscopy
Cells were fixed basically as described previously in Sun et al. (1992). Cells were frozen in a high pressure freezer (HPM010; Bal-Tec Inc.) and transferred to anhydrous acetone containing 2% OsO4 in an automatic freeze-substitution apparatus (EM AFS; Leica) in which the temperature was gradually sifted from −80 to 23°C. After washing three times with anhydrous acetone, the samples were infiltrated with increasing concentrations of Spurr's resin in anhydrous acetone, and finally with 100% Spurr's resin. After polymerization in capsules at 60°C, ultrathin sections were cut on a microtome (Ultracut UCT; Leica). The sections were immunostained with anti-HA antibody and 10 nm of gold immunogold conjugate EM goat anti–mouse IgG (Table S2) and stained with 3% uranyl acetate for 2 h. The sections were then examined with an electron microscope (H-7600; Hitachi) at 100 kV.
RNA and protein analyses of yeast cells
RNA extraction, Northern blotting, cell lysis for protein analyses, immunoprecipitation, and Western blotting were performed as described previously (Kimata et al., 2003, 2004). Detergent-free cell lysates used to obtain microsome fractions were prepared as described in Craven et al. (1996). Radioactive signal from Northern blots was detected using a phosphor imager (BAS-2500; Fuji). For SDS-PAGE, lysates from 107 cells and IPs from 3 × 107 cells were run on precast gels (Multigel II Mini; Daiichi Pure Chemicals; 7.5% acrylamide), unless otherwise noted. The chemiluminescent signal from Western blots was captured by a cooled CCD camera system (LAS-1000plus; Fuji) and quantified using imaging software (ImageGauge; Fuji). When obtaining the Ire1-FLAG/Ire1-HA values in Fig. 4 B, we confirmed the linear relation between the quantified chemiluminescent signal and the actual amount of the band protein. Antibodies used, including that against yeast BiP (Higashio et al., 2000), are listed in Table S2.
Bacterial expression and purification of MBP-CSSR and its mutant variants
To avoid artifactual disulfide bond formation, all versions of MBP-CSSR had all Cys residues changed to Ser because this amino acid replacement does not affect activity of Ire1 (Oikawa et al., 2005). An IRE1 gene partial fragment corresponding to the CSSR was PCR amplified from pRS315-IRE1(CS)-HA (Oikawa et al., 2005) using primer set P-5 and P-6, digested with BamHI and HindIII, and inserted into similarly digested pMAL-c2x (New England BioLabs, Inc.). To generate mutant variants, mutations were introduced by using the overlap PCR mutagenesis technique as described in Kimata et al. (2004). The PCR primers are listed in Table S1.
An Escherichia coli strain BL21 codon plus (DE3)–RIL (Strategene) was transformed with one of the resulting plasmids and cultured at 37°C in 400 ml of 2× YT medium. Expression of MBP-CSSR or its mutant variants was induced by addition of IPTG (0.3 mM final concentration) into the culture, followed by further incubation at 30°C for 1 h. After harvest, cells were suspended in 15 ml of E. coli lysis buffer (50 mM Hepes, pH 8.0, 300 mM KCl, 5 mM MgCl2, 10 mM imidazole, 1% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, 0.4 mg/ml benzamidine, 0.4 mg/ml pepstatin A, 0.4 mg/ml leupeptin, 0.3 mg/ml lysozyme, and 14 U/ml DNase I; Takara) and disrupted by ultrasonication. The lysate was clarified by centrifugation (SRX-201; Tomy; 8,200 rpm for 10 min) and incubated with 0.5 ml of HisLink protein purification resin beads (Promega) for 12 h. The beads were packed into an 8-mm-diam column and sequentially washed with 6 ml of 50 mM Hepes, pH 8.0, 1 M KCl, 5 mM MgCl2, and 0.1% Triton-X 100; 6 ml of 50 mM Hepes, 300 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, and 20 mM imidazole; 6 ml of 50 mM Hepes, 300 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, and 40 mM imidazole; 3 ml of 50 mM Hepes, 300 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, and 60 mM imidazole; 5 ml of 50 mM Hepes, 300 mM KCl, 5 mM MgCl2, and 10 mM ATP; and 3 ml of 20 mM Hepes, 100 mM KCl, 5 mM MgCl2, and 50% (vol/vol) glycerol. Bead-bound proteins were eluted with 50 mM Hepes, 100 mM KCl, 5 mM MgCl2, 200 mM imidazole, and 50% (vol/vol) glycerol, and elution fractions (1 ml each fraction) were analyzed by SDS-PAGE.
In vitro protein aggregation assay
Citrate synthase (Roche) was dialyzed against 20 mM Hepes, pH 7.0, 150 mM KCl, 2 mM MgCl2, and 10% (vol/vol) glycerol, and stored at −80°C. Luciferase (25 μM final concentration) or citrate synthase (50 μM final concentration) were denatured by incubation in guanidine HCl– denaturing solution (guanidine HCl [6 M for luciferase or 4 M for citrate synthase], 20 mM Hepes, 50 mM KCl, and 2 mM MgCl2) for 30 min at room temperature. The denaturing mixture was then diluted with assay buffer (20 mM Hepes, pH 7.2, 50 mM KCl, and 2 mM MgCl2) in the presence or absence of MBP-CSSR (wild type or mutant) or MBP, and aggregate formation was monitored by absorbance at 320 nm with a spectrophotometer (DU640; Beckman Coulter) at room temperature.
Online supplemental material
Table S1 lists Ire1 mutations analyzed in this study. PCR primers used for generation of these mutations are also listed. Table S2 lists antibodies used in this study. Fig. S1 shows that even when expressed from a multicopy plasmid, Ire1-HA was activated in an ER stress-dependent manner. In Fig. S2, Ire1-HA was expressed from a single copy plasmid, a multicopy plasmid, and the knocked in gene, and the expression levels were compared. Fig. S3 shows highly efficient intermolecular homo-cross-linking of ΔIΔV Ire1-HA, which supports high order oligomerization of this molecule. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704166/DC1.
Acknowledgments
We thank Junko Iida for technical assistance, Drs. Yoichi Noda and Koji Yoda for anti-Mnn9 antiserum, Dr. Kazutoshi Mori for plasmid pCZY1, and Drs. Akihiko Nakano and Ryogo Hirata for plasmid pSKY54-GFP-SEC12.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (14037240 to K. Kohno and 18050024 to Y. Kimata) and for 21st Century Center of Excellence Research from the Ministry of Education, Culture, Sports, Science and Technology and by a Grant-in-Aid from the Japanese Society for the Promotion of Science (KAKENHI 18570179 to Y. Kimata).
Abbreviations used in this paper: CSSR, core stress-sensing region; IP, immunoprecipitate; MBP, maltose binding protein; MHC, major histocompatibility complex; PERK, pancreatic ER kinase; Tun, Tunicamycin; UPR, unfolded protein response; UPRE, UPR element.
References
- Bertolotti, A., Y. Zhang, L.M. Hendershot, H.P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326–332. [DOI] [PubMed] [Google Scholar]
- Christianson, T.W., R.S. Sikorski, M. Dante, J.H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene. 110:119–122. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 87:391–404. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., C.E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. [DOI] [PubMed] [Google Scholar]
- Craven, R.A., M. Egerton, and C.J. Stirling. 1996. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of several protein precursors. EMBO J. 15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Credle, J.J., J.S. Finer-Moore, F.R. Papa, R.M. Stroud, and P. Walter. 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 102:18773–18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuRose, J.B., A.B. Tam, and M. Niwa. 2006. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol. Biol. Cell. 17:3095–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin, L., S. Vodala, C. Fraser, J. Ryan, M. Timms, S. Meusburger, B. Catimel, E.C. Nice, P.A. Silver, C.Y. Xiao, et al. 2006. The unfolded protein response transducer Ire1p contains a nuclear localization sequence recognized by multiple beta importins. Mol. Biol. Cell. 17:5309–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, H.P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 397:271–274. [DOI] [PubMed] [Google Scholar]
- Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 10:3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashio, H., Y. Kimata, T. Kiriyama, A. Hirata, and K. Kohno. 2000. Sfb2p, a yeast protein related to Sec24p, can function as a constituent of COPII coats required for vesicle budding from the endoplasmic reticulum. J. Biol. Chem. 275:17900–17908. [DOI] [PubMed] [Google Scholar]
- Kimata, Y., Y.I. Kimata, Y. Shimizu, H. Abe, I.C. Farcasanu, M. Takeuchi, M.D. Rose, and K. Kohno. 2003. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell. 14:2559–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata, Y., D. Oikawa, Y. Shimizu, Y. Ishiwata-Kimata, and K. Kohno. 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 167:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata, Y., Y. Ishiwata-Kimata, S. Yamada, and K. Kohno. 2006. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 11:59–69. [DOI] [PubMed] [Google Scholar]
- Kohno, K. 2007. How transmembrane proteins sense endoplasmic reticulum stress. Antioxid. Redox Signal. In press. [DOI] [PubMed]
- Kohno, K., K. Normington, J. Sambrook, M.J. Gething, and K. Mori. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson, K.L., S.G. Fonseca, S. Ishigaki, L.X. Nguyen, E. Foss, R. Bortell, A.A. Rossini, and F. Urano. 2006. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4:245–254. [DOI] [PubMed] [Google Scholar]
- Liu, C.Y., M. Schroder, and R.J. Kaufman. 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275:24881–24885. [DOI] [PubMed] [Google Scholar]
- Liu, C.Y., H.N. Wong, J.A. Schauerte, and R.J. Kaufman. 2002. The protein kinase/endoribonuclease IRE1alpha that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J. Biol. Chem. 277:18346–18356. [DOI] [PubMed] [Google Scholar]
- Ma, K., K.M. Vattem, and R.C. Wek. 2002. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J. Biol. Chem. 277:18728–18735. [DOI] [PubMed] [Google Scholar]
- Mori, K., A. Sant, K. Kohno, K. Normington, M.J. Gething, and J.F. Sambrook. 1992. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11:2583–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K., W. Ma, M.J. Gething, and J. Sambrook. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 74:743–756. [DOI] [PubMed] [Google Scholar]
- Nadanaka, S., T. Okada, H. Yoshida, and K. Mori. 2007. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol. Cell. Biol. 27:1027–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa, D., Y. Kimata, M. Takeuchi, and K. Kohno. 2005. An essential dimer- forming subregion of the endoplasmic reticulum stress sensor Ire1. Biochem. J. 391:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa, D., Y. Kimata, and K. Kohno. 2007. Self-association and BiP dissociation are not sufficient for activation of the ER stress sensor Ire1. J. Cell Sci. 120:1681–1688. [DOI] [PubMed] [Google Scholar]
- Oka, M., M. Nakai, T. Endo, C.R. Lim, Y. Kimata, and K. Kohno. 1998. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 273:29727–29737. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Y. Kimata, H. Higashio, A. Tsuru, and K. Kohno. 2000. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 279:445–450. [DOI] [PubMed] [Google Scholar]
- Papa, F.R., C. Zhang, K. Shokat, and P. Walter. 2003. Bypassing a kinase activity with an ATP-competitive drug. Science. 302:1533–1537. [DOI] [PubMed] [Google Scholar]
- Sato, K., M. Sato, and A. Nakano. 2003. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol. Biol. Cell. 14:3605–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu, C.E., and P. Walter. 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 3:99–111. [DOI] [PubMed] [Google Scholar]
- Sidrauski, C., and P. Walter. 1997. The transmembrane kinase Ire1p is a site- specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 90:1031–1039. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, G.H., A. Hirata, Y. Ohya, and Y. Anraku. 1992. Mutations in yeast calmodulin cause defects in spindle pole body functions and nuclear integrity. J. Cell Biol. 119:1625–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, K.J., C.K. Patil, L. Wodicka, D.J. Lockhart, J.S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. [DOI] [PubMed] [Google Scholar]
- Zhou, J., C.Y. Liu, S.H. Back, R.L. Clark, D. Peisach, Z. Xu, and R.J. Kaufman. 2006. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA. 103:14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]