Abstract
Viroids are small non-coding parasitic RNAs that are able to infect their host plants systemically. This circular naked RNA makes use of host proteins to accomplish its proliferation. Here we analyze the specific binding of the tomato protein Virp1 to the terminal right domain of potato spindle tuber viroid RNA (PSTVd). We find that two asymmetric internal loops within the PSTVd (+) RNA, each composed of the sequence elements 5′-ACAGG and CUCUUCC-5′, are responsible for the specific RNA–protein interaction. In view of the nucleotide composition we call this structural element an ‘RY motif’. The RY motif located close to the terminal right hairpin loop of the PSTVd secondary structure has an ∼5-fold stronger binding affinity than the more centrally located RY motif. Simultaneous sequence alterations in both RY motifs abolished the specific binding to Virp1. Mutations in any of the two RY motifs resulted in non-infectious viroid RNA, with the exception of one case, where reversion to sequence wild type took place. In contrast, the simultaneous exchange of two nucleotides within the terminal right hairpin loop of PSTVd had only moderate influence on the binding to Virp1. This variant was infectious and sequence changes were maintained in the progeny. The relevance of the phylogenetic conservation of the RY motif, and sequence elements therein, amongst various genera of the family Pospiviroidae is discussed.
INTRODUCTION
Viroids are subviral RNA pathogens that are able to infect some higher plants. They are unique for their small size and their propagation strategy [for review and general statements on viroids see (1–4)]. Their genome consists of a single-stranded, covalently closed RNA molecule, which adopts in most cases an unbranched rod-like secondary structure. Despite their simplicity, viroids are able to replicate autonomously. This capability is remarkable since there is no indication that the viroid RNA has the capacity to encode a peptide. Nevertheless, the non-coding viroid RNA supplies genetic information such as replicapability, pathogenicity, mobility and host-specificity. It is assumed that the RNA sequence folds in a specific secondary structure that provides binding signals for host (protein) factors. It is further assumed that these factors exert in combination with the viroid RNA the genetic program of this RNA replicon.
Potato spindle tuber viroid (PSTVd) is the type species of the genus Pospiviroid and in general of the family Pospiviroidae. Its circular genome consists of 359 nucleotides (5). Pospiviroidae are replicated in the nucleus by DNA-dependent RNA polymerase II via double-stranded RNA intermediates, in an asymmetric rolling-circle-like mechanism (6–10).
In agreement with the assumption that structural domains are responsible for the interaction with host factors, five sequence elements could be defined for PSTVd and related viroids almost two decades ago (11). These are the terminal left (TL) domain, the pathogenicity (P) domain, the less conserved variable (V) domain, the innermost central conserved region (CCR) and the terminal right (TR) domain. All these domains can also be used for phylogenetic classification (12). Each domain is described by a particular structure and is assumed to contribute to certain viroid functions in vivo. For example the CCR region has been shown to be relevant for processing of the linear oligomeric PSTVd RNA to the circular monomer (13–17). Further insights into the function of the different domains came from the construction of chimeric viroids (18–20). Even a single nucleotide change may decide whether PSTVd is able to replicate in Nicotiana tabacum (21,22), suggesting that this change could modulate interaction with one or several host factors.
Despite their functional importance for viroids, only a few host proteins have been reported to interact with this unique pathogenic RNA. For Pospiviroidae this includes histones and two uncharacterized proteins of 31 and 41 kDa (23), the ribosomal protein S25 (24), the large subunit for RNA polymerase II (10) and phloem protein 2 (25,26). Further, for two chloroplast RNA-binding proteins interaction was demonstrated with the RNA of avocado sunbotch viroid (ASBVd) (27), the type strain of chloroplast viroids of the family Asunviroidae. However, for none of these RNA–protein interactions a precise sequence element of a viroid RNA could be made responsible so far.
Using our RNA ligand-screening protocol (28) we have previously identified the viroid RNA binding protein (Virp1) from tomato by its capability to specifically interact with PSTVd (+) RNA (29). Virp1 contains, besides a nuclear localization signal (NLS), a bromodomain, which is found in a variety of proteins thought to play a role in dynamic chromatin (30). Virp1 is the first bromodomain-containing protein for which affinity for RNA has been reported. Moreover, we have previously demonstrated by immunoprecipitation and UV-cross-link experiments that Virp1 interacts with PSTVd (+) RNA in vivo (29). Virp1 is expressed in different tissues of healthy plants, including petals, which are normally not infected by PSTVd (22). We could also show that in PSTVd-infected plants Virp1 expression was down regulated. The potential role of Virp1 in healthy plants and during viroid infection has been discussed previously (29). Applying the yeast three-hybrid system, we could localize the viroid RNA region that is responsible for specific interaction with Virp1 in the TR domain of PSTVd (31). A partial PSTVd (+) RNA containing about 80 nucleotides of the TR domain was sufficient for specific binding. This analysis included also some variants of the TR domain, which had been described earlier by Hammond (32). We noticed a reduced binding affinity of Virp1 for the PSTVd-R+ mutant. This PSTVd variant contained a single nucleotide insertion and was only infectious when Agrobacterium-mediated inoculation was used, but delivered progeny with further, differently altered sequence variants. We also assayed those sequence variants and found that the affinity for Virp1 was partially restored (31). Based on these data we re-discovered a short conserved sequence element that is present twice in the TR of PSTVd. Already in 1987, Symons and co-workers pointed out that the sequence 3′-CUUCC-5′ (nucleotides 188–184) in the lower strand of the TR domain is highly conserved amongst viroids closely related to PSTVd (33). In the typical rod-like viroid secondary structure of PSTVd, the three 5′-terminal nucleotides of this motif—nucleotides 3′-UCC-5′ (nucleotides 186–184)—base-pair with the three nucleotides 5′-AGG-3′ (nucleotides 173–175) of the upper strand of the TR domain. The residual two nucleotides U187 and C188 of the conserved pentamer sequence form, together with C189, the longer single stranded region of an asymmetric internal loop that is closed by U190, which base-pairs with A171 (compare Fig. 1). The same loop motif re-occurs between A150 and G154, and U209 and C203. Within this structural motif, the lower strand is composed solely of pyrimidine residues and each pentamer in the top strand is composed of purines with the exception of the looped C residue. In view of this base composition we call this asymmetric internal loop structure an ‘RY motif’.
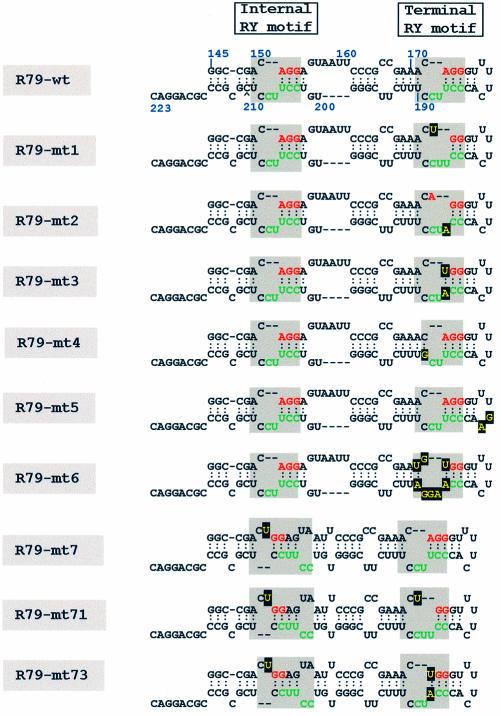
Figure 1.
Sequence and secondary structure of the 79 base PSTVd transcript R79-wt and its sequences variants. Secondary structures were determined by the RNA fold program (42) using the web tool http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1-2.3.cgi. Shaded boxes indicate the internal and terminal RY motifs; the sequence elements found in other Pospiviroidae are color-coded as in Figure 6. Nucleotides specifically introduced into the different sequence variants are boxed. The internal RY motif is found if the temperature is set to 11°C or lower.
In this context it is of interest that the sequence alterations in mutant PSTVd-R+ (32) prevented the formation of the typical structural interaction in the terminally located RY motif. The nucleotides exchanges in the progeny obtained after Agrobacterium-mediated inoculation of PSTVd-R+ restored the original base pair interaction despite further sequence alterations in the terminal right loop. Since the binding of Virp1 to the TR domain of PSTVd correlated with the potential to form this element of secondary structure (31) we decided to analyze its influence on the specific interaction of Virp1 with PSTVd (+) RNA in greater detail. Here we describe the introduction of several mutations in either of the two RY motifs located within the PSTVd TR domain and the influence of these sequence manipulations on the binding affinity of Virp1 and on the viroid infectivity in tomato plants.
MATERIALS AND METHODS
PSTVd RNAs
Two kinds of PSTVd (+) RNAs of isolate KF-440-2 (accession number X58388) were used in this study. The longer-than-unit-length PSTVd (+) RNA was obtained by in vitro transcription as described by Hammann et al. (34) using EcoRI-linearized plasmid pHa106 (13). The same plasmid was used for the synthesis of minus RNA after linearization with HindIII. The RNA transcript consisting of the 79 PSTVd-specific nucleotides 145–223 (R79-wt) was obtained after PCR amplification of pHa106 plasmid DNA with the two DNA oligonucleotides 19GEM (5′-CCGGAATTCGAGCTCGCCC) and T3Sau (5′-GGTCTAGAAATTAACCCTCACTAAAGGGCCGACAGGAGTAATTCC). The resulting PCR product was cloned in the T-easy vector system (Promega, Madison, WI), yielding plasmid pR79-wt. After digestion with AvaII, plasmid pR79-wt was transcribed with T3 RNA polymerase delivering transcript R79-wt. All mutations described were introduced into plasmid pHa106 and plasmid pR79-wt.
Depending on the distance of the mutations to be introduced, two in vitro mutagenesis protocols were used: first, the Gene Editor in vitro site-directed mutagenesis system (Promega), which is based on the simultaneous annealing of two oligonucleotides to the pHa106 DNA template. One of the two oligonucleotides was in each mutagenesis experiment the selection oligonucleotide ‘bottom strand’ (Promega), 5′-pCCGCGAGACCCACCCTTGGAGGCTCCAGATTTATC, which is specific for the region of the β lactamase gene contained in the pHa106 vector sequence. The second oligonucleotide carried the actual mutation and was one of the following ‘mutagenic’ oligonucleotides mt1, 5′-CCCGCCGAAACTGGGTTTTCACCCTTCCTTTCTTC; mt2, 5′-CCCGCCGAAACTGGGTTTTCACCCATCCTTTCTTC; mt3, 5′-CCCGCCGAAACTGGGTTTTCACCCATCCTTTCTTC; mt5, 5′-CCCGCCGAAACAGGGTTTGAACCCTTC (bold indicates altered nucleotides). After synthesis and ligation of the mutant strand the desired mutation had already been linked to the antibiotic selection marker. To avoid mismatch repair in vivo, two subsequent transformation events were performed according to the protocol provided by Promega. The Gene Editor system showed in our hands low efficiency and fidelity and therefore a second protocol for in vitro mutagenesis was applied. The megaprimer method (35) consisted of two amplification steps. In the first amplification step we used plasmid pHa106 as a template for PCR, in combination with a DNA oligonucleotide specific for the promoter region of T7 RNA polymerase (New England Biolabs, Beverly, MA) and one of the following ‘mutagenic’ DNA oligonucleotides, respectively: mt4, 5′-CACCCTTCGTTTCTTCG; mt6, 5′-CGCCGAATGTGGGTTTTCACCCAAGGAATTCTTCG; mt7, 5′-GCCCAGCGGCCGACT GGAGTAA (bold indicates altered nucleotides). The resulting DNA fragments were subjected to a second PCR reaction using again pHa106 as template in combination with DNA oligonucleotide 42pGEM, 5′-CGAGAGAGATGATAGGGTCTGCTTCAGTAAGCCAGATGCTAC. The amplified DNA was then cloned into the T-easy vector system (Promega) yielding plasmid pT-Ha106-mt4/mt6/mt7. About 1 µg of these plasmids was digested with EcoRI and used as templates for in vitro transcription with SP6 RNA polymerase. The two constructs Ha106-mt71 and Ha106-mt73 that carried two types of sequence variations were done in the same strategy, however, using pHa106-mt1, pHa106-mt3 as templates.
To generate the same sequence manipulations in the context of R79-wt we used the differently altered plasmids pHa106 as templates for PCR with the two DNA oligonucleotides 19GEM and T3Sau as described above. For generating the mutations in R79-mt7 instead of DNA oligonucleotide T3Sau the DNA oligonucleotide MT3Sau, 5′-GGTCTAGAAATTAACCCTCACTAAAGGGCCGACTGGAGTAATTCC was used. All resulting DNA fragments were subjected to sequence analysis, confirming the correct sequence manipulations.
Expression and purification of Virp1Δ
The plasmid pHis-Virp1Δ encoding Virp1Δ, an N-terminally truncated form of Virp1 with a His-tag contained all domains necessary for specific interaction with PSTVd (+) RNA and has been described previously (29). Cultivation of 1 l Escherichia coli (DE3) carrying the plasmid was done as described previously (36). The protein was purified on a Ni-NTA agarose column under native conditions according to a protocol provided by the manufacturer (Qiagen, Hilden, Germany). In brief: cells were thawed on ice for 15 min followed for each of six tubes by re-suspension in 1 ml lysis buffer (50 mM Tris–HCl, 300 mM NaCl, 10 mM imidazole, pH 8.0), supplemented with lysozyme to a final concentration of 1 mg/ ml. After incubation for 30 min on ice, the lysates were centrifuged at 4°C for 30 min at 18 000 g. The combined supernatants were mixed with equal volume of Ni-NTA agarose (Qiagen) from which the ethanol had been replaced by water. After incubation for 1 h at 4°C under slow rotation, the lysate was transferred to a small column with closed outlet. The column was washed twice with 10 ml washing buffer (50 mM Tris–HCl, 300 mM NaCl, 20 mM imidazole, pH 8.0). Finally, the protein was eluted by the addition of 5 ml elution buffer (50 mM Tris–HCl, 300 mM NaCl, 250 mM imidazole, pH 8.0). The sample was concentrated to about one-fifth of the volume in a Centricon Centrifugal Filter Device YM-30 (Millipore, Billerica, USA). After addition of 1 ml of dilution buffer containing Tris–HCl 20 mM pH 7.5, 150 mM NaCl, the sample was again concentrated to about half (further dilution of salt concentration might cause unspecific protein–protein association). The quality of the purified protein was analyzed on a 12% SDS–PAGE and its concentration was determined (37).
Electrophoresis mobility shift assay (EMSA)
All reactions of complex formation between Virp1Δ and PSTVd (+) RNA were carried out at room temperature for 30 min in a total volume of 10 µl binding buffer with a final concentration of 10 mM HEPES–KOH, 50 mM KCl, 100 µM EDTA pH 7.9 and 5% glycerol. All reactions contained 1 µg/µl of baker’s yeast tRNA (Roche Molecular Biochemicals, Switzerland). The concentration of Virp1Δ and PSTVd (+) RNA was varied as indicated in the Results. The chemical concentration of the radioactively labeled RNA was ∼1 fM and thus much lower than the lowest protein concentration tested. The RNA–protein complexes were separated by electrophoresis on a 6% non-denaturing polyacrylamide gel (acrylamide:bisacylamide 29:1) using as running buffer 0.5× TBE (50 mM Tris, 50 mM boric acid and 1 mM EDTA, pH 8.3) at ∼10 V/cm at room temperature. The gels were dried and visualized either by autoradiography or quantified by phosphorimager analysis (Storm 840, Molecular Dynamics) with the aid of Image Quant software.
Northwestern assay
Northwestern assays were performed as described by Martinez de Alba et al. (29). The northwestern blot was followed by immunoblot analysis. As a primary antibody we used rabbit polyclonal antibody raised against Virp1Δ, or a Penta-His antibody (Qiagen). As a secondary antibody we used an alkaline-phosphatase conjugated anti-rabbit antibody (Promega) and a HRP-anti-mouse antibody (Jackson Immuno Research Laboratories), respectively.
Infectivity assays
In vitro synthesized RNA transcripts of the variants of Ha106 carrying the engineered nucleotide exchanges were prepared. 100 ng of the RNA were assayed for infectivity on young tomato plants (cultivar Rentita) as described (38). Four weeks after inoculation total RNA was extracted from each plant as described (39). RNA samples (∼7 µg) were separated on a 5% denaturing gel (40). Samples were electroblotted and hybridized as described (39) using as probe in vitro synthesized PSTVd (–) RNA (pHa106, linearized with HindIII and transcribed with T7 RNA polymerase) using ∼2 × 105 c.p.m./ml.
Sequence analysis of viroid progeny
Samples with detectable signals of PSTVd were subjected to a reverse transcription kit (RT–PCR kit, Invitrogen, Carlsbad, USA), using DNA oligonucleotides pstvdbamrev: 5′-GGGGATCCCTGAAGCGCTCCTCCGAGC and pstvdbamfor: 5′-AGGGATCCCCGGGGAAACCTGGAGCGA, by using a temperature of 60°C for 50 min as annealing temperature for reverse transcription. After 2 min at 94°C 35 PCR amplification cycles were carried out consisting of: 30 s at 94°C, 30 s at 68°C and 2 min at 72°C. The PCR products were cloned into pGEM T-easy vector (Promega) and transformed into competent E.coli JM83 cells. Representative plasmids were sequenced (Genome lab at IMBB-FORTH, Heraklion, Crete).
RESULTS
The influence of the terminal RY motif on the binding of PSTVd RNA to Virp1
Previous analysis had shown that only the right domain of PSTVd had affinity for Virp1, while the left half of PSTVd did not show any specific interaction (31). Here we constructed a 79-base sub-fragment of PSTVd (+) RNA, called R79-wt, ranging from G145 to C223 (Fig. 1, top). This fragment contained the right terminal loop (TR domain) including two copies of the RY motif. To discriminate between the two RY motifs we call them the ‘terminal’ and ‘internal’ RY motif (Fig. 1).
R79-wt was fully sufficient for the specific interaction with Virp1 (Fig. 2A and data not shown). As a first step we tested the contribution of the terminal RY motif in the specific binding to Virp1 by substituting A173 with U (R79-mt1) and U186 with A (R79-mt2), and the combination of both (R79-mt3). Either of the single substitutions would disrupt the base pair of nucleotides 173/186, resulting in a slightly enlarged asymmetric internal loop. The double substitution R79-mt3 would restore the original type of internal loop, with the modification that the downstream helix is closed with U/A instead of A/U (Fig. 1).
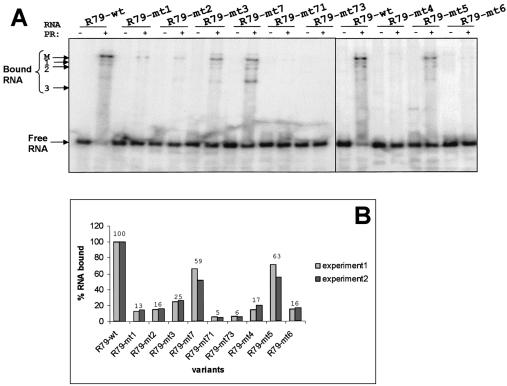
Figure 2.
Binding analysis of different 79 base PSTVd (+) RNA transcripts to Virp1Δ by EMSA. (A) Separation of complexes on a native 6% polyacrylamide gel. Concentrations used: RNA transcripts 100 nM (including the radioactively labeled transcript) and Virp1Δ 1 µM (in lanes with a ‘+’). All transcripts are shown in the absence or presence of Virp1Δ. The position of the free RNA is indicated, as well as the position of the main (M) complex and three additional complexes of higher mobility. (B) Quantitative analysis of RNA bound with the aid of a phosphorimager. The experiment was done in duplicate, experiment 1 corresponds to the gel shown in (A). The numbers on top indicate the average of the percentage of RNA bound.
We performed EMSAs of the R79-wt and its sequence variants with a truncated version of Virp1 (Virp1Δ), which has been shown to retain full and specific RNA-binding activity (29). For RNAs R79-mt1 and R79-mt2 we detected that the fraction of bound RNA dropped to 13% for R79-mt1 and to 16% for R79-mt2 as compared with the wild type RNA (Fig. 2B). The double-mutated RNA R79-mt3 with its restored base pair 173–186 showed slightly restored binding, approaching ∼25% of the R79-wt binding activity (Fig. 2B). The pattern of complex formation was similar to that observed with R79-wt, which is characterized by a predominating slow migrating complex (Fig. 2A, M) and at least three additional complexes of higher mobility (Fig. 2A, marked 1–3). Thus, none of the three mutations completely abolished RNA–protein interaction. In parallel, the same mutations were also introduced into the infectious longer-than-unit length transcript Ha106 (40) and the resulting RNA transcripts were bioassayed. Only the mt2 construct was able to establish infections after prolonged growth in two out of four tomato test plants, but analysis of the progeny revealed reversion to wild type sequence in both plants.
Next, we intended to disrupt the secondary structure of the RY motif. This was accomplished by changing C189 into G (R79-mt4), which converted the internal loop into a bulge loop with two single-stranded nucleotides. Again, this sequence change resulted in a strong impairment, but not a complete abolishment of binding capacity (Fig. 2). In parallel, we introduced two mutations into the right terminal loop (R79-mt5). Sequence variants within this loop have been described by Hammond (32), which were not infectious but could replicate after agro-infection, suggesting impairment of the transport function. While the mutation described by Hammond affected the secondary structure of the terminal RY motif, our mutation R79-mt5 was designed to leave the structure of the terminal RY motif unaffected. The binding assay showed that R79-mt5 retained significant binding activity, suggesting that the nucleotides in the right terminal loop play only a supportive role for specific recognition of viroid RNA by Virp1. Mutations R79-mt4 and R79-mt5 were also tested for infectivity in the context of a full-length PSTV (+) RNA transcript. Mutant Ha106-mt4 was not infectious, but Ha106-mt5 was. Analysis of progeny revealed that the sequence changes introduced into the loop region were tolerated and maintained, confirming that the nature of nucleotides 180/181 are of minor importance for amplification of PSTVd, provided that these changes do not influence the structure of neighboring domains.
Restoration of some binding activity in double-mutant R79-mt3 suggested that the overall structure of the RY motif might be of relevance rather than its actual sequence. To test this we constructed R79-mt6 in which we exchanged in the upper strand ACA (171–173) by UGU and in the lower strand UUCCU (186–190) by AAGGA. These transversions had no impact on the overall secondary structure, but destroyed the canonical terminal RY motif, converting it actually to a ‘YR motif’. The mobility shift assay showed that this RNA still had binding affinity at a similar level as mutations R79-mt1 and mt2, most likely reflecting the contribution of the unchanged internal RY motif to the specific interaction, so far not taken in consideration.
The influence of the internal RY motif on the binding of PSTVd RNA to Virp1
To study the influence of the internal RY motif on the specific binding activity we exchanged A152 for U (R79-mt7). This is an equivalent manipulation to the exchange of A173 to U (R79-mt1) within the terminal RY motif. Further, we combined R79-mt7 with R79-mt1 and R79-mt3 generating mutants R79-mt71 and R79-mt73, respectively. The latter two sequence variants were thus simultaneously mutated in the internal as well as in the terminal RY motif of PSTVd. The RNA R79-mt7 had significant binding affinity despite the structural re-arrangement of the internal RY motif (Fig. 1). In agreement with the assumption that the binding of R79-mt7 to Virp1 was due to the intact terminal RY motif, the double mutations R79-mt71 and R79-mt73 lost almost all their binding capacity (Fig. 2). This also reveals that the residual binding observed with mutants R79-mt1–4 and R79-mt6 was due to the internal RY motif. The mobility shift assay of Figure 2A not only shows that R79-mt7 retained binding potential, but also that the pattern of binding was changed. While the main complex that was formed with R79-wt was still present, the high mobility complex 3 was now predominating. None of these mutations was infectious when tested in the context of a longer-than-unit-length PSTVd RNA transcript.
The binding of mutated PSTVd RNAs to filter-bound Virp
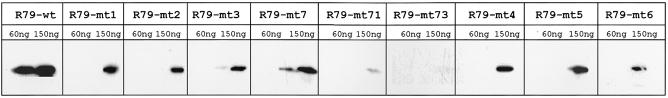
All the mutated PSTVd (+) RNAs mentioned so far were also subjected to a northwestern analysis (Fig. 3). We used two standardized protein concentrations on the filter, which in each case were quantified by immunoblotting (not shown). When the filter-bound Virp1Δ was exposed to radioactively labeled RNAs most mutant RNAs showed binding only at the higher concentration of protein, though at reduced level. Only R79-mt7 showed also some binding at the lower concentration, while the double mutants R79-mt71 and R79-mt73 showed hardly any binding, even at the higher concentration. Although the northwestern analysis generates rather qualitative than quantitative data, the nature and strength of binding detected in this way correlated well with the data obtained by mobility shift analysis.
Figure 3.
Northwestern analysis of Virp1Δ and different 79 base PSTVd (+) RNA transcripts. Virp1Δ was blotted in the two amounts indicated (60 and 150 ng). The RNA that was bound is indicated at the top. After RNA binding the filters were immunoblotted with Virp1Δ-specific antibodies to confirm the amounts of protein on the filter (not shown).
Analysis of the relative contribution of the two RY motifs for the binding of PSTVd RNA to Virp1
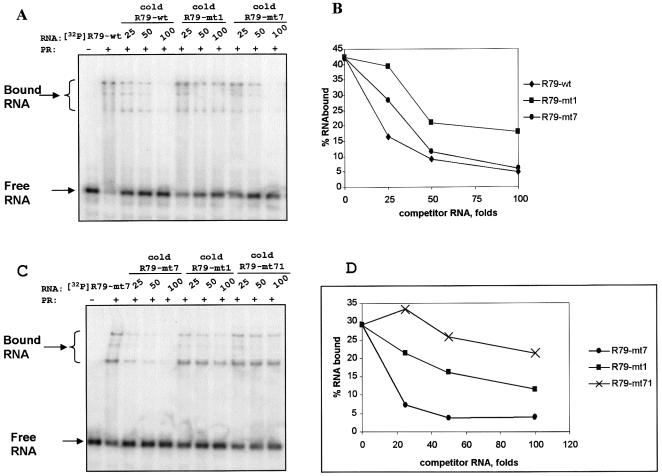
Collectively the mutation data suggested that both RY motifs contributed to the specific interaction of PSTVd (+) RNA with Virp1, although their contribution was of different quality. Inactivation of the terminal RY motif resulted in almost complete loss of binding capacity suggesting that it is more important than the internal RY motif. Yet, none of the five manipulations within the terminal RY motif (R79-mt1-4 and R79-mt6) completely eliminated Virp1 binding. Consistent with the strong influence of the terminal RY motif, the inactivation of the internal RY motif had only a moderate impact on the overall capacity of the PSTVd (+) RNA, but if it was combined with a mutation of the terminal RY motif, binding activity was lost. To verify the different contribution of the two RY motifs to binding we performed competition experiments (Fig. 4A). Radioactively labeled R79-wt (100 nM) was allowed to bind to Virp1Δ (1 µM) with either no competitor RNA or 25-, 50- or 100-fold excess of different competitor RNAs. Figure 4B shows that an ∼20-fold excess of unlabeled R79-wt competitor reduced complex formation to about half. If mutated RNAs R79-mt1 and R79-mt7 were used as competitors, an ∼50- or 38-fold excess was required to achieve the same reduction in complex formation. These competition data confirmed that the terminal RY motif was binding stronger than the internal RY motif.
Figure 4.
Competition experiments. (A) Binding of PSTVd-specific transcript R79-wt (100 nM) to Virp1Δ (1 µM) in the absence or presence of competitor RNA. The type of competitor RNA and its molar excess are indicated. (B) Quantitative analysis of (A) with the aid of a phosphorimager. The graph allows the calculation of D½ (concentration of competitor required to reduce binding by 50%). (C and D) Equivalent analysis with binding of R79-mt7 to Virp1Δ using the competitor as indicated and described in the text.
To study the contribution of these two domains to binding in greater detail we performed a second competition experiment (Fig. 4C), this time allowing radioactively labeled R79-mt7 to bind Virp1Δ (at the same concentrations as above). The three competitors R79-mt7, R79- mt1 and R79-mt71 required ∼25-, 75- or 100-fold excess, respectively, to reduce R79-mt7 binding to Virp1Δ to 50% (Fig. 4D). Again, this competition experiment confirmed that binding of the terminal RY motif was stronger than that of the internal RY motif.
In an attempt to quantify the relative strength of the two binding sites we performed a titration experiment (Fig. 5A). The RNA of R79-wt and the two mutants R79-mt3 and R79-mt7 were adjusted to 1 nM and allowed to bind to Virp1Δ at three different concentrations ranging from 0.1 to 3.3 µM. Quantification of the bound RNA allowed determination of the dissociation constants (Kd) for mutants R79-mt3 and R79-mt7 and the apparent Kd for the wild type RNA (Fig. 5B). This analysis revealed that binding of the terminal RY motif is about five times stronger than the internal RY motif. However, destruction of the internal RY motif seemed to influence the quality of binding at the terminal RY motif, favoring the formation of a different, fast migrating RNA–protein complex (see also Fig. 2A). It is therefore conceivable to assume that binding at the internal RY motif works efficiently only in the presence of an intact terminal RY motif. This is in agreement with the apparent Kd observed for the wild type sequence with the two intact RY motifs with a 2–3-fold increase in binding as compared to R79-mt7 (with inactivated internal RY motif), which is much higher than expected considering the weak binding of the internal motif in mutant R79-mt3.
Figure 5.
Titration experiments. (A) EMSA of R79-wt and the constructs R79-mt3 and R79-mt-7, which are mutated in the terminal and internal RY motif (compare Fig. 1). The RNAs (1 nM) were analyzed in the absence or presence of the three different concentrations of Virp1Δ as indicated. (B) Quantitative analysis of data from (A) plotted as bound RNA over free RNA (Virp1-R/R) versus protein concentration (47). The slope allows determination of the dissociation constant Kd.
DISCUSSION
Functional analysis of the RY motifs
Based on the differential interaction of previously described PSTVd mutants (32), we have previously suggested the 5′-AGG/CUUCC-5′motif as the determinant of the specific interaction between Virp1 and PSTVd (+) RNA (31). Here we substantiate this assumption by studying the interaction of Virp1 with a 79 base sub-fragment of PSTVd containing the TR domain. However, our analysis suggests that the motif that is recognized by Virp1 is slightly larger than the 5′-AGG/CUUCC-5′motif and that it includes the entire asymmetric internal loop that we termed ‘RY motif’. Moreover, we found that the second RY motif also contributes to the binding. We were primarily interested in a functional test rather than in demarcating the precise borders of the motif or analyzing the role of each nucleotide within the sequence pattern. Thus, we could show that a single nucleotide exchange introduced simultaneously into both RY motifs almost completely abolished the ability of the viroid RNA to interact with Virp1. By contrast, simultaneous exchanges of two nucleotides in the terminal right loop—leaving the terminal RY motif intact—had only a moderate effect on binding affinity and no effect on infectivity. The individual inactivation of the RY motifs revealed that each motif can bind Virp1 without the need of the other, which, however, does not preclude a cooperative effect. Compensatory mutations in the terminal RY motif that maintained the overall viroid secondary structure did not restore binding activity, which suggests that Virp1 recognizes a domain with a specific RNA motif (sequence plus structure) rather than just a secondary structure. Using different experimental approaches we could show that the terminal motif plays the most significant role in the specific interaction and has an ∼5-fold higher affinity than the internal RY motif. However, despite this, the internal motif also seems to be of importance. This is based not only on the bioassays where any nucleotide change in any RY motif resulted in loss of infectivity, but also in a change of the composition of complexes formed (Fig. 2A, see 79R-mt7). In general, the detection of several distinct RNA–protein complexes suggests a complex binding reaction, possibly involving two protein molecules per binding site, making a total of four proteins per RNA, which is in agreement with the number of complexes seen in EMSA experiments. Although unlikely, we cannot rule out that more than one RNA molecule participates in the reaction.
The RY motif in Pospiviroids
The terminal RY motif was the strongest in Virp1 binding. In accordance with this functionality it is the phylogenetically most conserved RY motif. In 1987 it had already been noticed that the sequence 3′-CUUCC-5′ was conserved in the TR domain of several viroids related to PSTVd (33), now grouped as members of different genera of Pospiviridae (3). This finding suggested a functional role, but the relevance of the conserved pentameric sequence pattern fell somewhat into oblivion, possibly because it was absent in many viroids from other further viroid genera discovered thereafter, so that is was not considered in ensuing discussions. Further support for a functional importance of the RY motif comes from the observation that in many Pospiviroids, including PSTVd, the motif is redundant (41). It should be stressed, however, that the formation of the internal RY motif may be temperature-dependent because using the Mfold folding program (42), the internal RY motif is present in the most stable secondary structure of PSTVd only if the temperature is set to 11°C or lower.
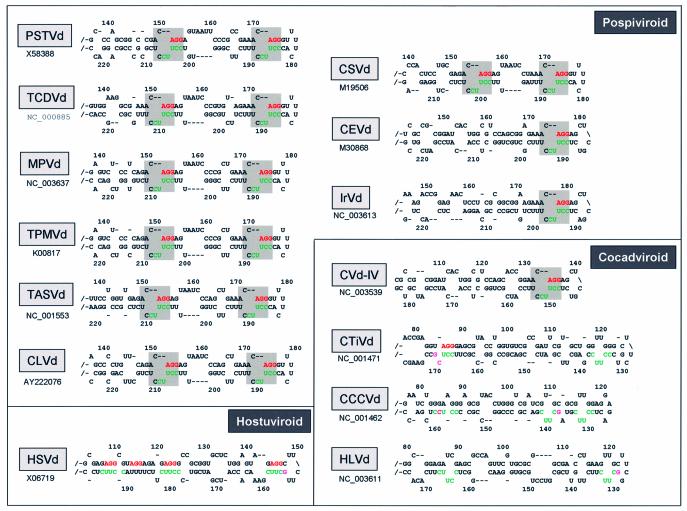
An internal RY motif can be found also for tomato chlorotic dwarf viroid (TCDVd), mexican papita viroid (MPVd), tomato planta macho viroid (TPMVd), columnea latent viroid (CLVd) and tomato apical stunt viroid (TASVd) (Fig. 6). The motifs are separated typically by 16 and 12 unrelated nucleotides in the upper and lower strand, respectively, while in TCDVd the distances are 17 and 14 nucleotides. Chrysanthemum stunt viroid (CSVd) has the same terminal RY motif as PSTVd; also here a second RY motif is present, although separated by only 11 and 7 intervening nucleotides (Fig. 6). However, in citrus exocortis viroid (CEVd), as well as in iresine viroid 1 (IrVd1), only the terminal RY motif is present. This difference in occurrence of the RY motif and the distance of the two copies most likely reflects the phylogenetic relationship within Pospiviroids (Fig. 6).
Figure 6.
The RY motifs and further sequence elements found in the TR domain of representative species of three different genera of Pospiviroidae. An overview of viroid sequences and their classification can be found at the subviral RNA database (48). All secondary structures were calculated using the web tool (42); temperatures were set to 10 or 20°C. The RY motifs are shaded. The AGG triplet is marked in red, the CUUCC-5′ element is marked in green and non-canonical nucleotides in magenta. The full names of the viroid acronyms are given in the text; accession numbers of the sequences used are indicated.
Elements of the RY motif in other genera of Pospiviroidae
Hop stunt viroid (HSVd) is the representative viroid species of the Hostuviroids and its (+) RNA has shown binding activity to Virp1 (31,43), although at reduced level. In accordance with this, some sequence elements of the RY motif occur frequently, i.e. the AGG triplet (marked in red in Figs 1 and 6) and the CUUCC-5′ pentamer (marked in green in Figs 1 and 6). However, even if one assumes structural re-arrangement, no proper RY motif can be formed. The region in question has been studied previously by Sano and Ishiguro who constructed chimeric viroids by exchange of the TR domain of CEVd and HSVd (20). Both chimeric viroids were infectious, though at greatly reduced levels and without generation of symptoms. This included the chimeric viroid CE/HS-TR, in which the canonical RY motif of the CEVd TR had been replaced by the non-canonical motif of HSVd. It is thus possible that HSVd possesses an alternative binding motif for the ortholog Virp1 protein from hop. For the region of HSVd close to the terminal hairpin, which we discuss as a binding site for Virp1 or related proteins, Amari et al. have proposed an intriguing hypothesis by suggesting a secondary structure resembling a mutated form of hammerhead domain (44). It remains to be seen whether there is a connection between these two observations. Cocadviroids are a more distantly related genus of the family Pospiviroidae. Here the situation is more complex. The citrus viroid IV (CVd-IV) has a canonical RY motif at terminal position and the coconut cadang-cadang viroid (CCCVd) and coconut tinangaja viroid (CTiVd) have the conserved 3′-CUUCC-5′ pentamer, also located within the first internal loop next to the closing right hairpin loop (Fig. 6). Both coconut viroids miss the matching AGG triplet in this region, but show between each other a closely related sequence motif, which most likely adopts the RY motif function. The 3′-CUUCC-5′ element is located within the domain that is found repeated in CCCVd subspecies. Similar to PSTVd, the redundancy of the motif may strengthen the interaction of CCCVd with a functional Virp1 ortholog in palm trees suggesting that this region could be responsible for host specificity. If one allows one mismatch (magenta coding in Fig. 6), more repetitions of the 3′-CUUCC-5′ pentamer can be found in the coconut viroids. In CTiVd there is even a match to an AGG motif, which is unlikely to be accidental. The fourth representative of the Cocadviroids is hop latent viroid (HLVd), which has a proper internal 3′-CUUCC-5′ element, plus a relaxed form of it, close to the terminal right hairpin (Fig. 6). The Apsca- and Coleviroids, which are further genera of Pospiviroidae, do not contain such sequence elements and in accordance with this, binding to Virp1 was not detected with either apple scar skin viroid (ASSVd), or with grapevine yellow speckle viroid (GYSSVd) (43). It should be added that both these viroids, however, have the element twice on their (–) RNA. Also Coleviroids do not have the structural elements, but they possess close to their terminal right loop a purine region that interacts with a pyrimidine region (not shown).
The function of the RY motif during viroid replication
Despite the characterization of the RNA-binding motif, the biological role of the interaction of PSTVd and Virp1 remains elusive. However, several observations support the notion that the RY motifs are essential for establishing a viroid infection. The phylogenetic conservation of the RY motif in several genera of Pospiviroidae argues for a functional role and the redundancy of the motif in PSTVd and related viroids suggests that a strong interaction is beneficial for propagation of the viroid RNA. The fact that Virp1 expression is impaired in PSTVd-infected tomato (29) does not argue against involvement in viroid proliferation, because at late stages of infection the viroid replication rate is also reduced. Assuming that the interaction of Virp1, or a homolog protein from another host, is required for viroid replication, the RY motif is likely to determine the host range of the viroid. In accordance with this assumption that the terminal domain may influence host range, it has been reported that the change of the experimental host Gynura aurantiaca to a hybrid tomato resulted in a sequence duplication in the TR domain of CEVd (45), also causing the duplication of the RY motif. As pointed out earlier, the observation that PSTVd variants with disordered terminal RY motif were able to replicate (32), but not to spread, argues for involvement of Virp1 in the movement of PSTVd RNA. Also the lack of infectivity for any of the mutations specifically introduced in any of the RY motifs does not exclude that such sequence variants might be able to replicate at a cellular level. In this context it is of interest that Gora-Sochacka et al. (46) could clone PSTVd cDNAs with sequence changes in the terminal RY motif (46). In the first variant, nucleotide A173 had been changed to G173; similar to our mutant U173 this cDNA was not infectious. But the authors also found a mutation U186 to C and another cDNA where U187 was changed to C. Similar to our A186 mutation, variant C186 (and also variant C187) was infectious, however, at a considerably reduced rate. Since the progeny originating from these mutated cDNAs were not analyzed, it is possible that a sequence reversion to wild type occurred as in our case. This is common for PSTVd sequences that are altered in a single nucleotide.
Besides the involvement in PSTVd proliferation, Virp1 should be expected to have a role in healthy plants. It is likely that the protein will interact with a cellular RNA that exhibits an RY motif in the proper secondary structure. The identification of this cellular RNA partner might be a key for the understanding of viroid pathogenicity.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Kriton Kalantidis and Alexandra Boutla for reading the manuscript. M.G. has been supported by a Marie Curie training fellowship (contract HPMT-CT-2000-00175) and by the National Institution of Scholarships, IKY, Athens (contract 669). The work was in part supported by the European Union (contract QLG2-CT-2002-01673 VIS) and in part by the General Secretariat for Research and Technology of the Hellenic Ministry of Development via the Bulgarian-Greek cooperation program (PN18/3-1-2003).
REFERENCES
- 1.Flores R., Di Serio,F. and Hernandez,C. (1997) Viroids: the noncoding genomes. Semin Virol., 8, 65–73. [Google Scholar]
- 2.Diener T.O. (2001) The viroid: biological oddity or evolutionary fossil? Adv. Virus Res., 57, 137–184. [DOI] [PubMed] [Google Scholar]
- 3.Flores R. (2001) A naked plant-specific RNA ten-fold smaller than the smallest known viral RNA: the viroid. C. R. Acad. Sci. III, 324, 943–952. [DOI] [PubMed] [Google Scholar]
- 4.Hull R. (2002) Matthews’ Plant Virology, 4th Edn. Academic Press, San Diego, CA. [Google Scholar]
- 5.Gross H.J., Domdey,H., Lossow,C., Jank,P., Raba,M., Alberty,H. and Sänger,H.L. (1978) Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203–208. [DOI] [PubMed] [Google Scholar]
- 6.Branch A.D. and Robertson,H.D. (1984) A replication cycle for viroids and other small infectious RNAs. Science, 223, 450–455. [DOI] [PubMed] [Google Scholar]
- 7.Schindler I.-M. and Mühlbach,H.-P. (1992) Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci., 84, 221–229. [Google Scholar]
- 8.Bonfiglioli R.G., Webb,D.R. and Symons,R.H. (1996) Tissue and intra-cellular distribution of coconut cadang cadang viroid and citrus exocortis viroid determined by in situ hybridization and confocal laser scanning and transmission electron microscopy. Plant J., 9, 457–465. [Google Scholar]
- 9.Harders J., Lukacs,N., Robert-Nicoud,M., Jovin,T.M. and Riesner,D. (1989) Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J., 8, 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warrilow D. and Symons,R.H. (1999) Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch. Virol., 144, 2367–2375. [DOI] [PubMed] [Google Scholar]
- 11.Keese P. and Symons,R.H. (1985) Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl Acad. Sci. USA, 82, 4582–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elena S.F., Dopazo,J., De La Pena,M., Flores,R., Diener,T.O. and Moya,A. (2001) Phylogenetic analysis of viroid and viroid-like satellite RNAs from plants: a reassessment. J. Mol. Evol., 53, 155–159. [DOI] [PubMed] [Google Scholar]
- 13.Tsagris M., Tabler,M. and Sänger,H.L. (1991) Ribonuclease T1 generates circular RNA molecules from viroid-specific RNA transcripts by cleavage and intramolecular ligation. Nucleic Acids Res., 19, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabler M., Tzortzakaki,S. and Tsagris,M. (1992) Processing of linear longer-than-unit-length potato spindle tuber viroid RNAs into infectious monomeric circular molecules by a G- specific endoribonuclease. Virology, 190, 746–753. [Published erratum appears in Virology (1993) 192, 397.]. [DOI] [PubMed] [Google Scholar]
- 15.Baumstark T. and Riesner,D. (1995) Only one of four possible secondary structures of the central conserved region of potato spindle tuber viroid is a substrate for processing in a potato nuclear extract. Nucleic Acids Res., 23, 4246–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumstark T., Schröder,A.R. and Riesner,D. (1997) Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J., 16, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrader O., Baumstark,T. and Riesner,D. (2003) A mini-RNA containing the tetraloop, wobble-pair and loop E motifs of the central conserved region of potato spindle tuber viroid is processed into a minicircle. Nucleic Acids Res., 31, 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond R.W., Diener,T.O. and Owens,R.A. (1989) Infectivity of chimeric viroid transcripts reveals the presence of alternative processing sites in potato spindle tuber viroid. Virology, 170, 486–495. [DOI] [PubMed] [Google Scholar]
- 19.Gora A., Candresse,T. and Zagorski,W. (1996) Use of intramolecular chimeras to map molecular determinants of symptom severity of potato spindle tuber viroid (PSTVd). Arch. Virol., 141, 2045–2055. [DOI] [PubMed] [Google Scholar]
- 20.Sano T. and Ishiguro,A. (1998) Viability and pathogenicity of intersubgroup viroid chimeras suggest possible involvement of the terminal right region in replication. Virology, 240, 238–244. [DOI] [PubMed] [Google Scholar]
- 21.Wassenegger M., Spieker,R.L., Thalmeir,S., Gast,F.U., Riedel,L. and Sänger,H.L. (1996) A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology, 226, 191–197. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y., Qi,Y., Xun,Y., Owens,R. and Ding,B. (2002) Movement of potato spindle tuber viroid reveals regulatory points of Phloem-mediated RNA traffic. Plant Physiol., 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff P., Gilz,R., Schumacher,J. and Riesner,D. (1985) Complexes of viroids with histones and other proteins. Nucleic Acids Res., 13, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner R., Mühlbach,H.P. and Guitton,M.C. (1995) Isolation of viroid-RNA-binding proteins from an expression library with nonradioactive-labeled RNA probes. Biotechniques, 19, 218–222. [PubMed] [Google Scholar]
- 25.Gomez G. and Pallas,V. (2001) Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant Microb. Interact., 14, 910–913. [DOI] [PubMed] [Google Scholar]
- 26.Owens R.A., Blackburn,M. and Ding,B. (2001) Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant Microb. Interact., 14, 905–909. [DOI] [PubMed] [Google Scholar]
- 27.Daros J.A. and Flores,R. (2002) A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J., 21, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sägesser R., Martinez,E., Tsagris,M. and Tabler,M. (1997) Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res., 25, 3816–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez de Alba A.E., Sägesser,R., Tabler,M. and Tsagris,M. (2003) A bromodomain-containing protein from tomato binds specifically potato spindle tuber viroid RNA in vitro and in vivo. J. Virol., 77, 9685–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filetici P., Ornaghi,P. and Ballario,P. (2001) The bromodomain: a chromatin browser? Front. Biosci., 6, D866–D876. [DOI] [PubMed] [Google Scholar]
- 31.Maniataki E., Martinez de Alba,A.E., Sägesser,R., Tabler,M. and Tsagris,M. (2003) Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA, 9, 346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond R.W. (1994) Agrobacterium-mediated inoculation of PSTVd cDNAs onto tomato reveals the biological effect of apparently lethal mutations. Virology, 201, 36–45. [DOI] [PubMed] [Google Scholar]
- 33.Keese P. and Symons,R.H. (1987) The structure of viroids and virusoids. In Semancik,J.S. (ed.), Viroids and Viroid-like Pathogens. CRC Press, Boca Raton, FL, pp. 1–47. [Google Scholar]
- 34.Hammann C., Hormes,R., Sczakiel,G. and Tabler,M. (1997) A spermidine-induced conformational change of long-armed hammerhead ribozymes: ionic requirements for fast cleavage kinetics. Nucleic Acids Res., 25, 4715–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke S.H. and Madison,E.L. (1997) Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res., 25, 3371–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denti M.A., Martinez de Alba,A.E., Sägesser,R., Tsagris,M. and Tabler,M. (2000) A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme. Nucleic Acids Res., 28, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 38.Tabler M. and Sänger,H.L. (1984) Cloned single- and double-stranded DNA copies of potato spindle tuber viroid (PSTV) RNA and co-inoculated subgenomic DNA fragments are infectious. EMBO J., 3, 3055–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papaefthimiou I., Hamilton,A.J., Denti,M.A., Baulcombe,D.C., Tsagris,M. and Tabler,M. (2001) Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res., 29, 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsagris M., Tabler,M., Mühlbach,H.-P. and Sänger,H.L. (1987) Linear oligomeric potato spindle tuber viroid (PSTV) RNAs are accurately processed in vitro to the monomeric circular viroid proper when incubated with a nuclear extract from healthy potato cells. EMBO J., 6, 2173–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maniataki E. (2000) Interactions of viroids with their hosts: two approaches. PhD Thesis (in Greek), University of Crete, Heraklion, Greece.
- 42.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez de Alba E. (2000) Isolation and characterisation of viroid-binding proteins. PhD Thesis, Universidad del Pais Vasco, Bilbao, Spain.
- 44.Amari K., Gomez,G., Myrta,A., Di Terlizzi,B. and Pallas,V. (2001) The molecular characterization of 16 new sequence variants of Hop stunt viroid reveals the existence of invariable regions and a conserved hammerhead-like structure on the viroid molecule. J. Gen. Virol., 82, 953–962. [DOI] [PubMed] [Google Scholar]
- 45.Semancik J.S., Szychowski,J.A., Rakowski,A.G. and Symons,R.H. (1993) Isolates of citrus exocortis viroid recovered by host and tissue selection. J. Gen. Virol., 74, 2427–2436. [DOI] [PubMed] [Google Scholar]
- 46.Gora-Sochacka A., Kierzek,A., Candresse,T. and Zagorski,W. (1997) The genetic stability of potato spindle tuber viroid (PSTVd) molecular variants. RNA, 3, 68–74. [PMC free article] [PubMed] [Google Scholar]
- 47.Black D.L., Chan,R.C., Min,H., Wang,J. and Bell,L. (1998) The electrophoretic mobility shift assay for RNA binding proteins. In Smith,C.W.J. (ed.), RNA–protein Interactions: A Practical Approach. The practical approach series 192. IRL Press, Oxford, UK, pp. 109–136. [Google Scholar]
- 48.Pelchat M., Rocheleau,L., Perreault,J. and Perreault,J.P. (2003) SubViral RNA: a database of the smallest known auto-replicable RNA species. Nucleic Acids Res., 31, 444–445. [DOI] [PMC free article] [PubMed] [Google Scholar]