Abstract
Skeletal muscle stem cell–derived myoblasts are mainly responsible for postnatal muscle growth and injury-induced muscle regeneration. However, the cellular signaling pathways controlling the proliferation and differentiation of myoblasts are not fully understood. We demonstrate that Janus kinase 1 (JAK1) is required for myoblast proliferation and that it also functions as a checkpoint to prevent myoblasts from premature differentiation. Deliberate knockdown of JAK1 in both primary and immortalized myoblasts induces precocious myogenic differentiation with a concomitant reduction in cell proliferation. This is caused, in part, by an accelerated induction of MyoD, myocyte enhancer–binding factor 2 (MEF2), p21Cip1, and p27Kip1, a faster down-regulation of Id1, and an increase in MEF2-dependent gene transcription. Downstream of JAK1, of all the signal transducer and activator of transcriptions (STATs) present in myoblasts, we find that only STAT1 knockdown promotes myogenic differentiation in both primary and immortalized myoblasts. Leukemia inhibitory factor stimulates myoblast proliferation and represses differentiation via JAK1–STAT1–STAT3. Thus, JAK1–STAT1–STAT3 constitutes a signaling pathway that promotes myoblast proliferation and prevents premature myoblast differentiation.
Introduction
In vertebrates, all skeletal muscles of trunk and limbs originate from somites, which are formed sequentially in a rostral-caudal direction through segmentation of the paraxial mesoderm during embryogenesis (Buckingham et al., 2003). In response to signals from the neural tube, notochord, and ectoderm, somites further differentiate into ventral-medially positioned sclerotome and dorsally located dermatome with the muscle-forming myotome sandwiched in between (Buckingham et al., 2003). In myotome, the muscle precursor cells establish their myogenic fate to form proliferating myoblasts by selectively expressing one or a few myogenic regulatory factors (MRFs). Under appropriate conditions, the myoblasts withdraw from the cell cycle to differentiate into mononucleated myocytes, which, in turn, align with each other and fuse to form multinucleated myotubes or myofibers.
Muscle stem cells, which are also called muscle satellite cells (MSCs), start to form at the late stage of vertebrate embryo development (Morgan and Partridge, 2003; Dhawan and Rando, 2005; Holterman and Rudnicki, 2005; Wagers and Conboy, 2005). In the adult, most of the MSCs are quiescent and uniquely located between basal lamina and the plasma membrane of the myofibers. Several molecular markers, including Pax7, c-Met, M-cadherin, and CD34, are expressed in quiescent MSCs. In contrast, MyoD is not expressed in quiescent MSCs. In response to muscle injury or exercise, these quiescent MSCs become activated, as indicated by the expression of MyoD, reenter the cell cycle, and actively proliferate to form myoblasts. Eventually, these proliferating myoblasts irreversibly withdraw from cell cycles, differentiate, and fuse with existing myofibers. Accumulating evidence indicates that myoblasts are the primary cell types responsible for muscle regeneration in vivo (Dhawan and Rando, 2005; Holterman and Rudnicki, 2005; Wagers and Conboy, 2005).
Elucidation of the molecular mechanisms underlying myogenic differentiation has been greatly facilitated by the availability of several immortalized myogenic cell lines, including C2C12 cells, which are derived from mouse MSCs (Yaffe and Saxel, 1977). Thus, C2C12 cells represent an excellent cell culture model to study the proliferation and differentiation of MSC-derived myoblasts. Two families of transcription factors play critical roles in myogenesis. MRFs consist of Myf5, MyoD, MRF4, and myogenin. MRFs normally heterodimerize with gene E2A products (i.e., E12/E47) and bind to the consensus sequence of CANNTG (E box) in the promoters of many muscle-specific genes (Molkentin and Olson, 1996; Sabourin and Rudnicki, 2000; Tapscott, 2005). Id, a negative regulator for myogenesis, represses myogenic differentiation by binding to and sequestering either MRFs or E proteins, thus preventing MRFs from binding to the E box (Benezra et al., 1990). In addition to MRFs, myocyte enhancer–binding factor 2 (MEF2), which consists of MEF2A, 2B, 2C, and 2D, is also essential for myogenesis (Black and Olson, 1998; McKinsey et al., 2002). MRFs and MEF2 physically interact with each other to synergistically activate many muscle-specific genes (Molkentin et al., 1995).
Many intracellular signaling molecules/pathways are known to modulate myogenic differentiation by regulating myogenin gene expression. Among them, the p38 MAPK, insulin-like growth factors/phosphatidylinositol 3-kinase/Akt, calcium/calmodulin-activated protein kinase, and calcineurin positively regulate myogenic differentiation (Coolican et al., 1997; Zetser et al., 1999; Friday et al., 2000; McKinsey et al., 2000; Tamir and Bengal, 2000; Wu et al., 2000; Xu and Wu, 2000; Xu et al., 2002), whereas extracellular signal-regulated kinase (ERK) has dual roles: it inhibits differentiation at the early stage of differentiation but promotes myocyte fusion at the late stage of differentiation (Bennett and Tonks, 1997; Coolican et al., 1997; Wu et al., 2000). The Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway represents one of the best-characterized cellular signaling pathways (O'Shea et al., 2002). Four JAKs (i.e., JAK1, 2, 3, and Tyk2) and seven STATs (i.e., STAT1, 2, 3, 4, 5a, 5b, and 6) have been identified in the mouse and human genomes. Although it is well established that the JAK–STAT pathway plays essential roles in hematopoiesis and antimicrobial immune response (O'Shea et al., 2002), it remains unclear whether the JAK–STAT pathway plays any essential role in myogenesis. Several lines of evidence suggest that the JAK–STAT pathway may have a role in myogenic differentiation. In regenerating rat muscles, proliferating myoblasts were found to contain higher levels of phosphorylated (i.e., active) STAT3 (Kami and Senba, 2002). In response to leukemia inhibitory factor (LIF), proliferating primary myoblasts grown in culture were also found to contain higher levels of phosphorylated STAT3 (Megeney et al., 1996; Spangenburg and Booth, 2002). Recently, MyoD was found to interact with STAT3 in an overexpression study (Kataoka et al., 2003). Nevertheless, the aforementioned evidence was mainly correlative in nature, and none of these studies has addressed the question of whether and how the JAK–STAT pathway is involved in myogenic differentiation.
In this study, we report that the JAK1–STAT1–STAT3 pathway plays dual roles in proliferating myoblasts: it is required for myoblast proliferation and also serves as a key checkpoint to prevent myoblasts from premature differentiation. Specific knockdown of either JAK1 or STAT1 (to a lesser extent) reduces cell proliferation and induces precocious myogenic differentiation. LIF engages the JAK1–STAT1–STAT3 pathway to promote proliferation and to prevent the premature differentiation of myoblasts.
Results
JAK1 knockdown induces precocious myogenic differentiation in both C2C12 and primary myoblasts
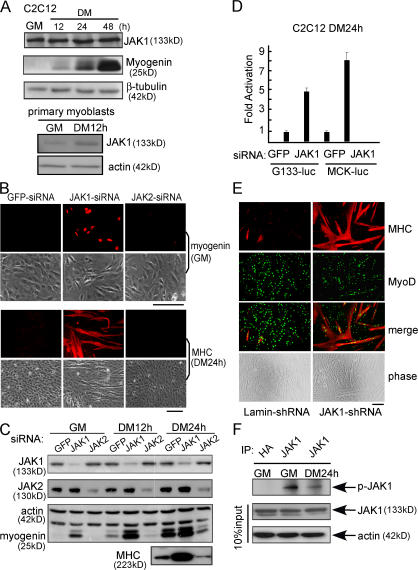
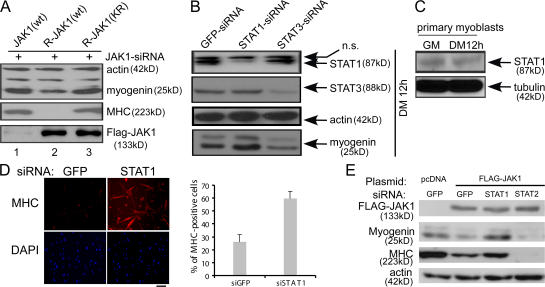
To explore the role of JAK1 in myogenic differentiation, we first examined its expression profiles in both immortalized C2C12 cells and primary myoblasts by Western blotting. As shown in Fig. 1 A, JAK1 was expressed in both C2C12 cells and primary myoblasts both before and after differentiation. To explore its functional role in myogenic differentiation, we first knocked down the endogenous JAK1 in C2C12 cells with siRNA. Surprisingly, compared with the control cells transfected with an siRNA against the gene encoding the jellyfish GFP, a JAK1-specific siRNA dramatically accelerated myogenic differentiation, as indicated by an increased number of myogenin-positive and myosin heavy chain (MHC)–positive cells and precocious formation of large multinucleated myotubes (Fig. 1 B). Western blotting confirmed that JAK1-siRNA was specific and effective and potently induced the expression of myogenin and MHC (Fig. 1 C). In contrast, a JAK2-specific siRNA inhibited C2C12 differentiation, as indicated by decreased myogenin and MHC expression levels compared with the GFP-siRNA control (Fig. 1 C). Consistently, JAK1-siRNA greatly activated two myogenic luciferase reporter genes driven by a fragment of the native myogenin promoter and muscle creatine kinase (MCK) promoter (i.e., G133-luc and MCK-luc, respectively; Fig. 1 D). To find out whether the JAK1 knockdown had a similar effect on primary myoblasts, we infected the primary myoblasts (i.e., MyoD+) with adenoviruses expressing either the JAK1-specific short hairpin RNA (shRNA) or human lamin-specific shRNA (control). Compared with cells infected with lamin-shRNA, those infected with JAK1-shRNA underwent potent and accelerated differentiation, as manifested by the considerably increased number and size of MHC-positive myotubes (Fig. 1 E). In addition, we determined the kinase activity of the endogenous JAK1 in C2C12 cells before and after differentiation. We found that the kinase activity of JAK1 decreased upon differentiation (Fig. 1 F).
Figure 1.
JAK1 knockdown accelerates myogenic differentiation. (A) WCEs from either C2C12 or primary myoblasts were subjected to Western blotting for various markers. (B and C) C2C12 cells were transfected with various siRNAs as indicated. (B) Cells were fixed at different time points as indicated and subjected to immunostaining for either myogenin or MHC. Phase-contrast images of the same field are also shown. (C) WCEs were prepared, and 20 μg WCE was subjected to immunoblotting. (D) Duplicate C2C12 cells were transfected with siRNAs and the luciferase reporter constructs as indicated. 24 h after transfection, cells were switched to DM for another 24 h. WCEs were prepared and subjected to luciferase assays. Fold activation was calculated as the ratio of the luciferase activity of the JAK1-siRNA–transfected cells over that of the GFP-siRNA–transfected cells. The results are presented as mean ± SD (error bars). (E) Primary myoblasts were infected with adenoviruses expressing either JAK1-shRNA or lamin-shRNA. 3 d after infection, cells were fixed and subjected to double immunostaining for MyoD and MHC. (F) WCEs were prepared from C2C12 cells grown in GM or DM for 24 h. The endogenous JAK1 was immunoprecipitated (IP) from 1 mg WCE and subjected to in vitro kinase assays. The HA antibody was used as a negative control. Bars, 100 μm.
JAK1 and the ERK pathway cooperate to repress myogenic differentiation
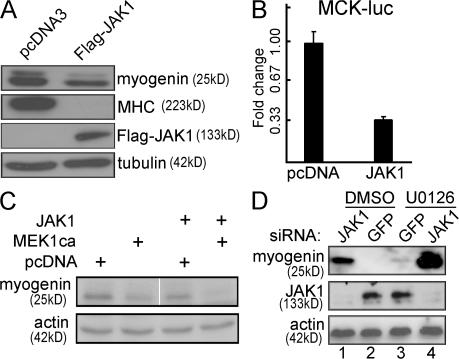
Because the knockdown of JAK1 stimulates myogenic differentiation (Fig. 1), we next asked what happens if we overexpress JAK1 in myogenic cells. We generated stable C2C12 cells expressing either the wild-type JAK1 or an empty vector and subjected these cells to differentiation. As shown in Fig. 2 A, upon differentiation, cells overexpressing JAK1 had much lower myogenin levels and undetectable MHC compared with those with an empty vector. Consistently, the overexpression of JAK1 inhibited the activity of MCK-luc (Fig. 2 B). The inhibitory role of JAK1 at the early phase of myogenic differentiation was reminiscent of that of the ERK pathway (Bennett and Tonks, 1997; Coolican et al., 1997; Wu et al., 2000). To study the potential cross talk between the two pathways, we first examined the effect of JAK1-siRNA on the levels of the active ERKs (i.e., phospho-ERK) in C2C12 cells. As a control, we also examined the status of the active p38 MAPK and Akt, both of which are indispensable for myogenic differentiation (Zetser et al., 1999; Wu et al., 2000; Xu and Wu, 2000). We found that the JAK1 knockdown did not affect the levels of the active ERKs, p38 MAPKs, and Akt in C2C12 cells (unpublished data), suggesting that the potent prodifferentiation effect of JAK1-siRNA was not mediated by these pathways. We then tested whether JAK1 cooperates with the ERK pathway to repress myogenic differentiation. Although cells overexpressing either MEK1ΔN4, which constitutively activates ERK (Mansour et al., 1996), or JAK1 already led to a reduced expression of myogenin, cells overexpressing both further repressed myogenin expression (Fig. 2 C). Conversely, cells treated with either JAK1-siRNA or U0126, a specific inhibitor for the ERK pathway (Favata et al., 1998), differentiated faster as indicated by the accelerated induction of myogenin (Fig. 2 D, compare lanes 1 and 3 with lane 2). Importantly, cells treated with both JAK1-siRNA and U0126 together had a more potent myogenin induction than either treatment alone (Fig. 2 D, lane 4). Our data suggest that JAK1 and the ERK pathway cooperate to repress early myogenic differentiation.
Figure 2.
JAK1 and the ERK pathway cooperate to repress myogenic differentiation. (A) C2C12 cells stably expressing either a wild-type Flag-JAK1 or an empty vector (pcDNA3) were allowed to differentiate in DM for various times. 20 μg WCE was subjected to immunoblotting using various antibodies as indicated. (B) Duplicate C2C12 cells were cotransfected with MCK-luc together with either an empty vector or the wild-type Flag-JAK1. 24 h after transfection, cells were switched to DM for another 24 h before harvest. WCEs were subjected to luciferase analysis. Fold change was calculated as the ratio of the luciferase activity of cells transfected with Flag-JAK1 over that with an empty vector. The results are presented as mean ± SD (error bars). (C and D) C2C12 cells were transfected with either cDNA expression vectors (C) or various siRNAs (D) as indicated. (C) Cells were grown in GM for 24 h and in DM for another 24 h. (D) 10 μM DMSO or U0126 was added when cells were switched to DM and kept for 12 h. Cells were harvested, and WCEs were subjected to immunoblotting. + denotes that cells were transfected with the construct indicated on the left; ca, constitutively active. The white line indicates that intervening lanes have been spliced out.
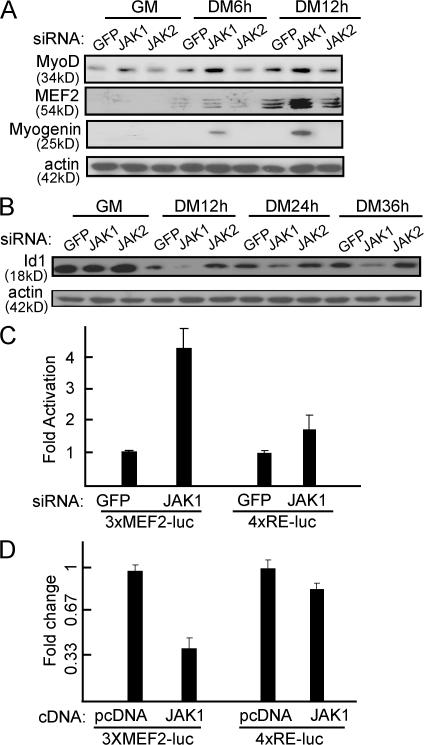
Knockdown of JAK1 induces the expression of MyoD and MEF2, accelerates the down-regulation of Id1, and activates MEF2-dependent gene transcription
To uncover the molecular mechanisms underlying the prodifferentiation effect of JAK1-siRNA, we examined several key players involved in early myogenic differentiation. As shown in Fig. 3 A, the protein levels of both MyoD and MEF2 were clearly induced by JAK1-siRNA at the early stage of differentiation. This induction mainly occurred at the transcription level, as the mRNA levels of MyoD, MEF2A, 2C, and 2D were also induced by JAK1-siRNA as judged by semiquantitative RT-PCR (unpublished data). In contrast, Id1, a known negative regulator for myogenic differentiation (Benezra et al., 1990), was down-regulated at a faster rate in the presence of JAK1-siRNA (Fig. 3 B). In addition, we also examined the impact of JAK1-siRNA on both MyoD and MEF2-dependent gene transcription. Although JAK1-siRNA had less effect on the activity of 4xRE-luc, a MyoD-dependent reporter, it substantially elevated the activity of 3xMEF2-luc, an MEF2-dependent reporter (Fig. 3 C). Consistently, the overexpression of JAK1 greatly inhibited the MEF2-dependent gene transcription but had less effect on MyoD-dependent gene transcription (Fig. 3 D).
Figure 3.
JAK1-siRNA induces the accelerated expression of MyoD and MEF2, promotes a faster down-regulation of Id1, and enhances MEF2-dependent gene transcription. (A and B) C2C12 cells were transfected with various siRNAs as indicated. 24 h after transfection, cells were induced to differentiate in DM for various times as indicated. WCEs were prepared and subjected to immunoblotting. (C and D) Duplicate C2C12 cells were cotransfected with the luciferase reporter constructs together with either siRNAs (C) or cDNAs (D) as indicated. Cells were induced to differentiate in DM for 12 h before harvest. WCEs were subjected to luciferase assays. Fold activation or change was calculated as described in Figs. 1 and 2. The results are presented as mean ± SD (error bars).
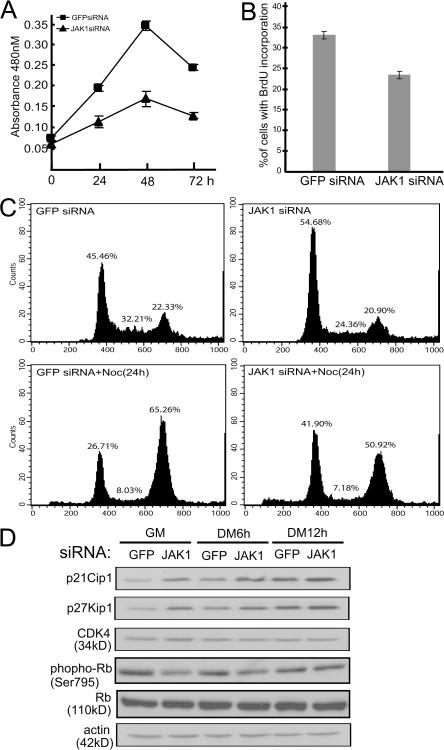
Knockdown of JAK1 inhibits myoblast proliferation
While we were studying the impact of JAK1-siRNA on myogenic differentiation, we also noticed that JAK1-siRNA considerably affected cell proliferation. In a time course experiment, we found that C2C12 cells transfected with JAK1-siRNA displayed a reduced proliferation rate compared with the control cells transfected with GFP-siRNA (Fig. 4 A). Similarly, a lower percentage of C2C12 cells treated with JAK1-siRNA incorporated BrdU compared with cells treated with GFP-siRNA (Fig. 4 B). To closely examine the effect of JAK1-siRNA on the cell cycle, we subjected the siRNA-treated C2C12 cells to flow cytometry analysis. We found that a higher proportion of cells transfected with JAK1-siRNA were arrested in G1 phase compared with the GFP-siRNA–transfected control cells no matter whether nocodazole was used or not (to reduce the in terference from either untransfected cells or cells already in S or early G2 phase; Fig. 4 C). To investigate the underlying molecular mechanisms, we examined several key cell cycle regulators. Although JAK1-siRNA had no obvious effect on the levels of CDK4 and total Rb, it induced p21Cip1 and p27Kip1, two prominent CDK2 inhibitors, in both proliferating cells as well as in cells undergoing early differentiation (Fig. 4 D). Consistently, the levels of Rb phosphorylation at Ser795, a site mainly phosphorylated by CDK2 and CDK4 (Ortega et al., 2003), also decreased in cells with elevated p21Cip1 and p27Kip1 (Fig. 4 D).
Figure 4.
JAK1-siRNA inhibits myoblast proliferation. C2C12 cells were transfected with either GFP-siRNA or JAK1-siRNA as indicated. (A) WST-1 reagent was added to cells at different times as indicated, and absorbance at 480 nm was measured by a plate reader. The experiment was performed in triplicate, and the results are presented as mean ± SD (error bars). (B and C) 30 h after siRNA transfection, 10 μM BrdU was added for 1 h (B). Cells were fixed and subjected to immunostaining using antibodies against BrdU. Nuclei were counterstained with DAPI, and images were taken and analyzed by fluorescent microscopy. The percentage of cells positive for BrdU staining was calculated based on cells from five randomly chosen fields (10× magnifications). The results are presented as mean ± SD. (C, top) Cells were trypsinized, fixed, and subjected to FACS analysis. (bottom) Cells were treated with nocodazole for 24 h followed by FACS analysis. (D) WCE was prepared at different times as indicated, and 30 μg WCE was subjected to immunoblotting analysis.
STAT1 specifically mediates the effect of JAK1 during myogenic differentiation
Although our aforementioned data indicated that JAK1 negatively regulates myogenic differentiation, it remained unclear whether its kinase activity is required in this process. To address this issue, we generated two siRNA-resistant JAK1 constructs: one encoding the wild-type protein and the other encoding a kinase-dead mutant. We then transfected C2C12 cells with JAK1-siRNA together with expression vectors encoding either the wild-type JAK1 (i.e., siRNA sensitive) or the siRNA-resistant JAK1. As shown in Fig. 5 A, although both forms of the siRNA-resistant JAK1 were expressed at high levels in the presence of JAK1-siRNA, only the one retaining the kinase activity effectively reversed the prodifferentiation effect of JAK1-siRNA, as indicated by reduced myogenin expression and a complete lack of MHC expression (lane 2). This suggested that the kinase activity of JAK1 is essential for its repressive effect during myogenic differentiation. To identify downstream mediators of JAK1 in myogenic differentiation, we focused on STATs that are well-established downstream targets and mediators of JAKs (O'Shea et al., 2002). We first designed siRNAs to individually knock down STATs that are present in C2C12 cells (i.e., STAT1, 2, 3, 5A, and 5B as judged by RT-PCR; unpublished data). Among them, only STAT1-siRNA led to a precocious induction of myogenin (Fig. 5 B), an effect similar to that of JAK1-siRNA (Fig. 1 C). In contrast, the siRNAs against STAT3 and other STATs inhibited myogenin expression (Fig. 5 B and unpublished data). Transfection of C2C12 cells with a second set of siRNAs targeting a different region of STAT1 and STAT3 generated similar results (unpublished data). To extend the study to primary myoblasts, we first confirmed that STAT1 was expressed in primary myoblasts both before and after differentiation (Fig. 5 C). Consistently, primary myoblasts transfected with STAT1-siRNA differentiated faster than those with GFP-siRNA, as indicated by a substantially increased number of MHC-positive cells (Fig. 5 D). Furthermore, we found that the repression of myogenin and MHC mediated by the overexpression of JAK1 could be rescued by STAT1-siRNA but not by STAT2-siRNA (Fig. 5 E).
Figure 5.
STAT1 mediates the antidifferentiation effect of JAK1. (A) C2C12 cells were transfected with JAK1-siRNA together with various forms of JAK1 cDNA. After 24 h in DM, cells were harvested, and WCEs were subjected to immunoblotting. R, siRNA resistant; KR, kinase-dead mutant of JAK1 with Lys896 replaced by Arg. (B) C2C12 cells were transfected with various siRNAs as indicated. WCEs were prepared from cells grown in DM for 12 h and subjected to immunoblotting. ns, nonspecific band. (C) 100 μg WCE prepared from primary myoblasts harvested at different times was subjected to immunoblotting. (D) Freshly isolated primary myoblasts were transfected with either GFP-siRNA or STAT1-siRNA. 24 h after transfection, cells were fixed and subjected to immunostaining for MHC (top). The nuclei of the cells were counterstained with DAPI (bottom). The percentage of MHC-positive cells was calculated based on cells from five randomly chosen fields. Error bars represent SD. (E) C2C12 cells were cotransfected with various siRNAs and cDNA expression vectors as indicated. After 36 h in DM, cells were harvested, and WCEs were subjected to immunoblotting. Bar, 100 μm.
LIF promotes proliferation and inhibits the differentiation of C2C12 cells via JAK1–STAT1–STAT3
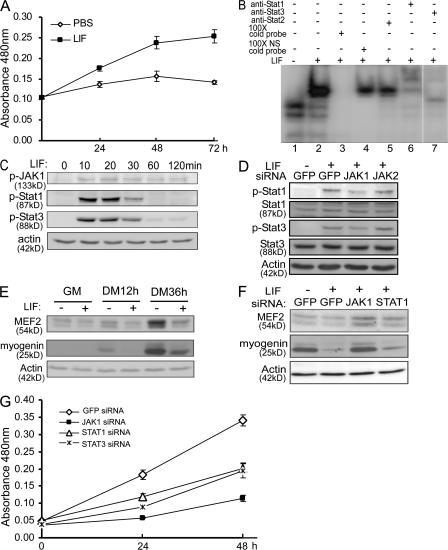
LIF is a known mitogen for both primary myoblasts and C2C12 cells (Austin and Burgess, 1991; Megeney et al., 1996; Spangenburg and Booth, 2002). Because JAK1 is also required for myoblast proliferation (Fig. 4), naturally, we wanted to test whether LIF promotes myoblast proliferation through JAK1 and STAT1. When C2C12 cells were exposed to LIF, cell proliferation was clearly accelerated as measured by WST-1 assays (Fig. 6 A). To reveal the composition of the STAT complex induced by LIF, we performed electrophoretic mobility shift assays (EMSAs) using an oligonucleotide containing a consensus STAT-binding site as a probe (O'Shea et al., 2002). As shown in Fig. 6 B, we did detect specific STAT complexes. Supershift assays with several STAT-specific antibodies revealed that the LIF-induced STAT complexes mainly consist of STAT1 and STAT3 but not STAT2 (Fig. 6 B, lanes 6 and 7). Furthermore, JAK1, STAT1, and STAT3 were all activated as early as 10 min after LIF treatment, which was evident by an increase in their tyrosine-phosphorylated (i.e., active) forms even though their total protein levels did not change much (Fig. 6 C and unpublished data). Moreover, the LIF-induced tyrosine phosphorylation of STAT1 and STAT3 could be reduced by JAK1-siRNA but not by JAK2-siRNA, suggesting that JAK1 mediates the LIF-induced phosphorylation of STAT1 and STAT3 in C2C12 cells (Fig. 6 D). In addition to its role in myoblast proliferation, LIF was also known to inhibit myogenin expression and myogenic differentiation (Jo et al., 2005). To uncover the underlying mechanisms, we examined the expression status of MyoD and MEF2. We found that LIF repressed the expression of MEF2 in both proliferating myoblasts and cells undergoing differentiation (Fig. 6 E). In contrast, LIF had less effect on the expression levels of MyoD (unpublished data). Importantly, LIF-mediated down-regulation of both MEF2 and myogenin was efficiently rescued by JAK1-siRNA and was partially rescued by STAT1-siRNA (Fig. 6 F). Furthermore, LIF-induced C2C12 proliferation was also greatly inhibited by JAK1-siRNA and, to a lesser extent, the siRNAs against either STAT1 or STAT3 (Fig. 6 G). Thus, our aforementioned data suggested that JAK1–STAT1–STAT3 act downstream of LIF and mediate its effect on myoblast proliferation and differentiation.
Figure 6.
LIF stimulates proliferation and represses differentiation via JAK1–STAT1–STAT3. (A) C2C12 cells were treated with either vehicle (i.e., PBS) or LIF. At different time points, cells in 96-well plates (five wells/time point/sample) were subjected to WST-1 assays. The absorbance at 480 nm was plotted against time. The results are presented as mean ± SD (error bars). (B) Cells were treated with LIF for 15 min before harvest. 20 μg WCE was subjected to EMSA. NS, nonspecific. (C) Cells were treated with LIF for various times as indicated. (D) Cells were first transfected with either GFP-siRNA or JAK1-siRNA followed by LIF treatment for 15 min. (E) C2C12 cells were treated with LIF for various times. (F) Cells were first transfected with various siRNAs as indicated followed by LIF treatment for 12 h in DM. WCEs from C–F were subjected to immunoblotting. (G) Cells in 96-well plates were transfected with siRNAs followed by LIF treatment for various times before being subjected to WST-1 assays. The results are plotted and presented the same way as in A. The plus and minus signs indicate that the reagents listed on the left were present and absent, respectively. The white line indicates that intervening lanes have been spliced out.
JAK1/STAT1/STAT3 are up-regulated and activated in cardiotoxin-induced regenerating muscles
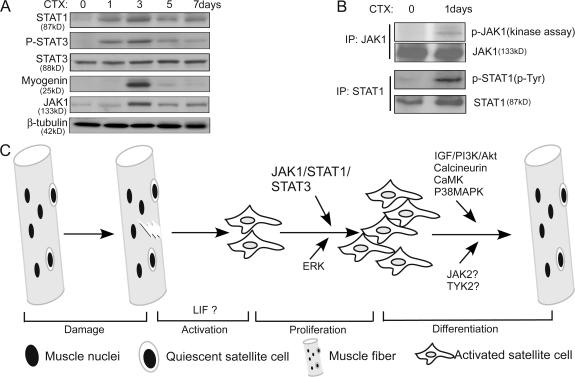
Because MSC-derived primary myoblasts are mainly responsible for injury-induced muscle regeneration, as an initial attempt, we examined the status of JAK1, STAT1, and STAT3 in regenerating tibialis anterior (TA) muscles in response to cardiotoxin-induced muscle injury (Charge and Rudnicki, 2004). In this injury-induced muscle regeneration model, the satellite cell– derived myoblasts actively proliferated in the first 2–3 d (Yan et al., 2003). A majority of myoblasts started to differentiate by day 3 as indicated by a peak expression of myogenin (Fig. 7 A). The damaged area was largely repaired by day 15 (Yan et al., 2003). Consistently, we found that the total levels of both JAK1 and STAT1 increased upon injury, peaked at day 3 after injury, and gradually decreased afterward (Fig. 7 A). Importantly, both the kinase activity of JAK1 and levels of the active STAT1 increased as early as 1 d after injury (Fig. 7 B). As to STAT3, although its total levels did not change much in response to the injury, the levels of active STAT3 substantially increased 1 d after injury and peaked at day 3 (Fig. 7 A).
Figure 7.
JAK1/STAT1/STAT3 were activated during cardiotoxin-induced muscle regeneration. (A) After the injection of cardiotoxin (CTX), TA muscles were isolated at different time points as indicated. WCEs from TA muscles were prepared, and 200 μg WCE was subjected to immunoblotting. (B) WCEs from control and regenerating TA muscles were separately subjected to immunoprecipitation (IP) with either the anti-JAK1 antibody (top two panels) or the anti-STAT1 antibody (bottom two panels). The immunoprecipitated JAK1 was subjected to both protein kinase assays (top) and immunoblotting (second panel), whereas the immunoprecipitated STAT1 was only subjected to immunoblotting (third and fourth panels). (C) A schematic on injury-induced muscle regeneration. Key molecules/pathways acting at different stages of regeneration are indicated.
Discussion
The role of the JAK1–STAT1–STAT3 pathway in both myoblast differentiation and muscle regeneration
In this study, we provide evidence showing that the JAK1– STAT1–STAT3 pathway has two distinct roles in myogenic differentiation: on one hand, it is required for myoblast proliferation as a result of its involvement in regulating the expression of p21Cip1, p27Kip1, and Id1. On the other hand, it prevents myoblasts from premature differentiation by actively repressing genes essential for differentiation (e.g., MyoD, MEF2, and myogenin). In this sense, the status of the JAK1–STAT1–STAT3 pathway can be viewed as a key checkpoint for differentiation, as shutdown of this pathway is a prerequisite for myoblasts to initiate the differentiation program. The diverse roles of the JAK1–STAT1–STAT3 pathway are especially important during injury-induced muscle regeneration. MSCs normally undergo three distinct phases during injury-induced regeneration: activation, proliferation, and differentiation (Fig. 7 C). In the activation phase, the quiescent MSCs are activated by an ill-defined mechanism in response to injury. Certain cytokines/chemokines released by inflammatory cells could potentially serve as the trigger (Tidball, 2005; Wagers and Conboy, 2005). In the proliferation phase, the activated MSCs actively proliferate to generate a sufficient number of myoblasts. Premature differentiation of myoblasts is undesired at this stage. In the final differentiation phase, myoblasts differentiate and fuse with existing myofibers to repair the damaged muscles. Based on the properties of the JAK1–STAT1–STAT3 pathway in myoblast cultures, we expect that the pathway mainly operates in the proliferation phase during muscle regeneration. Consistently, our preliminary study shows that JAK1, STAT1, and STAT3 are all activated in regenerating muscles at a time when myoblasts actively proliferate (Fig. 7, A and B). Because the invading inflammatory cells, including neutrophils, macrophages, and T cells, are also present in regenerating muscles, it remains to be further clarified whether the changes in JAK1–STAT1–STAT3 specifically occur in proliferating myoblasts.
The distinct roles of STAT1 and STAT3 in myoblast proliferation and differentiation
Although STAT3 is commonly associated with cell proliferation in many different cell types, STAT1 is rarely associated with proliferation (O'Shea et al., 2002). In fact, the activation of STAT1 often reduces cell proliferation (Chin et al., 1996; O'Shea et al., 2002). Therefore, it is quite unique that STAT1 is required for myoblast proliferation. Although it remains unclear what dictates these different outcomes, presumably, it is a cell context–dependent phenomenon, which implies that STAT1 has to cooperate with other molecules/pathways to bring about different effects on cell proliferation. The involvement of STAT3 in myoblast proliferation is supported by our following findings: (1) STAT3 and STAT1 form complexes in response to LIF stimulation and (2) the knockdown of STAT3 reduces LIF-induced myoblast proliferation. Consistently, an earlier study showed that LIF activates STAT3 in proliferating myoblasts (Megeney et al., 1996). Unexpectedly, the knockdown of STAT3 does not accelerate differentiation the same way as the knockdown of STAT1 does (Fig. 5 B). A possible explanation is that STAT3 may be required for both the proliferation and differentiation of myoblasts, which is supported by our findings that the levels of active STAT3 gradually increase during differentiation in C2C12 cells (unpublished data) and that the knockdown of STAT3 inhibits differentiation (Fig. 5 B). It is likely that STAT3 may function at different phases of differentiation by associating with different partners. Further investigation is needed to clarify the role of STAT3 in myogenic differentiation.
LIF utilizes the JAK1–STAT1–STAT3 pathway to regulate the proliferation and differentiation of myoblasts
So far, several cytokines and growth factors, including LIF, hepatocyte growth factor, and basic FGF, have been shown to stimulate the proliferation of myoblasts in vitro (Charge and Rudnicki, 2004; Dhawan and Rando, 2005). The only in vivo data came from the LIF knockout mice. It was shown that injury-induced muscle regeneration is delayed in LIF−/− mice and that the defect can be rescued by the injection of exogenous LIF (Kurek et al., 1997). Thus, LIF is essential for the proliferation of myoblasts both in vivo and in vitro. As a member of the IL-6 family of cytokines, LIF is known to exert its diverse functions mainly through the JAK–STAT pathway (Heinrich et al., 2003; Metcalf, 2003). Consistent with this notion, we demonstrate here that LIF specifically utilizes the JAK1–STAT1–STAT3 pathway to regulate the proliferation and differentiation of myoblasts. It remains to be seen whether other members of the IL-6 family are capable of regulating myogenic differentiation through similar mechanisms.
Multiple JAK–STAT pathways are operative in myoblasts with opposing effects
Although several different JAK family members are usually associated with the same receptor complex (e.g., JAK1 and JAK2 associate with the α and β chains of interferon-γ receptor, respectively) in cytokine signaling (Leonard and O'Shea, 1998), JAK1 seems to form homodimers during myogenic differentiation, as the knockdown of either JAK2 or Tyk2, the remaining two members of the JAK family present in both primary and immortalized myoblasts, has the opposite effect as compared with that of JAK1 (Fig. 1 C and unpublished data). This suggests that multiple JAK–STAT pathways function to control the proliferation and differentiation of myoblasts with opposing effects. In the future, it is essential for us to understand how these different JAK–STAT pathways coordinate with each other to control myogenic differentiation and how they cross talk with other signaling pathways. With such knowledge, it is possible to accelerate injury-induced muscle regeneration by differentially modulating different JAK–STAT pathways with small molecules. We believe that our work has provided a new direction in studying the biology of muscle stem cells.
Materials and methods
Cell lines, DNA constructs, and reagents
C2C12 cells were maintained in DME with 20% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin (also called growth medium [GM]) in a 37°C incubator with 5% CO2. To induce differentiation, cells were grown in DME containing 2% horse serum (also called differentiation medium [DM]). Flag-JAK1 (human) and two STAT1 constructs (i.e., the wild-type and Y701F mutant) were gifts from Z. Wen (Hong Kong University of Science and Technology, Hong Kong, China). G133-luc, MCK-luc, 4xRE-luc, and 3xMEF2-luc were described previously (Xu et al., 2002; Sun et al., 2004). Silent mutation was introduced into Flag-JAK1(wild type) and the kinase-dead Flag-JAK1(K896R) constructs to generate siRNA-resistant R-Flag-JAK1(wild type) and R-Flag-JAK1(K896R), respectively, using the oligonucleotide 5′-ACCCGAAAGCGGCGGCAATCACATAGCTGATCTGAAAAAG-3′ (top strand). U0126 and LIF were purchased from Calbiochem and Chemicon, respectively.
Antibodies, immunoblotting, immunostaining, and microscopic imaging
The sources of the antibodies used in this study are listed as follows: JAK1, JAK2, and STAT1 were purchased from Upstate Biotechnology; JAK1, Rb, CDK4, myogenin, MyoD, Id1, p21Cip1, p27Kip1, β-actin, and MEF2 were obtained from Santa Cruz Biotechnology, Inc.; anti-Flag was purchased from Sigma-Aldrich; phosphor-Rb (Ser795), phosphor-JAK1 (Tyr1022/1023), phopho-STAT1 (Tyr705), and phosphor-STAT3 (Tyr705) were obtained from Cell Signaling; and MHC was purchased from Developmental Studies Hybridoma Bank. Polyclonal STAT3 antibody was a gift from Z. Wen. Immunoblotting was performed according to standard procedures (Xu et al., 2002). For immunostaining, FITC- and rhodamine-conjugated secondary antibodies and DAPI were used to label selected molecules and to counterstain the nuclei, respectively. The images were acquired at room temperature by a CCD camera (Spot RT; Diagnostic Instruments) mounted on a fluorescent microscope (IX70; Olympus) using SPOT software (version 4.0.9; Diagnostic Instruments). UPlanFL 10× NA 0.3 and LCPlanFL 20× NA 0.4 objectives (Olympus) were used. The brightness and contrast of the images were adjusted by Photoshop 6.0 (Adobe).
Cell lysis and preparation of soluble WCEs
After removal of the culture medium, cells were washed once with PBS before addition of the lysis buffer (50 mM Hepes, pH 7.6, 1% vol/vol Triton X-100, 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 100 mM NaF, 20 mM p-nitrophenylphosphate, 20 mM β-glycerophosphate, 50 μM sodium vanadate, 2 mM DTT, 0.5 mM PMSF, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, and 0.7 μg/ml pepstatin). Cells were lysed for 10 min at 4°C followed by centrifugation to remove the insoluble cell debris. The concentration of the whole cell extracts (WCEs) was determined by a protein assay reagent (Bio-Rad Laboratories).
Preparation of primary myoblasts
Isolation of primary MSCs was performed as described previously with minor modifications (Allen et al., 1997). In brief, skeletal muscles of 2-mo-old C57BL/6J mice were isolated, minced, and digested in 1.25 mg/ml protease type XVII (Sigma-Aldrich) for 1.5 h in a 37°C water bath. Satellite cells were purified by discontinuous Percoll gradient centrifugation and collected from the interface of 20 and 60% Percoll. Purified satellite cells were then cultured in GM (DME containing 20% FBS and 2% chicken embryo extracts) in culture dishes coated with 4 mg/ml Matrigel (BD Biosciences) to generate primary myoblasts. Myoblasts were induced to differentiate in DM (DME with 5% horse serum).
Cardiotoxin-induced muscle injury and regeneration
TA muscles of 6–8-wk-old C57BL/6 mice were injected with 25 μl of 10 μM cardiotoxin (Sigma-Aldrich). At different time points after injury, mice were killed by cervical dislocation, and the TA muscles were surgically isolated and homogenized in the lysis buffer followed by centrifugation to remove insoluble debris. Uninjured TA muscles were used as a control.
siRNA transfection and adenovirus preparation and infection
To deliver oligonucleotide-based siRNA, 50–70% confluent C2C12 cells were transfected with 100 nM siRNA using LipofectAMINE 2000 (Invitrogen). The following siRNAs were synthesized at Dharmacon, Inc.: JAK1 (5′-GCCUGAGAGUGGAGGUAAC-3′), JAK2 (5′-GCAAACCAGGAAUGCUCAA-3′), STAT1 (5′-GCGUAAUCUCCAGGAUAAC-3′), STAT3 (5′-CTGGATAACTTCATTAGCA-3′), and enhanced GFP (5′-GCUGACCCUGAAGUUCAUC-3′). To generate shRNA from an adenoviral vector, we used the Block-iT RNAi expression kit (Invitrogen) according to the manufacturer's instructions. 20 μl of viruses (∼108 plaque-forming U/ml) was used to infect 60% confluent primary myoblasts in 35-mm plates.
In vitro immune complex kinase assays
JAK1 was first immunoprecipitated from 1 mg WCEs, washed extensively, and reconstituted in the kinase buffer (10 mM Hepes, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 100 μM Na3VO4, and 0.25 mCi/ml γ-[32P]ATP). The reaction was incubated at room temperature for 30 min and terminated by adding sample loading dye. The reaction was separated on 8% SDS-PAGE, and the gel was dried and subjected to autoradiography.
WST-1 assays
C2C12 cells were first seeded into 96-well plates in triplicate at a density of 103 cells/well followed by transfection with various siRNAs the next day. WST-1 reagent (Boehringer) was then added at fixed time points according to the manufacturer's instructions. The absorbance at 480 nm was measured using a microtiter plate reader.
EMSA
EMSA was performed as described previously (Chan et al., 2003). The top strand sequence of the probe is 5′-GTCGACATTTCCCGTAAATC-3′.
BrdU labeling and FACS analysis
An in situ cell proliferation kit (Roche) was used to measure BrdU incorporation. Cells were labeled with 10 μM BrdU for 1 h. Nuclei incorporating BrdU were visualized by fluorescein-conjugated monoclonal anti-BrdU antibody. For FACS analysis, cells were trypsinized, washed with PBS, fixed in 70% ethanol for 30 min on ice, harvested by centrifugation at 300 g for 5 min, and resuspended in 400 μl PBS. To stain the DNA, 5 μl propidium iodide (5 mg/ml) and 5 μl RNase A (10 mg/ml) were added to the solution. Cells were incubated at 37°C for 30 min before being subjected to FACS analysis.
Acknowledgments
We thank Drs. Zilong Wen, Xianghong Wang (Hong Kong University, Hong Kong, China), Terence Partridge (Imperial College, London, UK), Weimin Zhong, (Yale, New Haven, CT), and Yi Sun (University of California, Los Angeles, Los Angeles, CA) for reagents, protocols, and helpful comments.
This work was supported by a competitive earmarked research grant (project no. 663007) and a central allocation grant (CA06/07.SC02) from the Hong Kong Research Grant Council as well as an Area of Excellence Scheme grant (AOE/B-15/01).
L. Sun and K. Ma contributed equally to this paper.
L. Sun's and K. Ma's present address is The First Clinical Hospital, Jilin University, Changchun 130021, China.
Abbreviations used in this paper: DM, differentiation medium; EMSA, electro-phoretic mobility shift assay; ERK, extracellular signal-regulated kinase; GM, growth medium; JAK, Janus kinase; LIF, leukemia inhibitory factor; MCK, muscle creatine kinase; MEF2, myocyte enhancer–binding factor 2; MHC, myosin heavy chain; MRF, myogenic regulatory factor; MSC, muscle satellite cell; shRNA, short hairpin RNA; STAT, signal transducer and activator of transcription; TA, tibialis anterior; WCE, whole cell extract.
References
- Allen, R.E., C.J. Temm-Grove, S.M. Sheehan, and G. Rice. 1997. Skeletal muscle satellite cell cultures. Methods Cell Biol. 52:155–176. [DOI] [PubMed] [Google Scholar]
- Austin, L., and A.W. Burgess. 1991. Stimulation of myoblast proliferation in culture by leukaemia inhibitory factor and other cytokines. J. Neurol. Sci. 101:193–197. [DOI] [PubMed] [Google Scholar]
- Benezra, R., R.L. Davis, D. Lockshon, D.L. Turner, and H. Weintraub. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 61:49–59. [DOI] [PubMed] [Google Scholar]
- Bennett, A.M., and N.K. Tonks. 1997. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 278:1288–1291. [DOI] [PubMed] [Google Scholar]
- Black, B.L., and E.N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167–196. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J.K., L. Sun, X.J. Yang, G. Zhu, and Z. Wu. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278:23515–23521. [DOI] [PubMed] [Google Scholar]
- Charge, S.B., and M.A. Rudnicki. 2004. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84:209–238. [DOI] [PubMed] [Google Scholar]
- Chin, Y.E., M. Kitagawa, W.C. Su, Z.H. You, Y. Iwamoto, and X.Y. Fu. 1996. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 272:719–722. [DOI] [PubMed] [Google Scholar]
- Coolican, S.A., D.S. Samuel, D.Z. Ewton, F.J. McWade, and J.R. Florini. 1997. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J. Biol. Chem. 272:6653–6662. [DOI] [PubMed] [Google Scholar]
- Dhawan, J., and T.A. Rando. 2005. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 15:666–673. [DOI] [PubMed] [Google Scholar]
- Favata, M.F., K.Y. Horiuchi, E.J. Manos, A.J. Daulerio, D.A. Stradley, W.S. Feeser, D.E. Van Dyk, W.J. Pitts, R.A. Earl, F. Hobbs, et al. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623–18632. [DOI] [PubMed] [Google Scholar]
- Friday, B.B., V. Horsley, and G.K. Pavlath. 2000. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J. Cell Biol. 149:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, P.C., I. Behrmann, S. Haan, H.M. Hermanns, G. Muller-Newen, and F. Schaper. 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman, C.E., and M.A. Rudnicki. 2005. Molecular regulation of satellite cell function. Semin. Cell Dev. Biol. 16:575–584. [DOI] [PubMed] [Google Scholar]
- Jo, C., H. Kim, I. Jo, I. Choi, S.C. Jung, J. Kim, S.S. Kim, and S.A. Jo. 2005. Leukemia inhibitory factor blocks early differentiation of skeletal muscle cells by activating ERK. Biochim. Biophys. Acta. 1743:187–197. [DOI] [PubMed] [Google Scholar]
- Kami, K., and E. Senba. 2002. In vivo activation of STAT3 signaling in satellite cells and myofibers in regenerating rat skeletal muscles. J. Histochem. Cytochem. 50:1579–1589. [DOI] [PubMed] [Google Scholar]
- Kataoka, Y., I. Matsumura, S. Ezoe, S. Nakata, E. Takigawa, Y. Sato, A. Kawasaki, T. Yokota, K. Nakajima, A. Felsani, and Y. Kanakura. 2003. Reciprocal inhibition between MyoD and STAT3 in the regulation of growth and differentiation of myoblasts. J. Biol. Chem. 278:44178–44187. [DOI] [PubMed] [Google Scholar]
- Kurek, J.B., J.J. Bower, M. Romanella, F. Koentgen, M. Murphy, and L. Austin. 1997. The role of leukemia inhibitory factor in skeletal muscle regeneration. Muscle Nerve. 20:815–822. [DOI] [PubMed] [Google Scholar]
- Leonard, W.J., and J.J. O'Shea. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16:293–322. [DOI] [PubMed] [Google Scholar]
- Mansour, S.J., J.M. Candia, K.K. Gloor, and N.G. Ahn. 1996. Constitutively active mitogen-activated protein kinase kinase 1 (MAPKK1) and MAPKK2 mediate similar transcriptional and morphological responses. Cell Growth Differ. 7:243–250. [PubMed] [Google Scholar]
- McKinsey, T.A., C.L. Zhang, J. Lu, and E.N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 408:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T.A., C.L. Zhang, and E.N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40–47. [DOI] [PubMed] [Google Scholar]
- Megeney, L.A., R.L. Perry, J.E. LeCouter, and M.A. Rudnicki. 1996. bFGF and LIF signaling activates STAT3 in proliferating myoblasts. Dev. Genet. 19:139–145. [DOI] [PubMed] [Google Scholar]
- Metcalf, D. 2003. The unsolved enigmas of leukemia inhibitory factor. Stem Cells. 21:5–14. [DOI] [PubMed] [Google Scholar]
- Molkentin, J.D., and E.N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445–453. [DOI] [PubMed] [Google Scholar]
- Molkentin, J.D., B.L. Black, J.F. Martin, and E.N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 83:1125–1136. [DOI] [PubMed] [Google Scholar]
- Morgan, J.E., and T.A. Partridge. 2003. Muscle satellite cells. Int. J. Biochem. Cell Biol. 35:1151–1156. [DOI] [PubMed] [Google Scholar]
- O'Shea, J.J., M. Gadina, and R.D. Schreiber. 2002. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 109:S121–S131. [DOI] [PubMed] [Google Scholar]
- Ortega, S., I. Prieto, J. Odajima, A. Martin, P. Dubus, R. Sotillo, J.L. Barbero, M. Malumbres, and M. Barbacid. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35:25–31. [DOI] [PubMed] [Google Scholar]
- Sabourin, L.A., and M.A. Rudnicki. 2000. The molecular regulation of myogenesis. Clin. Genet. 57:16–25. [DOI] [PubMed] [Google Scholar]
- Spangenburg, E.E., and F.W. Booth. 2002. Multiple signaling pathways mediate LIF-induced skeletal muscle satellite cell proliferation. Am. J. Physiol. Cell Physiol. 283:C204–C211. [DOI] [PubMed] [Google Scholar]
- Sun, L., L. Liu, X.J. Yang, and Z. Wu. 2004. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 117:3021–3029. [DOI] [PubMed] [Google Scholar]
- Tamir, Y., and E. Bengal. 2000. Phosphoinositide 3-kinase induces the transcriptional activity of MEF2 proteins during muscle differentiation. J. Biol. Chem. 275:34424–34432. [DOI] [PubMed] [Google Scholar]
- Tapscott, S.J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 132:2685–2695. [DOI] [PubMed] [Google Scholar]
- Tidball, J.G. 2005. Inflammatory processes in muscle injury and repair. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288:R345–R353. [DOI] [PubMed] [Google Scholar]
- Wagers, A.J., and I.M. Conboy. 2005. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 122:659–667. [DOI] [PubMed] [Google Scholar]
- Wu, Z., P.J. Woodring, K.S. Bhakta, K. Tamura, F. Wen, J.R. Feramisco, M. Karin, J.Y. Wang, and P.L. Puri. 2000. p38 and extracellular signal- regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 20:3951–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., and Z. Wu. 2000. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 275:36750–36757. [DOI] [PubMed] [Google Scholar]
- Xu, Q., L. Yu, L. Liu, C.F. Cheung, X. Li, S.P. Yee, X.J. Yang, and Z. Wu. 2002. p38 Mitogen-activated protein kinase-, calcium-calmodulin-dependent protein kinase-, and calcineurin-mediated signaling pathways transcriptionally regulate myogenin expression. Mol. Biol. Cell. 13:1940–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, D., and O. Saxel. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 270:725–727. [DOI] [PubMed] [Google Scholar]
- Yan, Z., S. Choi, X. Liu, M. Zhang, J.J. Schageman, S.Y. Lee, R. Hart, L. Lin, F.A. Thurmond, and R.S. Williams. 2003. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 278:8826–8836. [DOI] [PubMed] [Google Scholar]
- Zetser, A., E. Gredinger, and E. Bengal. 1999. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 274:5193–5200. [DOI] [PubMed] [Google Scholar]