Abstract
The signaling systems of Notch and bone morphogenetic protein (BMP) are highly conserved from flies to mammals and have been shown to be important in the development of multiple organs. For instance, in the fate determination of mouse neuroepithelial cells, Notch signaling plays a role in keeping the progenitors from differentiating into neurons. BMP is also known to inhibit neuronal differentiation. In this paper, we show that BMP2 enhances Notch-induced transcriptional activation of Hes-5 and Hesr-1 in mouse neuroepithelial cells. BMP2 stimulation, in addition to the introduction of the intracellular domain of Notch (NIC), resulted in enhanced activation of the Hes-5 gene promoter. RBP-Jκ binding to its target sequence is important not only for Notch signaling, but also for BMP2 signaling, to activate the Hes-5 gene promoter. Smad1, a Smad species that is activated by BMP2, barely interacted with NIC, but did form a complex with NIC in the simultaneous presence of the coactivators P/CAF and p300. Recruitment of p300 to the NIC-containing complex was facilitated by activated Smad1, which is suggested to contribute to BMP2-mediated enhancement of Notch-induced Hes-5 expression. These data suggest a novel functional cooperation between Notch signaling and BMP signaling.
INTRODUCTION
The Notch signaling pathway is a highly conserved signaling mechanism and has been shown to be implicated in cell–cell communication in multiple developmental programs (1–4). In vertebrates, Notch signaling controls cell fate determination in a variety of cell types that include those of the nervous system, muscle, pancreas and the hematopoietic system (5). Four Notch family members (Notch 1–4), which are single transmembrane spanning proteins, have been identified in vertebrates (5). When the Notch ligands (Jagged-1, Jagged-2 and Delta-1 to Delta-3) are expressed by the neighboring cells and bind to Notch, it undergoes a series of proteolytic processes and the intracellular domain of the Notch receptor (NIC) is cleaved (6). The cleaved NIC then translocates to the nucleus, where it controls the expressions of its target genes via association with RBP-Jκ/CBF (7). The most established target genes for Notch signaling are Hairy/Enhancer of split (HES) genes, which are members of the genes possessing basic regions at their C-terminals and characteristic helix–loop–helix (HLH) motifs, that are referred to as basic HLH (bHLH) factors (8). The most investigated function of HES has been as a repressor for tissue-specific gene transcription. HES-1 and HES-5 have been shown to bind to their target DNA sequences, and to recruit histone deacetylase (HDAC) activity by associating with Groucho, resulting in transcriptional repression (9–11). Furthermore, they associate with ubiquitously expressed bHLH factors, such as E47, and prevent tissue-specific bHLH factors, such as Mash1, from forming functional complexes with E protein (12,13). In this manner, Notch represses the differentiation of cells to specific lineages.
Bone morphogenetic proteins (BMPs) are a family of cytokines belonging to the transforming growth factor-β (TGF-β) superfamily (14). BMPs are pleiotropic cytokines that are active in many tissues, including the CNS (15), and are also involved in the fate determination of cells in various organs. The action of BMPs is mediated by heterotetrameric serine/threonine kinase receptors and the downstream transcription factors Smad1, -5 or -8. After these transcription factors are phosphorylated on serine residues, they form a complex with a common mediator, Smad4, and the complex is translocated into the nucleus to activate the transcription of specific genes (16–18). Inhibitory Smad proteins, Smad6 and Smad7, repress the action of BMPs by inhibiting the receptor-mediated phosphorylation of Smad1, -5 or -8 or by competing with Smad4 for the binding to Smad1, -5 and -8 (16–18).
Recently, it has been demonstrated that BMP2 inhibits neurogenesis of mouse neuroepithelial cells (19–22). Although BMP genes are expressed mostly in the tectum, their receptors are expressed mainly in the ventricular zone where neural cell fate is thought to be determined. BMP proteins diffuse and function practically in the ventricular zone, where Notch signaling is also activated (23). The anti-neurogenic effect of BMP2 is thought to be at least partly mediated via induction of gene expression for Id1, Id3 and HES-5 in the neuroepithelial cells (19), among which HES-5 is also known to be induced by Notch activation. In the promoter region of the HES-5 gene, a consensus binding sequence for Smads and an RBP-Jκ binding site are found, and Smad7, an inhibitory Smad, inhibits BMP2-induced HES-5 gene transcription (19).
Since the transcription of HES-5 is induced by Notch as well as BMP2, we hypothesized that there might be a signaling cross-talk between the Notch and BMP signaling pathways. In the present study, we show that Notch and BMP signalings induce transcriptional activation of the HES-5 and Hesr-1 genes in a cooperative manner. We also show that Smad1 and NIC are able to form a complex containing P/CAF and p300. These results indicate a novel signaling cross-talk between Notch and BMP.
MATERIALS AND METHODS
Reagents and plasmid constructs
BMP2 (Yamanouchi Pharmaceutical) was dissolved in phosphate-buffered saline containing 0.1% albumin and stored at –80°C until use. H5-Luc containing a fragment of the Hes-5 promoter (nucleotides –179 to +72) and the luciferase gene was used (19). SMH5-Luc was constructed by introducing mutations (taaatcgcc) into a Smad-binding site (SBS) (–161 to –153: gcccgcgcc) of H5-Luc. RMH5-Luc was constructed by mutating the RBP-J binding site (–79 to –72: tgtggaa) of H5-Luc to tgtgctga. Hsr1-Luc containing the Hesr-1 gene promoter region (–609 to +130) and the luciferase gene was used. Flag-tagged Smad1 and a series of deletion mutants of Smad1 (MH1, MH1+Linker, Linker+MH2 and MH2) in pCDNA3, and an HA-tagged constitutively active form of ALK3 (CaALK3) in pCDNA3 were kindly provided by Dr K. Miyazono (Department of Molecular Pathology, Graduate School of Medicine, University of Tokyo). Flag-tagged and six repeats of Myc (6Myc)-tagged P/CAF in pCDNA3 were kind gifts from Drs P. ten Dijke and S. Itoh (The Netherlands Cancer Institute). Myc-tagged intracellular region of Notch1 (Myc NIC) was constructed by subcloning CMV-Myc N1IC (a kind gift from Dr U. Lendahl) into pEFBOS. Antibodies to Myc (Chemicon, Temecula, CA), Flag (Sigma) and HA (Santa Cruz) were used.
Animals and cell preparation
Time-pregnant ICR mice were used to prepare the neuroepithelial cells. Mice were treated according to the guidelines of the Kumamoto University Center for Animal Resources and Development. Neuroepithelial cells were prepared from the telencephalons of E14.5 mice and cultured as described previously (24). Briefly, the telencephalons were triturated in Hank’s balanced salt solution by mild pipetting with a 1 ml pipette tip (Gilson, Middleton, WI). Dissociated cells were cultured for 4 days in N2-supplemented DMEM-F12 containing 10 ng/ml basic FGF (R&D Systems, Minneapolis, MN) (N2/DMDM/F12/bFGF) on culture dishes which had been precoated with poly-l-ornithine (Sigma) and fibronectin (Life Technologies, Gaithersburg, MD). P19 cells were maintained in α-MEM media (Gibco) containing 10% fetal calf serum (FCS) and the media were replaced with α-MEM containing 1% FCS 30 min before cytokine stimulation.
Luciferase assay
Neuroepithelial cells replated on 12-well plates (Nunc) after a 4 day culture, or P19 cells cultured on 24-well plates (Nunc) overnight, were transfected with reporter vectors. Control transfection was performed with the sea pansy luciferase gene conjugated with the human elongation factor 1α promoter (R-Luc) (25). Transfection was performed using Trans-It LT1 (Mirus) according to the manufacturer’s procedures. Cells were stimulated with BMP2 (40 ng/ml) for 8 h on the following day and then solubilized. Luciferase activity was measured according to the recommended procedures for the Pikkagene Dual Luciferase Assay System (Tokyo Ink Inc., Tokyo, Japan). A Micro Lumat LB96B luminometer (Wallac Berthold) was used for the detection.
Reverse transcription–PCR (RT–PCR)
The first strand cDNAs were made from 5 µg of total RNA from E14.5 neuroepithelial cells and P19 cells using Superscript II (Invitrogen), and dissolved in 100 µl of TE. Then, 1 µl of the reverse transcription reaction was used as a template for PCR amplification (AmpliTaq Gold; Applied Biosystems) in a volume of 25 µl containing 2 µM gene-specific primers. The time cycle was 35 cycles of denaturation: 94°C for 30 s; annealing: 60°C for 30 s; extension: 72°C for 30 s; after an initial incubation at 95°C for 9 min. Ten microliters of the reaction were then separated in a 1.2% agarose gel and visualized by ethidium bromide staining. The gene-specific primers were as follows: mouse Notch1: NIC-S, 5′-atg tgg cag cca agc ctg ag-3′; NIC-AS, 5′-cac cag gtg agg ctg tgt tg-3′; mouse Notch3: NIC3-S, 5′-ctc acc acg gcc ttt cag tg-3′; NIC3-AS, 5′-gct ggg cta ggt gtt gag tc-3′; mouse RBP-Jκ: RBP-S, 5′-ggt tct tgt ggt cta agg ctg-3′; RBP-AS, 5′-att tta ccc tac ggg cac cat c-3′; mouse Delta1: Dl-S, 5′-act gtg gac tat aac ctc gtt c-3; Dl-AS, 5′-aca acc agc agg cag tcc ag-3′; mouse Jagged1: Jag1-S, 5′-atg att gac agc tgc act gtg-3′; Jag1-AS, 5′-tcc act tca tca tag cag gta c-3′; mouse Jagged2: Jag2-S, 5′-gga tgg ctt ccg ctg cca c-3′; Jag2-AS, 5′-ctg tga agc cgc tgt cac ag-3′; mouse BMP receptor (BMPR)-I: BMPR-I-S, 5′-cag act tgg acc aga aga agc c-3′; BMPR-I-AS, 5′-aca ttc tat tgt ctg cgt agc-3′; BMPR-II: BMPR-II-S, 5′-gct tcg cag aat caa gaa cg-3′; BMPR-II-AS, 5′-gtg gac tga gtg gtg ttg tg-3′; mouse Smad1: Smd1-S, 5′-gcg tgt aga act aga cca gcc gct-3′; Smd1-AS, 5′-agg aga gtt ggg gta gct gct-3′; Smad4: Smd4-S, 5′-gtt cag gta gga gag acg ttt a-3′; Smd4-AS, 5′-taa agg ctg tgg gtc cgc aat-3′; mouse G3PDH: G3P-S, 5′-acc aca gtc cat gcc atc ac-3′; G3P-AS, 5′-tcc acc acc ctg ttg ctg ta-3′.
Immunoblotting and immunoprecipitation
Cos7 cells were replated on 60 mm dishes (Nunc) and on the next day, cells were transfected with expression vectors using LT1, and cultured overnight. On the following day, cells were lysed in NP40 buffer [0.5% NP40, 10 mM Tris–HCl pH 7.4, 150 mM NaCl, 3 mM pAPMSF (Wako Chemicals, Osaka, Japan), 5 mg/ml aprotinine (Sigma), 2 mM sodium orthovanadate (Wako Chemicals), 5 mM EDTA]. Lysates were immunoprecipitated with an antibody against Myc using Protein A Sepharose (Amersham, Arlington Heights, IL). Precipitates and, in some cases, cell lysates were subjected to SDS–PAGE and subsequent immunoblotting with antibodies against Myc. Detection was performed with an ECL system (Amersham).
RESULTS
Enhanced activation of Hes-5 and Hesr-1 transcription by BMP2 and Notch signalings
We have previously demonstrated that BMP2 stimulation of neuroepithelial cells prepared from fetal mouse telencephalons induces expression of the Hes-5 gene (19), which has been shown to be induced by activation of Notch signaling (26). This encouraged us to examine whether BMP2 stimulation and Notch activation show cross-regulatory effects on the transcriptional activation of the Hes-5 gene.
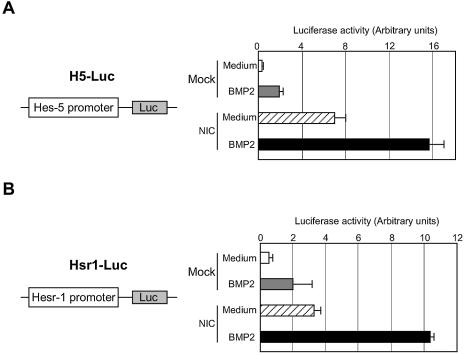
Hes-5 promoter activity was determined in the mouse neuroepithelial cells. As shown in Figure 1A, the Hes-5 promoter was activated when the cells were stimulated by BMP2. On activation by its ligands, the NIC is cleaved and translocated into nucleus where it induces target gene expression (6). Thereby, introduction of NIC into cells can mimic Notch activation. NIC introduction into neuroepithelial cells exhibited upregulation of the Hes-5 promoter activity (Fig. 1A). Interestingly, BMP2 stimulation of the NIC-expressing cells showed further enhancement of the Hes-5 promoter activation.
Figure 1.
Activation of Hes-5 and Hesr-1 gene promoters by BMP2 and Notch signalings. Neuroepithelial cells prepared from E14.5 mouse telencephalons were transfected with H5-Luc (A) or Hsr1-Luc (B) with or without NIC-pEF-BOS. On the following day, the cells were incubated with medium alone or BMP2 (40 ng/ml) for 8 h. The cells were then lysed and the luciferase activities were determined.
To ascertain whether or not the transcriptional enhancement by BMP2 and Notch was confined to the Hes-5 promoter, the effects of simultaneous activation of the two signaling pathways on another promoter were examined. Hesr-1 has recently been identified as one of the target genes of the Notch signaling pathway (27,28). We have also identified the same gene being expressed in neuroepithelial cells stimulated by bFGF (data not shown) and found that its expression was induced by BMP2 stimulation as well. As shown in Figure 1B, enhanced transcriptional activation by BMP2 stimulation and Notch expression was also observed in the case of the Hesr-1 promoter.
These results suggested that the cooperation of BMP2 and Notch signaling may generally take place in target genes of Notch signaling. However, the cooperative effect was not observed in the Hes-1 gene promoter (data not shown), whose expression is known to be induced by Notch activation but not by BMP2 stimulation.
Requirement of Smad and RBP-Jκ binding sites for the cooperative transactivation
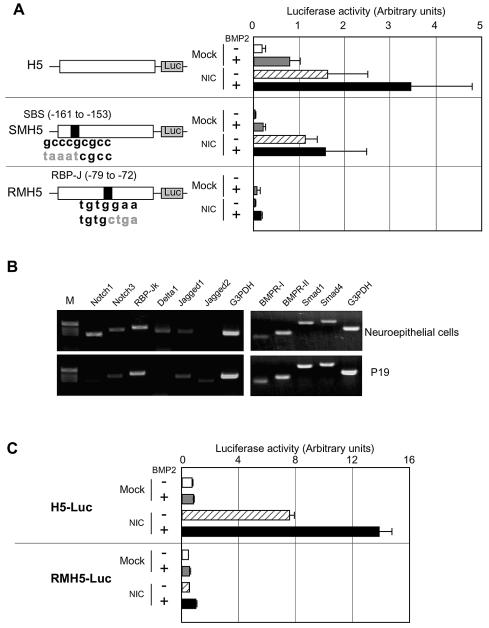
The failure of the enhancemnt of the Hes-1 gene promoter activation by BMP2 stimulation suggested a requirement for the SBS for the cooperation of BMP2 and Notch signalings. To ascertain this, nucleotide substitutions were introduced into a consensus SBS in the Hes-5 promoter (Fig. 2A). This mutation inhibited BMP2-induced Hes-5 gene promoter activation but not so significantly repressed the NIC-induced activation (Fig. 2A). Furthermore, the mutation led to the disappearance of the collaborative enhancement of the Hes-5 gene promoter activation induced by NIC and BMP2 (Fig. 2A).
Figure 2.
Requirement for a SBS and an RBP-Jκ binding site for the synergistic gene induction by BMP2 and Notch. (A) Requirement for a SBS and an RBP-Jκ binding site. Neuroepithelial cells were transfected with H5-Luc (top), SMH5-Luc (middle) or RMH5-Luc (bottom) with or without the NIC expression vector. On the next day, they were stimulated by medium or BMP2 (40 ng/ml) for 8 h and the luciferase activities were determined. (B) RT–PCR for Notch and Notch ligands in neuroepithelial cells and P19 cells. cDNA was synthesized from 5 µg of total RNA from neuroepithelial cells or P19 cells. PCR was performed using the cDNAs for the indicated genes. (C) P19 cells were transfected with H5-Luc (top) or RMH5-Luc (bottom) with or without the NIC expression vector, and then stimulated with BMP2 (40 ng/ml).
NIC can activate transcription of its target genes through interaction with RBP-Jκ which has DNA binding ability (29). To confirm the importance of the RBP-Jκ binding site for the cooperative gene activation by BMP2 and Notch, we constructed RMH5-Luc harboring nucleotide substitutions in the RBP-Jκ binding site in H5-Luc (Fig. 2A). RMH5-Luc was not activated by NIC expression and its cooperative activation by BMP2 and NIC was also not observed either (Fig. 2A). Interestingly, the BMP2-induced activation of the Hes-5 gene promoter was, unexpectedly, virtually abolished in RMH5-Luc. This indicated that the RBP-Jκ binding site is important not only for Notch signaling, but also for BMP2 signaling, to activate the Hes-5 gene promoter. It is, however, worth noting that in Figures 1 and 2A, H5-Luc could be activated by BMP2 stimulation alone. These contradicting observations made us hypothesize that Notch signaling might be activated endogenously in the neuroepithelial cells (23,30,31), thereby resulting in activation of the Hes-5 promoter by BMP2 stimulation alone. As shown in Figure 2B, neuroepithelial cells did indeed express Notch1, Notch3, and their ligands, Delta1 and Jagged1. They also expressed RBP-Jκ. In contrast, embryonic carcinoma P19 cells did not express detectable amounts of Notch1, Delta1, or Jagged2 compared with the neuroepithelial cells. P19 cells expressed RBP-Jκ, Notch3 and Jagged1, although the expression levels of Notch3 and Jagged1 were strongly lower than those in the neuroepithelial cells. These observations suggested that in P19 cells the Notch signaling pathway may not be endogenously activated. The components necessary for BMP2 to signal, such as BMPR-I, -II, Smad1 and Smad4, were similarly expressed in the both cells (Fig. 2B). Based on these observations, we studied the Hes-5 gene promoter activity in P19 cells. In support of our hypothesis, the Hes-5 gene promoter was not activated by BMP2 stimulation alone in the P19 cells (Fig. 2C), which is in marked contrast to the results obtained with the neuroepithelial cells. Expression of NIC activated the Hes-5 promoter and this was enhanced by BMP2 stimulation (Fig. 2C). RMH5-Luc did not respond to NIC expression or a combination of NIC expression and BMP2 stimulation. In addition to these observations, a promoter (pGa981-6) which possesses 12 repeats of RBP-Jκ binding sites (EBNA2RE), but no SBSs, responded to NIC expression but not to BMP2 stimulation in P19 cells, resulting in failure of the cooperative activation by Notch and BMP2 (data not shown). These results suggested that RBP-Jκ binding to its binding site is required for Hes-5 gene expression induced by Notch activation as well as BMP2 stimulation.
Molecular interaction between NIC, Smads, P/CAF and p300
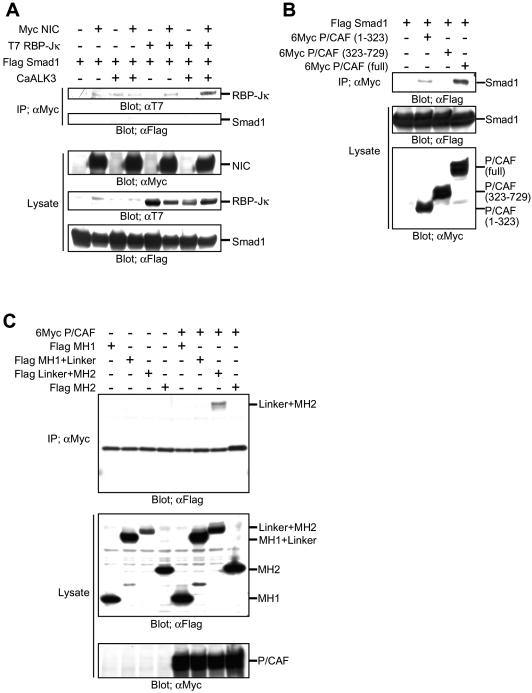
To explain the cooperative transcriptional enhancement induced by the Notch and BMP signalings observed above, we studied the molecular interaction between NIC and Smad1, a Smad protein species activated by BMP2 stimulation. A direct interaction between NIC and Smad1 was first investigated. They showed a barely detectable interaction, which is independent from the forced expression of RBP-Jκ (Fig. 3A).
Figure 3.
Molecular interactions between Smad1, NIC and P/CAF. (A–C) Cos7 cells were transfected with expression vectors for the indicated genes. On the next day, the cells were lysed and immunoprecipitated with an anti-Myc antibody. The precipitates and lysates were subjected to SDS–PAGE and blotted with the indicated antibodies.
Recently, two coactivators with histone acetyltransferase (HAT) activity, p300 and P/CAF (32,33), have been shown to interact with NIC, and they act cooperatively to mediate NIC-activated transcription (34). Smad1 has been shown to interact with one of these coactivators, p300 (35). Smad3, which is a Smad protein species activated downstream of TGF-β, can interact with P/CAF (36). Therefore, we speculated that Smad1 might also make a complex with P/CAF. As shown in Figure 3B, P/CAF could bind to Smad1 via its N-terminal region. The linker and MH2 regions of Smad1 were both required for the interaction with P/CAF (Fig. 3C). We then investigated whether NIC makes a complex with Smad1 in the presence of the coactivators. As shown in Figures 3A and 4A, NIC and Smad1 made a hardly detectable complex in the absence of p300 and P/CAF. Smad1 protein was observed in the NIC immunoprecipitates in the presence of either P/CAF or p300, although it was faint (Fig. 4A). In contrast, Smad1 was efficiently co-immunoprecipitated with NIC in the presence of both p300 and P/CAF. This complex formation was further enhanced by BMP stimulation. In this experiment, a constitutively active form of ALK3 (a type 2 BMPR) was used to mimic BMP stimulation. Taken together with the results shown in Figure 2, we next examined whether the presence of Smad1 enhanced the complex formation of NIC with p300 or P/CAF. As shown in Figure 4B, P/CAF, but not p300, was capable of binding to NIC. Simultaneous expression of p300 and P/CAF made NIC bind to P/CAF slightly more efficiently. Consistent with the results described above, when Smad1 and CaALK3 were expressed, a significantly larger amount of P/CAF was found in the NIC immunoprecipitates (Fig. 4B, right end lane). Surprisingly, p300 protein was also intensely observed in the NIC immunoprecipitates under this condition. Since Smad1 can only interact efficiently with NIC in the presence of both p300 and P/CAF (Fig. 4A), it is considered that a tetrameric complex of NIC, Smad1, p300 and P/CAF is formed upon Notch activation and BMP stimulation for the cooperative function of Notch and BMP signaling.
Figure 4.
Molecular interactions between NIC and Smad1 in the presence of p300 and P/CAF. (A and B) Cos7 cells were transfected with expression vectors for the indicated genes. Western blotting assay was performed as described in Figure 3.
p300 augments the cooperative enhancement of transcription induced by NIC and BMP2
The results of the efficient recruitment of p300 to a NIC complex in the presence of Smad1 encouraged us to study the effects of increased expression of p300 on the transactivation of the Hes-5 promoter by Smad1 and NIC. As shown in Figure 5, introduction of a p300 expression vector into P19 cells led to dose-dependent augmentation of the promoter activity when the cells were stimulated by both NIC and BMP2. In concordance with Figure 2C, luciferase activity was very weak in the absence of NIC and the activity was not enhanced by forced expression of p300 (data not shown). This result suggests the involvement of p300 in the coordinated transactivation by NIC and BMP2.
Figure 5.
Effect of additional expression of p300 on the cooperative transactivation by Notch and BMP2. P19 cells were transfected with H5-Luc and NIC with Mock or p300 expression vector. On the following day, the cells were stimulated with BMP2 (40 ng/ml) for 8 h and the luciferase activities were determined. Data are described as the percentage increase in luciferase activity compared with that obtained by NIC stimulation alone. The arbitrary units of luciferase activity for non- or NIC stimulation were 0.24 and 8.87, respectively.
DISCUSSION
A body of papers have demonstrated the importance of Notch signaling in the development of multicellular organisms. In mammals, several genes have been shown to be targets of Notch signaling (37). Among these genes, Hes-1 and Hes-5 have been well characterized (9,26). Recently, Hesr-1 has also been isolated as a target of Notch signaling (27). The current study shows that among these genes, the expression of Hes-5 and Hesr-1, but not Hes-1, could also be induced by BMP2 stimulation, a soluble signaling molecule distinct from Notch. Furthermore, we have demonstrated that BMP2 stimulation cooperates with Notch activation to enhance the expression of the Hes-5 and Hesr-1 genes, and proposed a molecular mechanism explaining the cooperation between these signalings, in which the transcriptional coactivators p300 and P/CAF are efficiently recruited to the transcriptional complex formed by NIC and RBP-Jκ.
Notch signaling has been shown to be activated in the ventricular zone of the mouse telencephalon, as its target genes are expressed in this area (38,39). Among the target genes of Notch, Hes-1 gene expression was less evident in the mouse telencephalon than Hes-5 and Hesr-1 (data not shown). We have previously shown that Hes-1 transcripts are not induced in telencephalic neuroepithelial cells by BMP2 (19). In this study, we found that the Hes-1 gene promoter did not respond to BMP2 stimulation. Receptors for BMP2 are intensely expressed in the ventricular zone (40) and BMP2 proteins per se exist there (41). It is thus thought that endogenous BMP signaling is also activated in the ventricular zone. The more intense expression of Hes-5 and Hesr-1 than Hes-1 is likely to partly reflect the cooperative effect of Notch and BMP2 signalings on the expression of Hes-5 and Hesr-1 but not Hes-1.
In the neural cells, BMP signaling appears to act as an amplifier of transcription. We demonstrated that BMP2 and leukemia inhibitory factor (LIF) could synergistically induce astrocyte differentiation from neuroepithelial cells prepared from the E14.5 mouse telencephalon (24). Smad1 and a transcription factor activated by LIF stimulation, i.e. STAT3, form a complex bridged by p300 and cooperate in synergy to induce the expression of the gene for glial fibrillary acidic protein (GFAP), an astrocyte-specific marker (24). Interestingly, mice deficient in gp130, a receptor component prerequisite for LIF signaling, display a decreased number of GFAP-expressing astrocytes (35). Furthermore, the gp130-deficient neuroepithelial cells did not respond to BMP2 stimulation to express GFAP (42). Another study demonstrated that BMP stimulation enhances the transcriptional activity of neurogenin1 (Ngn1), a neuronal differentiation-associated gene, resulting in the acceleration of neuronal differentiation but not astrocytes (43). In Ngn1-expressing cells, following BMP stimulation, a complex of Smad1 and p300 is preferentially recruited to Ngn1 but not to STAT3 (43). In addition, the fact that BMP stimulation does not induce neuronal differentiation in cells which do not express Ngn1, suggests that Ngn1 is a prerequisite for Smad-augmented neuronal differentiation (43). In the current study, we further demonstrated that BMP2 requires both an RBP-Jκ binding motif and active NIC to induce Hes-5 transcription. The situation seems to be comparable with the above-mentioned report that BMP requires transcriptional activity of its partners to enhance transcription of target genes.
In the two studies mentioned above, SBSs were not pin-pointed in the promoter regions of the target genes (24,43). The present data show that introduction of point mutations into a consensus SBS in the Hes-5 promoter led to a decrease in the promoter activity, and the Hes-1 gene promoter or the promoter construct with the RBP-Jκ binding motif but no Smad binding consensus sequences did not respond to BMP2 stimulation. Taken together, these results suggest that, in the case of cooperation with Notch signaling, BMP required specific binding sites for Smads in the promoter region of target genes.
NIC has been shown to form a complex with p300 through a three amino acid motif, resulting in the transactivation of its target genes (44). Another study demonstrated that P/CAF interacts predominantly with the NIC ankyrin repeat region to activate transcription (45). Recently, a study using an in vitro transcription system revealed that p300 and P/CAF can physically interact with NIC at the same time in the presence of Mastermind-like 1 (MAML1), and act to mediate transcriptional activation in a cooperative manner (34). In this complex, it was suggested that p300 plays a much more critical role as a HAT than P/CAF (34). In the current study, we did not study whether or not p300, P/CAF, or a combination of p300 and P/CAF was required for NIC-mediated transcription, but our results obviously suggest that Smad1 activated by BMP2 stimulation is likely to play a role in the efficient recruitment of p300 to a NIC-containing transcription complex, and that P/CAF is required for this complex formation. In this study, we showed that forced expression of p300 in cells leads to increased transactivation induced by a combination of NIC and BMP2, which suggests that the amount of endogenously present p300 in the nucleus may not be sufficient. Functional interaction between p300 and P/CAF has been demonstrated for FKLF2-induced transactivation (46). Hepatocyte nuclear factor-1 (HNF-1) requires synergism between CREB-binding protein (CBP; a molecule highly homologous to p300) and P/CAF to exert its transcriptional activity (47). Interaction of CBP with the N-terminal domain of HNF-1 increased the binding affinity of P/CAF with the C-terminal activation domain of HNF-1 (47). These previous observations indicate that a complex containing multiple coactivators is required for a variety of transcription factors for transactivation. Our current study suggests that this is the case with cooperative transactivation by two distinct transcription factors.
In conclusion, the data presented support the concept of functional cooperation between Notch signaling and BMP signaling. NIC and Smad1 form a complex with p300 and P/CAF in the specific promoter region, which possesses both an RBP-Jκ binding site and a SBS. In the complex, Smad1 enhances the recruitment of p300, which causes cooperative transactivation by Notch and BMP2 stimulation.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Drs K. Miyazono, T. Honjo, R. Kageyama, P. ten Dijke, S. Itoh and U. Lendahl for kindly providing plasmid constructs. We also thank Yamanouchi Pharmaceutical Co. Ltd for providing the human recombinant BMP2. We are very grateful to Ms Y. Noguchi for her excellent secretarial assistance. We also thank Ms K. Kaneko for technical assistance. This work has been supported by a Grant-in-Aid for Scientific Research on Priority Areas and a Grant-in-Aid for 21st Century COE Research from the Ministry of Education, Culture, Sports, Science and Technology, Human Frontier Science Program, and the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim.
REFERENCES
- 1.Artavanis-Tsakonas S., Matsuno,K. and Fortini,M.E. (1995) Notch signaling. Science, 268, 225–232. [DOI] [PubMed] [Google Scholar]
- 2.Bray S. (1998) A Notch affair. Cell, 93, 499–503. [DOI] [PubMed] [Google Scholar]
- 3.Nye J.S. and Kopan,R. (1995) Developmental signaling. Vertebrate ligands for Notch. Curr. Biol., 5, 966–969. [DOI] [PubMed] [Google Scholar]
- 4.Simpson P. (1995) Developmental genetics. The Notch connection. Nature, 375, 736–737. [DOI] [PubMed] [Google Scholar]
- 5.Lundkvist J. and Lendahl,U. (2001) Notch and the birth of glial cells. Trends Neurosci., 24, 492–494. [DOI] [PubMed] [Google Scholar]
- 6.Schroeter E.H., Kisslinger,J.A. and Kopan,R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–386. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh J.J., Henkel,T., Salmon,P., Robey,E., Peterson,M.G. and Hayward,S.D. (1996) Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol. Cell. Biol., 16, 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarriault S., Le Bail,O., Hirsinger,E., Pourquie,O., Logeat,F., Strong,C.F., Brou,C., Seidah,N.G. and Isra l,A. (1998) Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol., 18, 7423–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasai Y., Kageyama,R., Tagawa,Y., Shigemoto,R. and Nakanishi,S. (1992) Two mammalian helix–loop–helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev., 6, 2620–2634. [DOI] [PubMed] [Google Scholar]
- 10.Akazawa C., Sasai,Y., Nakanishi,S. and Kageyama,R. (1992) Molecular characterization of a rat negative regulator with a basic helix–loop–helix structure predominantly expressed in the developing nervous system. J. Biol. Chem., 267, 21879–21885. [PubMed] [Google Scholar]
- 11.Ohtsuka T., Sakamoto,M., Guillemot,F. and Kageyama,R. (2001) Roles of the basic helix–loop–helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem., 276, 30467–30474. [DOI] [PubMed] [Google Scholar]
- 12.Kageyama R. and Nakanishi,S. (1997) Helix–loop–helix factors in growth and differentiation of the vertebrate nervous system. Curr. Opin. Genet. Dev., 7, 659–665. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.E. (1997) Basic helix–loop–helix genes in neural development. Curr. Opin. Neurobiol., 7, 13–20. [DOI] [PubMed] [Google Scholar]
- 14.Hogan B.L. (1996) Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev., 10, 1580–1594. [DOI] [PubMed] [Google Scholar]
- 15.Mehler M.F., Mabie,P.C., Zhang,D. and Kessler,J.A. (1997) Bone morphogenetic proteins in the nervous system. Trends Neurosci., 20, 309–317. [DOI] [PubMed] [Google Scholar]
- 16.Massague J. and Chen,Y.G. (2000) Controlling TGF-beta signaling. Genes Dev., 14, 627–644. [PubMed] [Google Scholar]
- 17.Heldin C.H., Miyazono,K. and ten Dijke,P. (1997) TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- 18.Derynck R., Zhang,Y. and Feng,X.H. (1998) Smads: transcriptional activators of TGF-beta responses. Cell, 95, 737–740. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima K., Takizawa,T., Ochiai,W., Yanagisawa,M., Hisatsune,T., Nakafuku,M., Miyazono,K., Kishimoto,T., Kageyama,R. and Taga,T. (2001) BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc. Natl Acad. Sci. USA, 98, 5868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagisawa M., Takizawa,T., Ochiai,W., Uemura,A., Nakashima,K. and Taga,T. (2001) Fate alteration of neuroepithelial cells from neurogenesis to astrocytogenesis by bone morphogenetic proteins. Neurosci. Res., 41, 391–396. [DOI] [PubMed] [Google Scholar]
- 21.Nakashima K. and Taga,T. (2002) Mechanisms underlying cytokine-mediated cell-fate regulation in the nervous system. Mol. Neurobiol., 25, 233–244. [DOI] [PubMed] [Google Scholar]
- 22.Lim D.A., Tramontin,A.D., Trevejo,J.M., Herrera,D.G., Garcia-Verdugo,J.M. and Alvarez-Buylla,A. (2000) Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron, 28, 713–726. [DOI] [PubMed] [Google Scholar]
- 23.Irvin D.K., Zurcher,S.D., Nguyen,T., Weinmaster,G. and Kornblum,H.I. (2001) Expression patterns of Notch1, Notch2 and Notch3 suggest multiple functional roles for the Notch-DSL signaling system during brain development. J. Comp. Neurol., 436, 167–181. [PubMed] [Google Scholar]
- 24.Nakashima K., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Kawabata,M., Miyazono,K. and Taga,T. (1999) Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- 25.Takizawa T., Nakashima,K., Namihira,M., Ochiai,W., Uemura,A., Yanagisawa,M., Fujita,N., Nakao,M. and Taga,T. (2001) DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell, 1, 749–758. [DOI] [PubMed] [Google Scholar]
- 26.Takebayashi K., Akazawa,C., Nakanishi,S. and Kageyama,R. (1995) Structure and promoter analysis of the gene encoding the mouse helix–loop–helix factor HES-5. Identification of the neural precursor cell-specific promoter element. J. Biol. Chem., 270, 1342–1349. [DOI] [PubMed] [Google Scholar]
- 27.Kokubo H., Lun,Y. and Johnson,R.L. (1999) Identification and expression of a novel family of bHLH cDNAs related to Drosophila hairy and enhancer of split. Biochem. Biophys. Res. Commun., 260, 459–465. [DOI] [PubMed] [Google Scholar]
- 28.Iso T., Sartorelli,V., Poizat,C., Iezzi,S., Wu,H.Y., Chung,G., Kedes,L. and Hamamori,Y. (2001) HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol., 21, 6080–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K., Taniguchi,Y., Minoguchi,S., Sakai,T., Tun,T., Furukawa,T. and Honjo,T. (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol., 5, 1416–1423. [DOI] [PubMed] [Google Scholar]
- 30.de la Pompa J.L., Wakeham,A., Correia,K.M., Samper,E., Brown,S., Aguilera,R.J., Nakano,T., Honjo,T., Mak,T.W., Rossant,J. et al. (1997) Conservation of the Notch signalling pathway in mammalian neurogenesis. Development, 124, 1139–1148. [DOI] [PubMed] [Google Scholar]
- 31.Lindsell C.E., Boulter,J., diSibio,G., Gossler,A. and Weinmaster,G. (1996) Expression patterns of Jagged, Delta1, Notch1, Notch2 and Notch3 genes identify ligand–receptor pairs that may function in neural development. Mol. Cell. Neurosci., 8, 14–27. [DOI] [PubMed] [Google Scholar]
- 32.Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- 33.Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- 34.Wallberg A.E., Pedersen,K., Lendahl,U. and Roeder,R.G. (2002) p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol., 22, 7812–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakashima K., Wiese,S., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Yoshida,K., Kishimoto,T., Sendtner,M. and Taga,T. (1999) Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci., 19, 5429–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itoh S., Ericsson,J., Nishikawa,J., Heldin,C.H. and ten Dijke,P. (2000) The transcriptional co-activator P/CAF potentiates TGF-beta/Smad signaling. Nucleic Acids Res., 28, 4291–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iso T., Kedes,L. and Hamamori,Y. (2003) HES and HERP families: multiple effectors of the notch signaling pathway. J. Cell. Physiol., 194, 237–255. [DOI] [PubMed] [Google Scholar]
- 38.Ohtsuka T., Ishibashi,M., Gradwohl,G., Nakanishi,S., Guillemot,F. and Kageyama,R. (1999) Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J., 18, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura Y., Sakakibara,S., Miyata,T., Ogawa,M., Shimazaki,T., Weiss,S., Kageyama,R. and Okano,H. (2000) The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci., 20, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panchision D.M., Pickel,J.M., Studer,L., Lee,S.H., Turner,P.A., Hazel,T.G. and McKay,R.D. (2001) Sequential actions of BMP receptors control neural precursor cell production and fate. Genes Dev., 15, 2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mabie P.C., Mehler,M.F. and Kessler,J.A. (1999) Multiple roles of bone morphogenetic protein signaling in the regulation of cortical cell number and phenotype. J. Neurosci., 19, 7077–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakashima K., Yanagisawa,M., Arakawa,H. and Taga,T. (1999) Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS Lett., 457, 43–46. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y., Nadal-Vicens,M., Misono,S., Lin,M.Z., Zubiaga,A., Hua,X., Fan,G. and Greenberg,M.E. (2001) Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell, 104, 365–376. [DOI] [PubMed] [Google Scholar]
- 44.Oswald F., Tauber,B., Dobner,T., Bourteele,S., Kostezka,U., Adler,G., Liptay,S. and Schmid,R.M. (2001) p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol., 21, 7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurooka H. and Honjo,T. (2000) Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem., 275, 17211–17220. [DOI] [PubMed] [Google Scholar]
- 46.Song C.Z., Keller,K., Murata,K., Asano,H. and Stamatoyannopoulos,G. (2002) Functional interaction between coactivators CBP/p300, PCAF and transcription factor FKLF2. J. Biol. Chem., 277, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soutoglou E., Viollet,B., Vaxillaire,M., Yaniv,M., Pontoglio,M. and Talianidis,I. (2001) Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J., 20, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]