Abstract
The DNA-dependent protein kinase (DNA-PK) is one of the central enzymes involved in DNA double-strand break (DSB) repair. It facilitates proper alignment of the two ends of the broken DNA molecule and coordinates access of other factors to the repair complex. We discuss the latest findings on DNA-PK phosphorylation and offer a working model for the regulation of DNA-PK during DSB repair.
DNA double-strand break repair and nonhomologous end joining
DNA double-strand breaks (DSBs) can be accidentally introduced in cells by the action of ionizing radiation or certain reactive radicals. These agents have the ability to initiate a series of chemical reactions that ultimately sever the DNA backbone, resulting in chromosome breakage and fragmentation of genes (Hoeijmakers, 2001). Because such corruption of genetic material inevitably leads to problems with replication and cell division, it is of the utmost importance that cells have a mechanism to counteract DSBs. In addition, DSBs are generated in developing B and T cells during normal V(D)J recombination, implying that a working DSB repair system is not only necessary for an effective defense against DNA-modifying agents but also for a functional immune system in higher organisms (for review see Weterings and van Gent, 2004). As a result, two highly efficient DSB repair pathways have evolved in eukaryotic cells: homologous recombination (HR) and nonhomologous end joining (NHEJ).
The HR process mediates DSB repair by using a homologous DNA sequence as a template to guide proper restoration of the break. Because homologous templates are found on sister chromosomes, HR is thought to be active during the S and G2 cell cycle phases. Contrasting, NHEJ is characterized by its ability to directly ligate the two ends of the broken DNA molecule. This process does not have the need for a homologous template and is therefore theoretically not restricted to a certain phase of the cell cycle.
The general principle of NHEJ
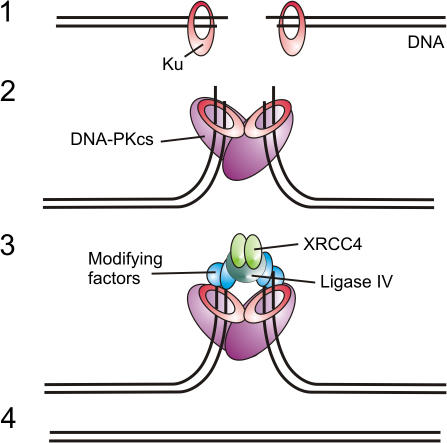
The NHEJ pathway utilizes several enzymes that capture both DNA ends, bring them together in a synaptic complex, and facilitate direct ligation of the DNA break (Fig. 1). The process is initiated by the association of DNA ends with the Ku 70/80 heterodimer, a protein with a ring-shaped structure that displays an extraordinary affinity for open DNA ends (Walker et al., 2001). The DNA–Ku 70/80 complex then functions as a scaffold to assemble the other key NHEJ proteins at the DNA termini.
Figure 1.
Schematic overview of the NHEJ process. (1) Recognition of a DSB by Ku 70/80. (2) Tethering of DNA ends by DNA-PKCS. (3) Processing of DNA ends. (4) Ligation of DNA ends.
One of the first enzymes to be attracted to the DNA–Ku 70/80 scaffold is a large, 460-kD serine/threonine kinase called DNA-dependent protein kinase catalytic subunit (DNA-PKCS). The protein complex that is formed after the association of both Ku 70/80 and DNA-PKCS at the DNA ends is generally referred to as DNA-dependent protein kinase (DNA-PK). The large DNA-PKCS molecule forms a distinct structure at the DNA termini, which is likely to play an active role in the formation of a synaptic complex that holds the two ends of the broken DNA molecule together (Fig. 1; for review see Weterings and van Gent, 2004).
The positioning of the two DNA ends in a synaptic complex sets the stage for religation of the broken DNA molecule. Before this can take place effectively, it is necessary that noncomplementary ends are processed. In the case of single-stranded overhangs, DNA termini can be made ligatable by either filling of the missing nucleotides or by resection of the overhang. Polymerases μ and λ, terminal deoxynucleotidyltransferase, polynucleotide kinase, and several nucleases have been shown to play a role in this processing. The best-characterized processing nuclease is the endonuclease Artemis, whose activities include the removal of single-strand overhangs (Ma et al., 2002). After correct processing, the two tethered DNA termini can finally be ligated. Ligation is mediated by the ligase IV–XRCC4 complex, possibly in conjunction with the recently discovered XLF (XRCC4-like factor)/Cernunnos protein (Ahnesorg et al., 2006), although at this point, little is known about the mechanism by which XLF stimulates ligation.
The DNA-dependent protein kinase catalytic subunit in NHEJ
After the introduction of a DSB, the DNA-PKCS enzyme is quickly recruited to the DNA–Ku scaffold. The serine/threonine kinase activity of DNA-PKCS is not activated until the DNA-PKCS molecule is associated with both the Ku 70/80 heterodimer and a DNA terminus. This latter requirement makes the DNA-PKCS protein kinase truly DNA dependent. Many targets for the DNA-PKCS kinase have been identified in vitro, including XRCC4, Ku 70/80, Artemis, p53, and even DNA-PKCS itself (autophosphorylation). This plethora of possible targets suggests that DNA-PKCS may be involved in several aspects of the NHEJ process, including activation of components of the synaptic complex and signal transduction to cell cycle regulators (Burma and Chen, 2004). Surprisingly, however, little evidence has been found for the biological relevance of these activities. To the contrary, much more convincing data are available that argue for a regulatory role for DNA-PKCS at the synapsis of the two DNA ends during NHEJ.
Information on the shape and conformation of the DNA–Ku–DNA-PKCS complex is scarce and provides little detail as a result of technical difficulties in the production of DNA-PKCS crystals for crystalographic structure studies. The best available information on the three-dimensional structure of the DNA–Ku–DNA-PKCS heterotrimer was obtained by single- particle electron microscopy at a 25-Å resolution (Spagnolo et al., 2006). These images clearly showed dimeric structures in which DNA ends were brought into close proximity by the formation of a synaptic complex that consisted of two DNA ends, two Ku 70/80 molecules, and two DNA-PKCS molecules (Fig. 1). In addition, both atomic force microscopy studies and biochemical experiments have shown that DNA-PKCS not only assembles at DNA ends but also facilitates the tethering of multiple DNA ends (for review see Weterings and van Gent, 2004). Collectively, these findings suggest that DNA-PKCS is responsible for the formation of a synaptic bridge between the two termini of the broken DNA molecule.
It has been well established that the presence of the large DNA-PKCS molecule at DNA ends effectively blocks access of either processing nucleases or ligases (Weterings et al., 2003; Block et al., 2004). Although this capping of DNA ends may have an important function in protection of the DNA termini against degradation or premature and incorrect ligation, it is clear that the DNA-PKCS cap has to be removed or altered before religation of the DNA ends and repair of the DSB can take place. Several authors have demonstrated that autophosphorylation of DNA-PKCS results in release of the cap and accessibility of the DNA termini for either processing enzymes or ligases (Ding et al., 2003; Weterings et al., 2003; Block et al., 2004). Recent research has demonstrated that DNA-PKCS autophosphorylation can occur in trans during the synapsis of two DNA-bound DNA-PKCS molecules (Meek et al., 2007). These findings clearly argue for a model in which DNA-PKCS protects the termini of a broken DNA molecule by capping it until the DNA ends find each other and are properly aligned in a synaptic complex. DNA-PKCS trans-autophosphorylation then liberates the DNA ends for proper processing and ligation by other NHEJ factors like Artemis (Goodarzi et al., 2006) and ligase IV–XRCC4 (Block et al., 2004). In this model, DNA-PKCS functions as a gatekeeper of the NHEJ process, regulating access to the DNA ends by autophosphorylation (Fig. 2).
Figure 2.
Regulation of DNA end accessibility by DNA-PKCS phosphorylation. DNA-PKCS protects the DNA ends until trans-autophosphorylation induces a conformational change that enables DNA end processing and ligation. In addition to autophosphorylation, ATM-mediated phosphorylation of DNA-PKCS may play a role in this process.
At present, it is not clear how we should envision the conformational changes in the DNA–DNA-PKCS complex that take place during autophosphorylation. It is also not unequivocally known which autophosphorylation sites need to be phosphorylated to facilitate the accessibility of DNA termini. Thus far, 16 amino acid residues that can be phosphorylated by the action of the DNA-PKCS kinase have been identified within the DNA-PKCS molecule (Fig. 3). The importance of DNA-PKCS autophosphorylation during NHEJ is demonstrated by numerous observations, showing that mutation of phosphorylation sites causes increased radiosensitivity and less efficient DSB repair (Chan et al., 2002; Ding et al., 2003; Chen et al., 2005; Douglas et al., 2007) and that an active DNA-PKCS kinase is required for NHEJ (Kurimasa et al., 1999).
Figure 3.
Phosphorylation sites within the DNA-PKCS molecule. The 2609 cluster (Chan et al., 2002; Douglas et al., 2002), 2056 cluster (Chen et al., 2005; Cui et al., 2005), 3205 residue (Douglas et al., 2002), 3950 residue (Douglas et al., 2007), and 3821, 4026, and 4102 residues (Ma et al., 2005) are included. S, serine; T, threonine.
Inhibiting phosphorylation of the entire 2609 cluster (Fig. 3) leads to a severe increase in radiosensitivity and diminished processing of DNA ends, whereas mutation of single residues within this cluster has a less severe (but still present) effect on radiosensitivity (Chan et al., 2002; Ding et al., 2003). In addition, we found that mutation of either the DNA-PKCS kinase domain or the 2609 cluster and the 2056 residue results in a rigid binding of DNA-PKCS to DNA ends in vivo, which most likely interferes with the NHEJ process (Uematsu et al., 2007). Collectively, these results demonstrate that autophosphorylation of the 2609 cluster influences the DNA–DNA-PKCS interaction in a manner that facilitates end joining.
In contrast, the 2056 cluster (Fig. 3) has been reported to inhibit DNA end processing upon phosphorylation, suggesting that the 2609 and 2056 clusters work in an opposite direction (Cui et al., 2005). The latter finding has at present not been confirmed, and it is not entirely transparent how this double regulation mechanism cooperates during DSB repair.
Phosphorylation of the recently discovered 3950 residue, which is located in the C-terminal kinase domain, most likely plays a role in regulation of the DNA-PKCS kinase activity. Mutation of this site with phosphomimic aspartic acid results in deficient V(D)J recombination and increased radiation sensitivity (Douglas et al., 2007). This finding suggests that the 3950 residue may be involved in the regulation of DNA-PKCS autophosphorylation by mediating kinase activity. Three other autophosphorylation sites have been identified in the C-terminal region of DNA-PKCS (3821, 4026, and 4102), although no in vivo functionality has been reported for these sites at present (Ma et al., 2005).
Other factors involved in DNA-PKCS phosphorylation
Recent experiments have shown that in vivo phosphorylation of the DNA-PKCS 2609 and 2647 residues can still occur at DSB sites in the absence of DNA-PKCS kinase activity (Chen et al., 2007; Uematsu et al., 2007). Clearly, in these cases, the DNA-PKCS molecule is not responsible for the phosphorylation events, and another kinase (or kinases) must be involved. These studies raise doubts as to whether all phosphorylation sites within the DNA-PKCS molecule are genuine autophosphorylation sites or whether they can be targets for other kinases as well.
One study reports that phosphorylation of the 2609 and 2647 residues (but not the 2056 residue) of DNA-PKCS is dependent on the ataxia telangiectasia mutated (ATM) protein (Chen et al., 2007). ATM is a damage-responsive protein kinase and a member of the phosphatidylinositol 3-kinase–like kinase group (PIKK), to which DNA-PKCS also belongs. Another study finds no absolute requirement for the presence of ATM but does acknowledge that several DNA-PKCS residues can be phosphorylated in the absence of a functional DNA-PKCS kinase, leaving open the possibility that ATM contributes to the phosphorylation of DNA-PKCS residues in vivo (Meek et al., 2007). A third study suggests that yet another member of the PIKK group, ataxia telangiectasia related protein (ATR), is involved in the regulation of DNA-PKCS phosphorylation at the 2609 and 2647 residues after the onset of UV damage (Yajima et al., 2006). At present, it is not clear whether the ATR-mediated phosphorylation of DNA-PKCS is relevant for DSB repair via the NHEJ pathway.
At this point, it is not known whether ATM (or ATR)- mediated phosphorylation of the DNA-PKCS 2609 and 2647 residues is redundant with autophosphorylation, but it is tempting to speculate that both routes of DNA-PKCS phosphorylation have their own biological significance. ATM is activated at an early stage after the introduction of a DSB, is present at DSB sites, and is involved in signal transduction and phosphorylation of a plethora of proteins that mediate cell cycle arrest during DSB repair (Lobrich and Jeggo, 2005). It is not unlikely that the ATM kinase, with its central role in the regulation of DSB repair and its physical presence at DSB sites, would be involved in regulation of the DNA-PKCS phosphorylation status and, thus, in regulation of the accessibility of DNA ends during the repair process.
Conclusions and speculations
It is becoming increasingly clear that DNA-PKCS plays a central role during the NHEJ process. This enzyme not only captures and tethers the two ends of a broken DNA molecule but also regulates the access of modifying enzymes and ligases to the DNA termini. A body of evidence supports a model in which regulation of the accessibility of DNA ends is mediated by autophosphorylation of DNA-PKCS in trans over the synaptic cleft between the two tethered DNA molecules (Fig. 2).
The latest findings that suggest the involvement of ATM in the phosphorylation of at least two DNA-PKCS residues open a new perspective on the regulation of DNA-PKCS in the synaptic complex. The codependence of DNA-PKCS phosphorylation on ATM would ensure that at least two signals need to be present before DNA-PKCS alters its conformation at the DNA ends: (1) phosphorylation of the ATM-responsive sites within the DNA-PKCS molecule and (2) autophosphorylation in trans over the synaptic cleft. Such a mechanism would tightly control DNA-PKCS activity, not allowing access to the DNA termini until two criteria have been met: a defined DNA damage response (as indicated by activation of the ATM kinase) and a proper alignment of the two DNA ends. Until these criteria are met, the DNA termini will be protected by the DNA-PKCS molecule: a lock with multiple keys.
Acknowledgments
We are grateful to Drs. Chaitanya S. Nirodi, Michael D. Story, Anthony J. Davis, and Benjamin P.C. Chen for critical revision of the manuscript.
This work was supported by National Institutes of Health grants 5-R37-CA050519-16 and PO1-CA92584.
Abbreviations used in this paper: ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia related protein; DSB, double-strand break; HR, homologous recombination; NHEJ, nonhomologous end joining.
References
- Ahnesorg, P., P. Smith, and S.P. Jackson. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 124:301–313. [DOI] [PubMed] [Google Scholar]
- Block, W.D., Y. Yu, D. Merkle, J.L. Gifford, Q. Ding, K. Meek, and S.P. Lees-Miller. 2004. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 32:4351–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma, S., and D.J. Chen. 2004. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst.). 3:909–918. [DOI] [PubMed] [Google Scholar]
- Chan, D.W., B.P. Chen, S. Prithivirajsingh, A. Kurimasa, M.D. Story, J. Qin, and D.J. Chen. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.P., D.W. Chan, J. Kobayashi, S. Burma, A. Asaithamby, K. Morotomi-Yano, E. Botvinick, J. Qin, and D.J. Chen. 2005. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 280:14709–14715. [DOI] [PubMed] [Google Scholar]
- Chen, B.P., N. Uematsu, J. Kobayashi, Y. Lerenthal, A. Krempler, H. Yajima, M. Lobrich, Y. Shiloh, and D.J. Chen. 2007. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 282:6582–6587. [DOI] [PubMed] [Google Scholar]
- Cui, X., Y. Yu, S. Gupta, Y.M. Cho, S.P. Lees-Miller, and K. Meek. 2005. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 25:10842–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Q., Y.V. Reddy, W. Wang, T. Woods, P. Douglas, D.A. Ramsden, S.P. Lees-Miller, and K. Meek. 2003. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23:5836–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, P., G.P. Sapkota, N. Morrice, Y. Yu, A.A. Goodarzi, D. Merkle, K. Meek, D.R. Alessi, and S.P. Lees-Miller. 2002. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA- dependent protein kinase. Biochem. J. 368:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, P., X. Cui, W.D. Block, Y. Yu, S. Gupta, Q. Ding, R. Ye, N. Morrice, S.P. Lees-Miller, and K. Meek. 2007. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 27:1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi, A.A., Y. Yu, E. Riballo, P. Douglas, S.A. Walker, R. Ye, C. Harer, C. Marchetti, N. Morrice, P.A. Jeggo, and S.P. Lees-Miller. 2006. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 25:3880–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers, J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374. [DOI] [PubMed] [Google Scholar]
- Kurimasa, A., S. Kumano, N.V. Boubnov, M.D. Story, C.S. Tung, S.R. Peterson, and D.J. Chen. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich, M., and P.A. Jeggo. 2005. The two edges of the ATM sword: co-operation between repair and checkpoint functions. Radiother. Oncol. 76:112–118. [DOI] [PubMed] [Google Scholar]
- Ma, Y., U. Pannicke, K. Schwarz, and M.R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 108:781–794. [DOI] [PubMed] [Google Scholar]
- Ma, Y., U. Pannicke, H. Lu, D. Niewolik, K. Schwarz, and M.R. Lieber. 2005. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 280:33839–33846. [DOI] [PubMed] [Google Scholar]
- Meek, K., P. Douglas, X. Cui, Q. Ding, and S.P. Lees-Miller. 2007. Trans autophosphorylation at DNA-PK's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell Biol. 27:3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo, L., A. Rivera-Calzada, L.H. Pearl, and O. Llorca. 2006. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 22:511–519. [DOI] [PubMed] [Google Scholar]
- Uematsu, N., E. Weterings, K. Yano, K. Morotomi-Yano, B. Jakob, G. Taucher-Scholz, P.O. Mari, D.C. van Gent, B.P. Chen, and D.J. Chen. 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double- strand breaks. J. Cell Biol. 177:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.R., R.A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 412:607–614. [DOI] [PubMed] [Google Scholar]
- Weterings, E., and D.C. van Gent. 2004. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst.). 3:1425–1435. [DOI] [PubMed] [Google Scholar]
- Weterings, E., N.S. Verkaik, H.T. Bruggenwirth, J.H. Hoeijmakers, and D.C. van Gent. 2003. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 31:7238–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima, H., K.J. Lee, and B.P. Chen. 2006. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 26:7520–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]