Abstract
Eukaryotic cells have a surveillance mechanism that identifies aberrantly processed pre-mRNAs and prevents their flow to the cytoplasm by tethering them near the site of transcription. Here we provide evidence that mRNA release from the transcription site requires the heptad repeat structure of the C-terminal domain (CTD) of RNA polymerase II. The mammalian CTD, which is essential for normal co-transcriptional maturation of mRNA precursors, comprises 52 heptad repeats. We show that a truncated CTD containing 31 repeats (heptads 1–23, 36–38, and 48–52) is sufficient to support transcription, splicing, cleavage, and polyadenylation. Yet, the resulting mRNAs are mostly retained in the vicinity of the gene after transcriptional shutoff. The retained mRNAs maintain the ability to recruit components of the exon junction complex and the nuclear exosome subunit Rrp6p, suggesting that binding of these proteins is not sufficient for RNA release. We propose that the missing heptads in the truncated CTD mutant are required for binding of proteins implicated in a final co-transcriptional maturation of spliced and 3′ end cleaved and polyadenylated mRNAs into export-competent ribonucleoprotein particles.
Introduction
In eukaryotic cells, messenger precursor molecules must undergo a series of maturation events that include 5′ capping, splicing, 3′ end cleavage, and polyadenylation. During processing, nascent mRNA assembles together with RNA binding proteins into ribonucleoprotein particles (mRNPs; Aguilera, 2005; Moore, 2005). Mature particles are exported to the cytoplasm and several lines of evidence indicate that mRNPs move from the sites of transcription to the nuclear pores by random Brownian motion. As diffusion cannot be regulated, traffic control of newly synthesized mRNA molecules is thought to rely on retention at dedicated sites within the nucleus (Gorski et al., 2006). According to the current view, any failure compromising the integrity of an mRNA may cause its retention in the nucleus and trigger its degradation. There is evidence suggesting that such a surveillance mechanism operates in close proximity to the gene template (Jensen et al., 2003) and, at least in yeast, at the nuclear pore (Galy et al., 2004).
A key connection between transcription and mRNP biogenesis is provided by the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNA Pol II LS), which binds several proteins essential for pre-mRNA processing (Bentley, 2005). The CTD of RNA Pol II LS is highly conserved, increasing in length and diversifying in structure with the complexity of organisms (Stiller and Hall, 2002). Contrasting with yeast, which contains 26 repeats of a conserved heptapeptide with the consensus sequence YSPTSPS, the mammalian CTD has 52 repeats, of which 21 obey the conserved consensus while the remainder display a variety of substitutions. Most of these nonconsensus repeats are located in the C-terminal part of the CTD (heptads 27–52; Fig. 1 A), and the last repeat (CTD52) is essential for cell viability and Pol II stability (Chapman et al., 2004). At the very C terminus, the mammalian CTD further comprises a specific 10-amino acid motif. CTD deletion analysis has shown that heptad repeats 1–15 or 1–25 support capping but not splicing or 3′ end formation, whereas heptads 27–52 plus the C-terminal 10 residues can support efficient capping, splicing, and 3′ end formation (Fong and Bentley, 2001). More recent studies have demonstrated that scrambling the 10 residues that lie C-terminal of heptad 52 impairs efficient release of RNA from the site of transcription (Bird et al., 2005). However, this mutation also reduces splicing and 3′ end cleavage (Fong et al., 2003), arguing that the CTD requirement for RNA release may be a consequence of its role in promoting pre-mRNA processing.
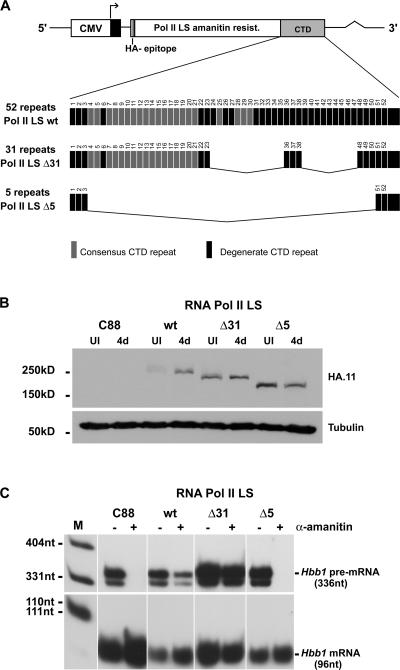
Figure 1.
A large deletion of the CTD abolishes LCR-dependent Hbb1 transcriptional activation. (A) Schematic representation of the RNA Pol II LS constructs. (B) Western blotting analysis. Total protein extracts were prepared from untransfected MEL C88 and from cells transfected with the indicated constructs, both before (UI) and after 4 d of erythroid differentiation (4d). The blot was incubated with antibodies anti-HA and anti-tubulin. (C) S1-nuclease protection assay. MEL C88 cells and cells transfected with the indicated constructs were induced for 4 d either with (+) or without (−) treatment with α-amanitin for 17 h. Hbb1 expression was analyzed with a probe that produces protected fragments of 336 and 96 nt for the pre-mRNA and mRNA, respectively. Due to its long half-life, high levels of mRNA synthesized before treatment with α-amanitin persist in the treated cells.
To further investigate the role of the CTD in transcript release, we generated murine erythroleukemia (MEL) cell lines that express α-amanitin–resistant RNA Pol II LS with either full-length or truncated forms of the CTD. Our results reveal that deleting 21 C-terminal heptads of the CTD causes transcript retention at the site of transcription but without inhibiting splicing or 3′ end formation. This implies a previously unsuspected involvement of the CTD in mRNP maturation events that occur after splicing, cleavage, and polyadenylation have taken place.
Results and discussion
Deletion of the CTD to 5 heptads abolishes LCR-dependent transcriptional activation of the Hbb1 gene
MEL cells were stably transfected with an α-amanitin–resistant form of the RNA polymerase II largest subunit (RNA Pol II LS; Bartolomei and Corden, 1987) containing either wild-type or deletion mutants of the CTD with 31 (Δ31) or 5 heptad repeats (Δ5) (Fig. 1 A; Bartolomei et al., 1988; Gerber et al., 1995). Each of these plasmids was cotransfected with a second plasmid containing the human β-globin gene (HBB) micro-locus control region (βLCR) and a puromycin resistance gene (Collis et al., 1990; Millevoi et al., 2002). Given the tendency of multiple copies of plasmid transgenes to co-integrate as tandem arrays, we reasoned that this cotransfection procedure should, in many cases, result in the positioning of the βLCR-bearing plasmid upstream of the α-amanitin–resistant RNA Pol II LS constructs. As the βLCR is able to activate heterologous, nonerythroid promoters (Blom van Assendelft et al., 1989; Collis et al., 1990) with the minimum requirements being a CAAT and CACCA or GC-rich (e.g., Sp1) elements (Antoniou and Grosveld, 1990; and unpublished data), this configuration should confer erythroid-specific induced transcription on the human cytomegalovirus (CMV) promoter linked to the α-amanitin–resistant RNA Pol II LS cassettes. Stably transfected clones were selected with puromycin and screened by an S1-nuclease protection assay for expression of the transfected RNA Pol II LS gene. We selected clones that showed low levels of exogenous, transgene-derived α-amanitin–resistant RNA Pol II LS expression in preinduced cells and high levels after 4 d of differentiation. Expression of the exogenous RNA Pol II LS was confirmed by Western blotting analysis with an antibody that recognizes the haemagglutinin (HA) epitope (Fig. 1 B), as previously described (Custódio et al., 2006).
We next confirmed that the exogenous RNA Pol II LS was functional. The endogenous RNA Pol II LS is degraded upon binding of α-amanitin (Nguyen et al., 1996), but the exogenous transgene-derived protein is resistant due to a single amino acid substitution that decreases its affinity for the toxin (Bartolomei and Corden, 1987). We therefore determined the ability of the different clones to transcribe endogenous murine βmajor-globin gene (Hbb1) in the presence of α-amanitin. After 17 h of α-amanitin treatment, S1-nuclease protection assays revealed no signal for Hbb1 pre-mRNA in untransfected MEL C88 cells, confirming that transcription by the endogenous RNA Pol II was abolished (Fig. 1 C, lane C88 +). In marked contrast, Hbb1 pre-mRNA was present in clones expressing the α-amanitin–resistant forms of RNA Pol II LS containing either the full-length CTD (Fig. 1 C, lane wt +) or the Δ31 truncation (Fig. 1 C, lane Δ31 +), but not the Δ5 variant (Fig. 1 C, lane Δ5 +). The finding that none of the clones expressing RNA Pol II Δ5 were able to support transcription of Hbb1 was surprising, taking into account that this mutant was previously shown to transcribe HBB under control of the SV40 promoter (McCracken et al., 1997b) and a rat homeobox reporter gene stably integrated into the genome of HeLa cells (Misteli and Spector, 1999). Additional studies indicated that the truncated variant of the CTD with only 5 heptad repeats did not affect TATA-box–mediated transcription (Gerber et al., 1995; McCracken et al., 1997b). However, this same form of the CTD in yeast and humans abolished activator- dependent induction of transcription of specific genes (Allison and Ingles, 1989; Gerber et al., 1995; Meininghaus and Eick, 1999). Furthermore, nuclear run-on experiments in mammalian cells suggested a global defect in transcription of endogenous genes (Meininghaus et al., 2000). Because endogenous murine Hbb1 is under control of the LCR (Collis et al., 1990), our observation that RNA Pol II Δ5 fails to support LCR-dependent transcription is consistent with previous data indicating a requirement of the CTD for enhancer-driven transcription (Allison and Ingles, 1989; Gerber et al., 1995; Meininghaus and Eick, 1999).
Deletion of the CTD to 31 heptads causes mRNA retention at the site of transcription
Having selected MEL cell clones that express functional α-amanitin–resistant wild-type or Δ31 RNA Pol II LS, we next super-transfected these cells with an HBB transgene that was either wild type (βWT) or a mutant variant possessing a GT to AC mutation at the 5′ splice site of the second intron (βSM) (Fig. 2 A). As this mutation inhibits splicing and causes retention of the RNA at the transcription site (Custódio et al., 1999), we were interested in determining whether the CTD is involved in recognition of the resulting aberrantly processed βSM pre-mRNA. Cells were induced for 3 d and analyzed by FISH. As previously described (Custódio et al., 1999), the transcription site of the βWT or βSM transgene is detected as a focus in the nucleus (Fig. 2 B). The wild-type mRNA is exported from the nucleus and accumulates in the cytoplasm (Fig. 2 B, a), in contrast to the mutant RNA, which is not detected in the cytoplasm (Fig. 2 B, e). Treatment of cells devoid of exogenous transgene-derived RNA Pol II with α-amanitin results in the disappearance of nuclear foci (Fig. 2 B, b and f). Contrastingly, nuclear foci remain clearly visible in cells transfected with α-amanitin–resistant forms of RNA Pol II LS (Fig. 2 B, c, d, g, and h).
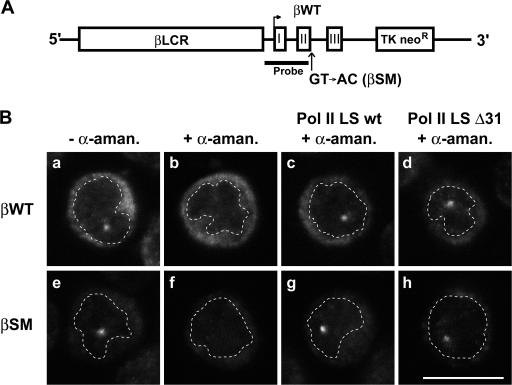
Figure 2.
Visualization of nascent RNA transcribed by transgene-derived RNA Pol II LS. (A) Schematic representation of wild-type (βWT) and splice mutant (βSM) human β-globin (HBB) constructs. Exons in the HBB gene are numerated (I, II, III). The probe used for FISH is complementary to exon I, intron I, and 205 nt of exon II. (B) Detection by FISH of βWT and splice mutant βSM RNA synthesized by either endogenous RNA Pol II or transgene-derived constructs, as indicated. All cells were induced for 3 d and either treated or untreated with α-amanitin as indicated. Dashed lines indicate the periphery of the nucleus. Bar, 10 μm.
We have previously shown that treating MEL cells with the transcription inhibitor actinomycin D for a short period of time (5–15 min) causes a dramatic, rapid reduction in the relative number of cells that contain a detectable nuclear focus of βWT transcripts within the nucleus, whereas the percentage of cells harboring βSM RNA foci remained largely unaltered, suggesting that these mutant RNAs were not being released from the site of transcription (Custódio et al., 1999). We therefore performed the same assay using cells that express the α-amanitin–resistant forms of RNA Pol II LS in an effort to gain insight into the possible role of the CTD in the process of mRNA release from the transcription site. The results show that actinomycin D treatment of cells expressing either endogenous RNA Pol II LS or the α-amanitin–resistant full-length CTD transgene product resulted in a significant decrease in the percentage of cells with βWT transcription foci (Fig. 3 A, a–d; and Fig. 3 C, a), as well as in the intensity of the remaining signal (Fig. 3 D). However, when transcription is dependent on the α-amanitin–resistant RNA Pol II LS containing the truncated Δ31 CTD, actinomycin D treatment does not cause any significant reduction in the percentage of cells with a visible focus (Fig. 3 A, e, f; and Fig. 3 C, a) or in the mean fluorescence intensity of each focus (Fig. 3 D). We therefore conclude that the transcripts are not being efficiently released. Parallel experiments performed with cells expressing the mutant HBB transgene showed that treatment with actinomycin D causes no significant change in the percentage of cells with a visible nuclear focus of βSM RNA (Fig. 3 B), regardless of whether this gene is transcribed by full-length (Fig. 3 B c, d; and Fig. 3 C, b) or truncated (Fig. 3 B, e, f; and Fig. 3 C, b) CTD versions of α-amanitin–resistant RNA Pol II LS. Thus, reducing the CTD to 31 heptad repeats is sufficient to prevent release of RNA transcribed from a normal gene while it does not interfere with the ability to retain transcripts derived from a gene with a severe splice mutation.
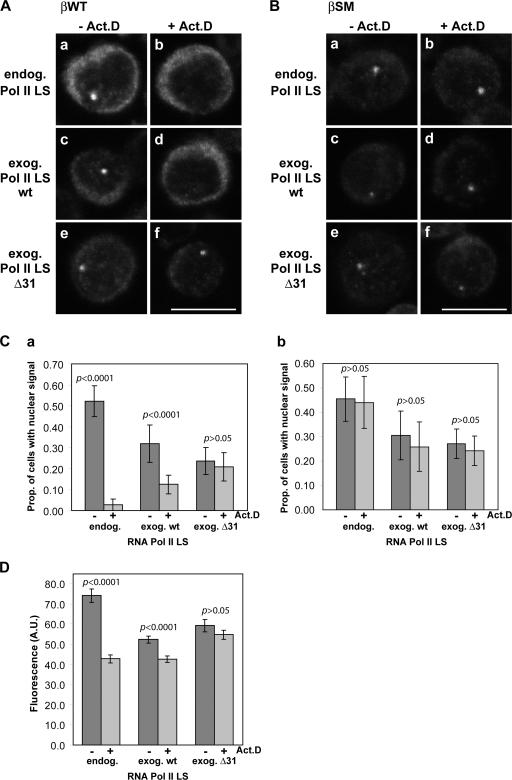
Figure 3.
The CTD Δ31 mutation causes mRNA retention at the site of transcription. RNA transcribed from βWT (A) and splice mutant βSM (B) transgenes was visualized by FISH. Transcription was by endogenous or exogenous RNA Pol II LS, as indicated. Treatment with actinomycin D was for 15 min. Bar, 10 μm. (C) The proportion of cells with a nuclear RNA focus, before and after actinomycin D. A total of 500–700 cells were counted in each experiment. Results are presented as means ± SD for at least three independent experiments; P values relative to nontreated cells (t test) are indicated. Due to a combination of the asynchronous nature of the cell cultures and position-effect variegation, not all cells contain a visible nuclear focus. As the fraction of cells with a nuclear focus varies between cell lines, the interpretation of the data relies on the comparison of the fraction of cells from the same line that contain a nuclear focus before and after treatment with actinomycin D. (D) Mean fluorescence intensity (arbitrary units, AU) of the nuclear βWT RNA focus, before and after actinomycin D. A total of 100 nuclear RNA foci were quantified in each experiment. Results are presented as mean fluorescence intensity ± SE for two independent experiments; P values relative to nontreated cells (t test) are indicated.
RNA transcribed by RNA Pol II LS Δ31 is spliced, cleaved, and polyadenylated
Previous studies have shown that deletion mutants of the CTD induce defects in splicing and 3′ end cleavage (McCracken et al., 1997b; Fong and Bentley, 2001; Fong et al., 2003). We therefore analyzed wild-type HBB (βWT) transcripts produced by endogenous or exogenous transgene RNA Pol II LS by RNase protection assays to monitor splicing of introns 1 and 2 as well as cleavage at the poly(A) addition site (Fig. 4 A). As expected, no signal for unspliced (US; Fig. 4 B, lane 2) and uncleaved (UC; Fig. 4 B, lane 4) HBB RNA was detected after inactivation of the endogenous RNA Pol II LS with α-amanitin (Fig. 4 B). However, bands corresponding to spliced (SP) and cleaved (CL) RNA were still present, most probably due to the long half-life of HBB mRNA.
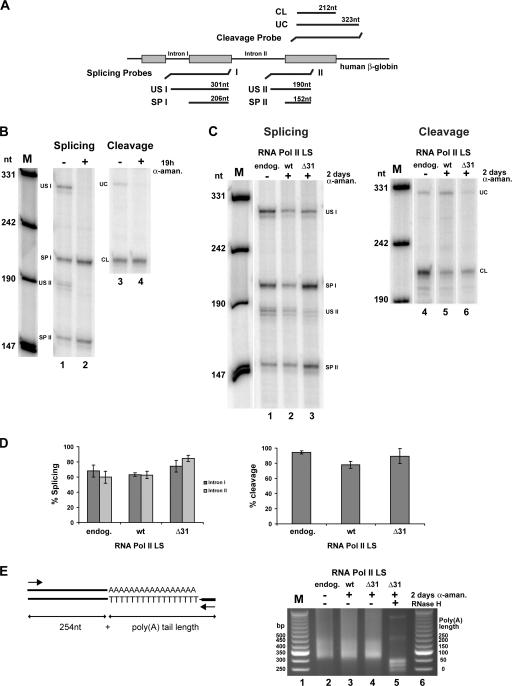
Figure 4.
The CTD Δ31 mutant supports splicing, cleavage, and polyadenylation. (A) Schematic representation of the RNase protection assay probes. (B) Nuclear RNA (2 μg) from MEL cells transfected with βWT (Fig. 2) induced to differentiate for 3 d and untreated (−) or treated (+) with α-amanitin for 19 h were analyzed by an RNase protection assay using the indicated probes; intron I unspliced (US I), intron II unspliced (US II), intron I spliced (SP I), intron II spliced (SP II), uncleaved (UC), and cleaved (CL). (C) Nuclear RNA fractions from cells transcribing the βWT transgene either by endogenous Pol II LS (endog.; 2.5 μg) or by exogenous RNA Pol II LS wild-type (wt; 5 μg) or RNA Pol II LS Δ31 (Δ31; 5 μg) that were induced to differentiate for 3 d and untreated (−) or treated (+) with α-amanitin for 2 d were analyzed using the same probes. (D) Quantification of splicing and cleavage efficiencies. The amount of unspliced (US), spliced (SP), uncleaved (UC), and cleaved (CL) RNA from three independent experiments was determined from PhosphorImager data corrected for [32P]U content of the RNase-protected fragment. Percentages of splicing and cleavage were calculated by dividing the value of spliced or cleaved product by the sum of the values of spliced and unspliced products or cleaved and uncleaved products, respectively. (E) Poly(A) tail length analysis of HBB transcripts. The diagram on the left illustrates the PCR-amplified products. Primers were designed to amplify a 254-nt product if the mRNA is not polyadenylated and any length over 254 nt is contributed by the poly(A) tail. On the right, an ethidium bromide–stained agarose gel with the products obtained from cells transcribing the βWT transgene either by endogenous Pol II LS (endog.; lane 2) or by exogenous RNA Pol II LS wild-type (wt; lane 3) or RNA Pol II LS Δ31 (Δ31; lanes 4 and 5) that were induced to differentiate for 3 d and untreated (−) or treated (+) with α-amanitin for 2 d. RNA from lane 5 was treated with RNase H in the presence of oligo dT12–18. Lanes 1 and 6 contain a 50-bp ladder. Similar results were obtained in two independent experiments.
To avoid complications caused by the presence of HBB mRNA synthesized by the endogenous RNA Pol II LS and which had accumulated before α-amanitin treatment, cells were exposed to this toxin immediately after the first day of induced erythroid differentiation. In MEL cells containing the βWT transgene and which rely exclusively on the endogenous RNA Pol II for transcription, this resulted in massive cell death, while there was survival of cells transfected with the α-amanitin–resistant forms of RNA Pol II LS. In these cells, after inactivation of the endogenous RNA Pol II LS, bands corresponding to both unspliced and spliced (Fig. 4 C, lanes 2 and 3) as well as uncleaved and cleaved (Fig. 4 C, lanes 5 and 6) products were clearly detected. In a control experiment, we analyzed RNA extracted from MEL cells containing the βWT transgene that had undergone the same period (3 d) of differentiation but not exposed to α-amanitin (Fig. 4 C, lanes 1 and 4). The results (Fig. 4 D) revealed no substantial reduction in the percentage of spliced and 3′ cleaved βWT mRNA transcribed by RNA Pol II LS Δ31 compared with that synthesized by either RNA Pol II LS wild type or endogenous RNA Pol II. We further observed that poly(A) tail length of HBB mRNA was similar in transcripts synthesized by either endogenous RNA Pol II RNA (Fig. 4 E, lane 2), RNA Pol II LS wild type (Fig. 4 E, lane 3), or Pol II LS Δ31 (Fig. 4 E, lane 4). Thus, a CTD with heptad repeats 1–23, 36–38, and 48–52 followed by the unique terminal 10-amino acid motif is sufficient to support efficient pre-mRNA processing, as predicted from previous studies (Fong et al., 2003; Rosonina and Blencowe, 2004).
Collectively, our data reveal that βWT transgene mRNA synthesized by the RNA Pol II Δ31 mutant is efficiently spliced, cleaved, and polyadenylated and yet remains in close vicinity to the gene after inhibition of transcription. This strongly suggests that the CTD is required for mRNA release from the site of transcription in a manner independent of splicing and 3′ formation. A possible splicing-independent involvement of the CTD in RNA release was also noted by Bentley and colleagues, who observed that intron-less pre-mRNA synthesized by a terminal 10-amino acid motif mutant CTD remained at the site of transcription, whereas synthesis by a wild-type CTD resulted in RNA release (Bird et al., 2005). However, the use of an intron-less reporter gene in these studies precluded definitive conclusions to be drawn.
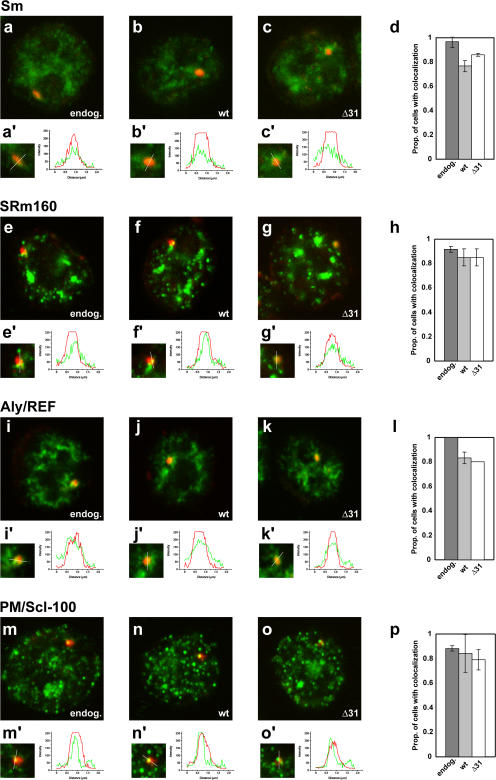
EJC proteins and the PM/Scl-100 exosome subunit are recruited to nascent transcripts synthesized by CTD mutant RNA Pol II
We have previously shown that exon junction complex (EJC) proteins and core spliceosome components (U snRNPs) accumulate on nascent wild-type HBB transcripts, but fail to associate with mutant transcripts that are not released from the transcription site (Custódio et al., 2004). To assess if the presence of truncation mutants of the RNA Pol II LS CTD affected this process, we conducted double-labeling (FISH plus immunocytochemical staining) of MEL cells that contained the βWT transgene transcribed by either endogenous or exogenous RNA Pol II (full-length or the Δ31 CTD mutant) after treatment with α-amanitin as before (Fig. 5). We used a probe to detect βWT transcripts and antibodies to detect snRNP Sm proteins (Fig. 5, a–c), and EJC components SRm160 (Fig. 5, e–g) and Aly/REF (Fig. 5, i–k). Using a previously described quantitative single-cell assay (Misteli et al., 1998; Mabon and Misteli, 2005), we detected all three proteins recruited to nascent transcripts irrespective of CTD length (Fig. 5, a′–k′ and d, h, and l). Moreover, we observed that colocalization of EJC proteins with transcripts synthesized by RNA Pol II LS Δ31 CTD persists after inhibition of transcription by actinomycin D, which adds additional evidence that RNA transcribed by RNA Pol II harboring a truncated Δ31 CTD and retained at the transcription site is normally spliced.
Figure 5.
EJC proteins and the exosome subunit PM/Scl-100 are recruited to the HBB transcription site. MEL cells were induced for 3 d and treated with α-amanitin. The βWT transgene (Fig. 2) was transcribed by endogenous RNA Pol II (a, e, i, and m), RNA Pol II LS wt (b, f, j, and n), or RNA Pol II LS Δ31 (c, g, k, and o). Cells were double-labeled for HBB RNA (red staining) and for protein (green staining) using the indicated antibodies. Relative fluorescence intensity (in arbitrary units) was measured along a line across the βWT RNA focus. We considered a positive recruitment when the intensity of the protein signal at the focus was more than twofold higher than the global nucleoplasmic signal (Mabon and Misteli, 2005). Representative results are shown for the indicated proteins. The proportion of cells exhibiting recruitment was quantified. Values represent average from 45 transcription sites analyzed in two independent experiments ± SD.
In the yeast Saccharomyces cerevisiae, retention of defective mRNA at the site of transcription requires Rrp6p and other components of the nuclear exosome, suggesting that this complex is part of a quality control checkpoint that monitors for correct processing of pre-mRNA (Hilleren et al., 2001; Jensen et al., 2001; Libri et al., 2002; Dunn et al., 2005). We therefore investigated whether the mammalian orthologue of Rrp6p associates with nascent βWT transcripts by immunofluorescence using a specific antibody (Brouwer et al., 2001). This protein was readily detectable throughout the nucleoplasm with a high concentration at the site of βWT transgene transcription (Fig. 5, m–o). Similar results were observed in cells that express exogenous RNA Pol II LS with either a wild-type (Fig. 5, n, n′, and p) or truncated Δ31 (Fig. 5, o, o′, and p) CTD.
Collectively, these data indicate that recruitment of neither EJC proteins nor nuclear exosome Rrp6 class of proteins to nascent mRNA is sufficient for its release from the site of transcription.
Conclusions
Accumulation of nascent mRNA in close proximity to their transcription site is thought to represent a surveillance mechanism that prevents defectively processed transcripts from entering the flow to the cytoplasm (Saguez et al., 2005; Gorski et al., 2006). A major player in cotranscriptional pre-mRNA maturation is the CTD of RNA Pol II, which acts by facilitating specific interactions between processing factors while the transcript is still attached to the polymerase (for review see Bentley, 2005). Previous work revealed that a mutation of the terminal 10-amino acid motif of the CTD inhibited splicing, 3′ end cleavage (Fong et al., 2003), and RNA release from the site of transcription (Bird et al., 2005). Based on these observations it was proposed that the CTD is required for transcript release as a consequence of its role in splicing and 3′ end cleavage (Bird et al., 2005). However, in this report we show that a partial truncation of the CTD (Δ31) containing heptads 1–23, 36–38, and 48–52 including the terminal 10-amino acid motif is sufficient to support transcription, splicing, 3′ end cleavage, and polyadenylation, but the newly synthesized mRNA fails to be efficiently released (Figs. 3 and 4). These novel observations imply that the CTD is involved in processes that control the release of transcripts by a mechanism independent from splicing and cleavage. Collectively with previously reported data (Bird et al., 2005), our results further suggest that different segments of the CTD play distinct roles in pre-mRNA processing and mRNA release from the vicinity of the gene template.
Although the mechanism via which the RNA Pol II LS CTD is involved in the release of mRNA from the transcription site is unknown, we speculate that the Δ31 CTD truncation mutant used in this study fails to bind and therefore recruit protein factors required to complete maturation of spliced and 3′ end cleaved/polyadenylated mRNA into export-competent mRNPs. We further propose that this defect in production of fully mature mRNA activates a quality control checkpoint or surveillance mechanism that prevents diffusion of mRNPs to the nuclear pores by tethering them near the gene template, with stalled mRNA being subsequently degraded by the exosome present at the transcription site. A prediction from this mechanistic model is that most mRNA transcribed by the Δ31 CTD mutant should not translate into protein. In good agreement with that prediction is the observation that the Δ31 CTD mutation cannot support long-term cell viability (Meininghaus et al., 2000).
Materials and methods
MEL cell culture and stable transfections
The maintenance, induction of erythroid differentiation, and stable transfection of the MEL cell line C88 were described elsewhere in detail (Antoniou, 1991). The cells were cotransfected as previously described (Custódio et al., 2006) with an α-amanitin–resistant RNA Pol II LS gene with either a full-length CTD (52 heptad repeats, wild type) or a CTD with 31 (Δ31) or 5 (Δ5) repeats (Fig. 1 A; Bartolomei et al., 1988; Gerber et al., 1995; plasmids provided by W. Schaffner, University of Zürich, Zürich, Switzerland) and a plasmid containing the βLCR (Collis et al., 1990) modified to carry a puromycin resistance gene under the control of a phosphoglycerate kinase promoter (Millevoi et al., 2002). Before transfection the RNA Pol II LS wild-type plasmid was linearized with MluI, the Δ31 and Δ5 plasmids were linearized with ClaI and the micro-βLCR plasmid was linearized with PvuI. Stable transfected clones were obtained by culture in the presence of 2.5 μg/ml puromycin (Sigma-Aldrich). Clones selected for further studies were then super-transfected with either the wild-type (βWT) or mutant (βSM) HBB genes in the micro-βLCR expression vector (Fig. 2 A; Custódio et al., 1999) with stably transfected cells isolated in the presence of 800 μg/ml G418. The function of endogenous RNA Pol II LS was inhibited by adding α-amanitin (Sigma-Aldrich) to the cell culture medium to a final concentration of 2.5 μg/ml.
In situ hybridization and immunofluorescence analysis
HBB transcripts were visualized by FISH (Custódio et al., 1999) and double labeling for RNA and protein was as previously described (Custódio et al., 2004). The probe used for FISH was a 740-bp fragment of the human β-globin gene extending from the SnaBI site at −265 bp from the transcriptional start point to the BamHI site at +475 bp. The fragment was labeled by nick-translation with either digoxigenin-11-dUTP (Roche) or Cy3-AP3-dUTP (GE Healthcare). The following primary antibodies were used for immunofluorescence: rabbit polyclonal directed against SRm160 (1:500; Blencowe et al., 1998; provided by B. Blencowe, University of Toronto, Ontario, Canada), mouse monoclonal directed against Aly/REF (1:100; clone 11G5; AbCam), human autoantiserum C45, specific for Sm proteins (1:75; provided by W. van Venrooij, University of Nijmegen, Nijmegen, Netherlands) and rabbit serum against PM/Scl-100 (1:75; provided by Ger Pruijn, University of Nijmegen). The secondary antibodies used were: AlexaFluor 488–conjugated goat anti–rabbit IgG (1:200; Jackson ImmunoResearch Laboratories, Inc.), AlexaFluor 488–conjugated goat anti–mouse IgG (1:200; Jackson ImmunoResearch Laboratories, Inc.), and FITC-conjugated donkey anti–human IgG (1:100; Jackson ImmunoResearch Laboratories, Inc.).
Microscopy and image quantification
Images were acquired on a laser scanning confocal microscope (LSM 510 or LSM 510 META; Carl Zeiss MicroImaging, Inc.) using the PlanApochromat 63×/1.4 objective. FITC and AlexaFluor 488 fluorescence was detected using the 488-nm line of the argon ion laser. Cy3 was excited with the 543-nm line of the helium-neon laser on the Zeiss LSM 510 and with the DPSS 561-10 laser on the Zeiss LSM 510 META confocal microscope. To quantify the intensity of the nuclear foci, single-cell images were acquired with no saturated pixels, always using the same settings. The mean intensity of fluorescence in the nuclear RNA focus was determined using ImageJ (http://rsb.info.nih.gov/ij/). Line profiles were obtained from unprocessed images using the LSM 510 software.
Western blotting
Total cell extracts were prepared as described in Custódio et al. (2006). Volumes of total extract equivalent to 106 cells were fractionated on a 7% polyacrylamide-SDS gel and proteins were transferred to nitrocellulose in 24 mM Tris, 193 mM glycine, and 20% methanol for 16 h at 30 mA. Western blotting with mouse monoclonal against HA epitope (HA.11; Covance) and mouse monoclonal against α-tubunin (clone B-5-1-2; Sigma-Aldrich) was as previously described (Custódio et al., 2006).
S1-nuclease and RNase protection assays
The levels of endogenous murine βmajor-globin gene (Hbb1) pre-mRNA were analyzed by an S1-nuclease protection assay as described (Custódio et al., 2006). RNase protection assays were performed as described (McCracken et al., 2002; Custódio et al., 2004). HBB RNase protection probes (McCracken et al., 1997a) were prepared by in vitro transcription with T7 RNA polymerase in the presence of α-[32P]UTP (GE Healthcare). In each reaction nuclear RNA, prepared as described (Custódio et al., 2004), was incubated with excess of the antisense RNA probe overnight at 50°C, and the hybridization products were digested with a mixture of RNase T1 and A (Ambion) at 37°C for 1 h. The RNase-protected fragments were resolved on a 6% denaturing polyacrylamide gel and the intensity of the bands was quantified using a PhosphorImager (Molecular Dynamics). After quantification of each gel band, background was subtracted and the values were normalized for different U residue contents of the protected probe fragments.
Poly(A) tail length analysis
The poly(A) tail length analysis of HBB transcripts was performed by PCR using the ligase-mediated poly(A) test (LM-PAT) described by Sallés et al. (1999). For cDNA synthesis we used 3 μg of total RNA, 50 ng of phosphorylated oligo dT12, and 1 μg of oligo(dT)-anchor primer. As a control, RNA was deadenylated by digestion with RNase H in the presence of oligo dT12–18. For PCR amplification, 1 μl of the template LM-PAT cDNA was added to a standard 25-μl PCR reaction containing 12.5 pmol of oligo(dT)-anchor primer and 12.5 pmol of an HBB specific primer that hybridizes 254 nt upstream from the polyadenylation signal (5′-GCAACGTGCTGGTCTGTGTGCTG-3′). The amplified products were resolved by electrophoresis on a 2% agarose gel stained with ethidium bromide.
Supplementary Material
Acknowledgments
We are grateful to Elmar Wahle for advice on the polyadenylation assay. We further thank the following groups for generously providing reagents: W. Schaffner for the CTD constructs; W. Van Venrooij for the human autoantiserum C45; B. Blencowe for the antibody against SRm160; and Ger Pruijn for the rabbit serum against PM-Scl100.
This work was supported by Fundação para a Ciência e Tecnologia, Portugal, the European Science Foundation (EuroDYNA Programme), and the European Commission (EURASNET, LSHG-CT-2005-518238).
M. Vivo's present address is Dipartimento di Biologia Strutturale e Funzionale, Università di Napoli “Federico II”, Napoli 80126, Italy.
Abbreviations used in this paper: βLCR, HBB micro-locus control region; CMV, cytomegalovirus; CTD, C-terminal domain; EJC, exon junction complex; HA, haemagglutinin; HBB, human β-globin gene; Hbb1, murine βmajor-globin gene; MEL, murine erythroleukemia; RNA Pol II LS, largest subunit of RNA polymerase II.
References
- Aguilera, A. 2005. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 17:242–250. [DOI] [PubMed] [Google Scholar]
- Allison, L.A., and C.J. Ingles. 1989. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc. Natl. Acad. Sci. USA. 86:2794–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou, A. 1991. Induction of erythroid-specific expression in murine erythroleukemia (MEL) cells. In Methods in Molecular Biology: Gene Transfer and Expression Protocols. Vol. 7. E.J. Murray, editor. The Human Press Inc., Clinfton, NJ. 421–434. [DOI] [PubMed]
- Antoniou, M., and F. Grosveld. 1990. β-globin dominant control region interacts differently with distal and proximal promoter elements. Genes Dev. 4:1007–1013. [DOI] [PubMed] [Google Scholar]
- Bartolomei, M.S., and J.L. Corden. 1987. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 7:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei, M.S., N.F. Halden, C.R. Cullen, and J.L. Corden. 1988. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 8:330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D.L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17:251–256. [DOI] [PubMed] [Google Scholar]
- Bird, G., N. Fong, J.C. Gatlin, S. Farabaugh, and D.L. Bentley. 2005. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol. Cell. 20:747–758. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J., R. Issner, J.A. Nickerson, and P.A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom van Assendelft, G., O. Hanscombe, F. Grosveld, and D.R. Greaves. 1989. The β-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell. 56:969–977. [DOI] [PubMed] [Google Scholar]
- Brouwer, R., C. Allmang, R. Raijmakers, Y. van Aarssen, W.V. Egberts, E. Petfalski, W.J. van Venrooij, D. Tollervey, and G.J. Pruijn. 2001. Three novel components of the human exosome. J. Biol. Chem. 276:6177–6184. [DOI] [PubMed] [Google Scholar]
- Chapman, R.D., B. Palancade, A. Lang, O. Bensaude, and D. Eick. 2004. The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res. 32:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis, P., M. Antoniou, and F. Grosveld. 1990. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J. 9:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio, N., M. Carmo-Fonseca, F. Geraghty, H.S. Pereira, F. Grosveld, and M. Antoniou. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18:2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio, N., C. Carvalho, I. Condado, M. Antoniou, B.J. Blencowe, and M. Carmo-Fonseca. 2004. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 10:622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio, N., M. Antoniou, and M. Carmo-Fonseca. 2006. Abundance of the largest subunit of RNA polymerase II in the nucleus is regulated by nucleo-cytoplasmic shuttling. Exp. Cell Res. 312:2557–2567. [DOI] [PubMed] [Google Scholar]
- Dunn, E.F., C.M. Hammell, C.A. Hodge, and C.N. Cole. 2005. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 19:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, N., and D.L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, N., G. Bird, M. Vigneron, and D.L. Bentley. 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 22:4274–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V., O. Gadal, M. Fromont-Racine, A. Romano, A. Jacquier, and U. Nehrbass. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 116:63–73. [DOI] [PubMed] [Google Scholar]
- Gerber, H.P., M. Hagmann, K. Seipel, O. Georgiev, M.A. West, Y. Litingtung, W. Schaffner, and J.L. Corden. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 374:660–662. [DOI] [PubMed] [Google Scholar]
- Gorski, S.A., M. Dundr, and T. Misteli. 2006. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18:284–290. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T.H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 413:538–542. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 7:887–898. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., K. Dower, D. Libri, and M. Rosbash. 2003. Early formation of mRNP: license for export or quality control? Mol. Cell. 11:1129–1138. [DOI] [PubMed] [Google Scholar]
- Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T.H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabon, S.A., and T. Misteli. 2005. Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo. PLoS Biol. 3:e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D.L. Bentley. 1997. a. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S.D. Patterson, M. Wickens, and D.L. Bentley. 1997. b. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 385:357–361. [DOI] [PubMed] [Google Scholar]
- McCracken, S., M. Lambermon, and B.J. Blencowe. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22:148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meininghaus, M., and D. Eick. 1999. Requirement of the carboxy-terminal domain of RNA polymerase II for the transcriptional activation of chromosomal c-fos and hsp70A genes. FEBS Lett. 446:173–176. [DOI] [PubMed] [Google Scholar]
- Meininghaus, M., R.D. Chapman, M. Horndasch, and D. Eick. 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J. Biol. Chem. 275:24375–24382. [DOI] [PubMed] [Google Scholar]
- Millevoi, S., F. Geraghty, B. Idowu, J.L. Tam, M. Antoniou, and S. Vagner. 2002. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 3:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and D.L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 3:697–705. [DOI] [PubMed] [Google Scholar]
- Misteli, T., J.F. Caceres, J.Q. Clement, A.R. Krainer, M.F. Wilkinson, and D.L. Spector. 1998. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science. 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.T., F. Giannoni, M.F. Dubois, S.J. Seo, M. Vigneron, C. Kedinger, and O. Bensaude. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24:2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina, E., and B.J. Blencowe. 2004. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA. 10:581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez, C., J.R. Olesen, and T.H. Jensen. 2005. Formation of export-competent mRNP: escaping nuclear destruction. Curr. Opin. Cell Biol. 17:287–293. [DOI] [PubMed] [Google Scholar]
- Sallés, F.J., W.G. Richards, and S. Strickland. 1999. Assaying the polyadenylation state of mRNAs. Methods. 17:38–45. [DOI] [PubMed] [Google Scholar]
- Stiller, J.W., and B.D. Hall. 2002. Evolution of the RNA polymerase II C-terminal domain. Proc. Natl. Acad. Sci. USA. 99:6091–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.