Abstract
Actin assembly at the leading edge of migrating cells depends on the availability of high-affinity free barbed ends (FBE) that drive actin filament elongation and subsequent membrane protrusion. We investigated the specific mechanisms through which the Rac1 and Rac2 small guanosine triphosphatases (GTPases) generate free barbed ends in neutrophils. Using neutrophils lacking either Rac1 or Rac2 and a neutrophil permeabilization model that maintains receptor signaling to the actin cytoskeleton, we assessed the mechanisms through which these two small GTPases mediate FBE generation downstream of the formyl-methionyl-leucyl-phenylalanine receptor. We demonstrate here that uncapping of existing barbed ends is mediated through Rac1, whereas cofilin- and ARP2/3-mediated FBE generation are regulated through Rac2. This unique combination of experimental tools has allowed us to identify the relative roles of uncapping (15%), cofilin severing (10%), and ARP2/3 de novo nucleation (75%) in FBE generation and the respective roles played by Rac1 and Rac2 in mediating actin dynamics.
Introduction
Actin cytoskeletal dynamics require the cycling of actin between monomeric and polymeric pools, a transition that is mediated in large part by the actin-binding and filament-severing protein cofilin (DesMarais et al., 2004; Kiuchi et al., 2007). Actin assembly is regulated through the generation of free high-affinity actin filament ends, also known as free barbed ends (FBEs). Three mechanisms regulate the availability of FBEs in vivo (Condeelis, 2001). In the first, de novo nucleation mediated by the Arp2/3 complex results in new actin FBEs (ARP2/3-dependent FBEs [ARPFBE]). Severing of noncovalent bonds of existing F-actin (Ichetovkin et al., 2002) is the second mechanism described for generating FBEs. This mechanism is known to be mediated by members of the actin depolymerizing factor/cofilin family, and so results in cofilin-mediated FBEs (COFFBE; Huang et al., 2006). The third mechanism for actin assembly involves uncapping of existing actin filament barbed ends. The uncapping mechanism, which results in the removal of actin filament capping proteins such as CapZ and gelsolin, leads to the exposure of FBEs, called uncapping-mediated FBEs (UNCAPFBE) that drive the polymerization of existing filaments.
Members of the Rho family of small GTPases have been demonstrated to play key roles in the regulation of the actin cytoskeleton (Bokoch, 1995a; Glogauer et al., 2000; Pestonjamasp et al., 2006). Rac and Cdc42 have been shown to regulate Arp2/3 de novo nucleation through Wiskott-Aldrich syndrome protein and the Wiskott-Aldrich syndrome protein family verprolin-homologous protein (Hall, 1992; Edwards et al., 1999; Bokoch, 2000; 2005; Pollard et al., 2000). Using a permeabilized neutrophil model we have previously shown that although Cdc42 regulates the Arp2/3 complex, it is unclear how Rac, and more specifically Rac1 and Rac2, regulate actin assembly (Glogauer et al., 2000). The development of Rac1 and Rac2 neutrophil-knockout models has allowed us to dissect the specific regulatory roles of these proteins in neutrophil functions (Glogauer et al., 2003). Previous studies have shown that the Rac1 and Rac2 isoforms carry out distinct roles in the regulation of neutrophil functions, including chemotaxis compass regulation (Rac1 specific) and actin polymerization (predominantly Rac2; Bokoch, 1995b; Roberts et al., 1999; Sun et al., 2004).
In the present investigation, we used a previously described cell permeabilization technique (Barkalow et al., 1996; Glogauer et al., 2000) in Rac1 and Rac2 knockout neutrophils to further dissect the specific roles of these small GTPases in generating FBEs. We describe here that Rac1 and Rac2 differentially regulate the generation of actin FBE through separate pathways: Rac1 mediates FBE generation during neutrophil chemotaxis through uncapping of existing filaments; Rac2 is responsible for the majority of FBE formation through its mediation of cofilin activation and the ARP2/3 complex.
Results and discussion
Rac1 and Rac2 differentially regulate actin FBE formation in neutrophils
To assess the relative roles of Rac1 and Rac2 in FBE generation downstream of the formyl-methionyl-leucyl-phenylalanine (fMLP) receptor, wild-type (WT), Rac1 null (Rac1N), and Rac2N neutrophils were subjected to a previously described pyrene-actin nucleation assay that measures FBE generation after neutrophil activation (Glogauer et al., 2000). Analysis of the pyrene-actin polymerization curves, where the slope is proportional to the FBE numbers, demonstrate that both Rac1N and Rac2N neutrophils display a defect in fMLP-mediated FBE generation compared with WT neutrophils (Fig. 1 A). Although Rac1N cells displayed a modest 30% defect in FBE generation, Rac2N cells exhibited a 70% defect in fMLP-mediated FBE generation, which was consistent with our previous work demonstrating that Rac2N cells are unable to migrate because of an inability to assemble actin during chemotaxis (Glogauer et al., 2003).
Figure 1.
Rac1 and Rac2 are both required for FBE formation downstream of fMLP stimulation. (A) Time series analysis of the fluorescence increase associated with FBE formation. One million neutrophils were permeabilized with 0.2% OG buffer, stimulated with 1 μM fMLP for 60 s, and then monitored for pyrene-actin incorporation. The slope of the fluorescence increase curve was used to determine the proportional increase in barbed ends. The relative contribution of FBEs was determined by adding 2 μM CD and converting the cytochalasin-sensitive assembly rate into the number of nucleation sites as described in Glogauer et al. (2000; CD inhibited >97% of all FBEs). Representative results of 18 observations are shown. (B) Rac2 is the primary regulator of FBE formation in neutrophils. Histogram shows the mean FBE increase compared with the nonstimulated WT control. Pyrene-actin incorporation was evaluated for 180 s and the ratio of the fMLP-stimulated to control slope was calculated to determine the increase in FBEs after stimulation. n = 18; P < 0.001. Error bars represent ± SEM.
Rac1 is the primary mediator of filament uncapping downstream of the fMLP receptor
To investigate the mechanisms through which Rac1 and Rac2 regulate actin assembly we used a partial permeabilization assay (Glogauer et al., 2000). This assay allows us to assess actin filament barbed end uncapping by measuring the release of actin filament capping proteins from existing barbed ends after neutrophil activation (Barkalow et al., 1996; Glogauer et al., 2000). As shown in Fig. 2 (A and B), Rac1N neutrophils demonstrated no release of CapZα after fMLP stimulation, whereas WT and Rac2N neutrophils showed a clear release of this protein after fMLP stimulation. Similarly, Rac1 defective mutants failed to release the barbed end capping proteins gelsolin (Fig. 2, C and D) and adducin (Fig. 2 E). The release of these two additional uncapping proteins was similar to the CapZα release kinetics (not depicted). Interestingly, Rac2N neutrophils showed a small reduction in gelsolin release compared with the WT cells, suggesting that Rac2 may have a minor role in the regulation of gelsolin uncapping. These results demonstrate that Rac1 is the Rac small GTPase responsible for efficient actin filament uncapping after neutrophil activation (Rac1→UNCAPFBE).
Figure 2.
Rac1 is the primary regulator of filament uncapping downstream of fMLP stimulation. (A) Western blot analysis of CapZα release after fMLP stimulation. Capping protein's release from barbed ends after fMLP stimulation was assessed in neutrophils lacking either Rac1 or Rac2 as indicated in Materials and methods. CapZα released from the cytoskeleton was measured in the supernatant of permeabilized cells and compared with total CapZα levels in the cell lysates. Immunoblot shown is representative of three independent experiments. (B) Analysis of CapZα release after fMLP stimulation. Immunoblots of CapZα in supernatant and total cell lysates were analyzed by densitometry and compared with the nonstimulated control. WT and Rac2N neutrophils released CapZα after fMLP stimulation. Rac1N cells failed to release CapZ after fMLP stimulation (P > 0.90). Error bars represent ± SEM. (C) Rac1 is the key mediator of gelsolin uncapping downstream of fMLP stimulation. The release of gelsolin was determined by immunoblotting of the supernatant of permeabilized neutrophils and total cell lysates after 60 s of fMLP stimulation. The immunoblot shown is representative of four independent experiments and illustrates the failure of Rac1N cells to release gelsolin after fMLP stimulation. (D) Densitometry analysis of gelsolin in the supernatant of permeabilized neutrophils. WT and Rac2N neutrophils significantly increase the supernatant levels of gelsolin after 60 s of fMLP stimulation (n = 4; *, P < 0.001). Rac1N cells failed to release gelsolin after fMLP stimulation (n = 4; P > 0.90). Error bars represent ± SEM. (E) Rac1 is the key mediator of adducin uncapping downstream of fMLP stimulation. The release of adducin was determined by immunoblotting of the supernatant of permeabilized neutrophils after 60 s of fMLP stimulation. The Western blot shown is representative of three independent experiments.
Rac2 mediates cofilin activation downstream of the fMLP receptor
It is clear from several studies that the actin binding protein cofilin has an essential role in the actin-remodeling process and is an essential element in cells undergoing rapid actin cytoskeletal turnover (Ichetovkin et al., 2002; Falet et al., 2005; Huang et al., 2006). It is clear that cofilin generates FBEs while generating free actin monomers that add on to FBEs at the leading edge of migrating cells (Zebda et al., 2000; DesMarais et al., 2004; Mouneimne et al., 2006). To determine if the Rac small GTPases regulate cofilin activity, cofilin phosphorylation at serine 3 was assessed before and after fMLP stimulation in WT, Rac1N, and Rac2N neutrophils. We observed that although WT and Rac1N neutrophils were able to efficiently activate cofilin downstream of fMLP, Rac2N neutrophils failed to dephosphorylate cofilin (activate) after fMLP stimulation (Fig. 3, A and B). These data demonstrate that Rac2 is the small GTPase responsible for cofilin dephosphorylation/activation in neutrophils (Rac2→COFFBE).
Figure 3.
Rac2 is the small GTPase regulator of cofilin dephosphorylation. (A) Western blot analysis of cofilin dephosphorylation after fMLP stimulation. Cofilin activation was assessed by the measurement of P-cofilin (Ser3) in cell lysates before and after 60 s of fMLP stimulation. Immunoblot shown is representative of three independent experiments. (B) Rac2N neutrophils failed to dephosphorylate P-cofilin after fMLP stimulation. Densitometry analysis of P-cofilin immunoblots in Rac1N, Rac2N, and WT cells after 60 s of fMLP stimulation. WT and Rac1N neutrophils significantly dephosphorylate and activate cofilin after 60 s of fMLP stimulation (n = 3; P < 0.001). Rac2N cells failed to dephosphorylate cofilin (n = 3; P > 0.4). Error bars represent ± SEM. (C) CIN expression rescues cofilin activation in Rac2N neutrophils. Rac2N murine neutrophils were transfected with GFP-CIN-WT or an empty GFP plasmid. Total cell lysates were immunoblotted for P-cofilin and total cofilin. Immunoblot shown is representative of three independent experiments. (D) CIN expression increases FBE formation in neutrophils. Murine neutrophils were transfected either with GFP-CIN-WT or GFP alone (control) and subjected to a pyrene-actin nucleation assay to quantify the relative FBE formation. Neutrophils transfected with CIN-WT had a significant increase in FBE formation both in the resting state and after fMLP stimulation (n = 4; P < 0.05). Error bars represent ± SEM.
To verify that cofilin dephosphorylation was required for generating FBE as demonstrated previously (Zebda et al., 2000; DesMarais et al., 2004), we used a novel approach, which we have recently described (Magalhaes et al., 2007), to transfect primary murine neutrophils with chronophin (CIN). CIN is a haloacid dehydrogenase–type phosphatase that directly dephosphorylates cofilin (Gohla et al., 2005). Considering that Rac2N neutrophils had an impaired dephosphorylation of cofilin, we asked whether an overexpression of CIN would rescue the Rac2N phenotype by dephosphorylating cofilin and generating FBEs. Cells were transfected with the constitutively active CIN-WT (GFP-CIN-WT) construct and analyzed for the formation of FBE (Gohla et al., 2005; Magalhaes et al., 2007). The overexpression of GFP-CIN-WT led to increased FBE formation both in fMLP-stimulated cells and in resting cells (Fig. 3 D). Importantly, Rac2N cells transfected with GFP-CIN-WT significantly increased FBE formation (P < 0.03), displaying levels comparable to the fMLP-stimulated WT-nontransfected group. To verify that the transfection protocol did not influence FBE generation, neutrophils were transfected with a GFP vector control and displayed no significant differences when compared with their nontransfected counterparts (P > 0.05). The overexpression of active CIN confirms that cofilin dephosphorylation (Fig. 3 C) leads to increased FBE generation in neutrophils (Fig. 3 D).
Rac2 is the regulator of COFFBE generation at the leading edge during chemotaxis
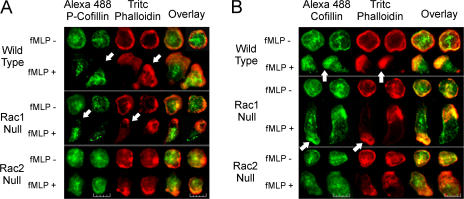
Previous studies have demonstrated the importance of cofilin in FBE formation at the leading edge and in generation of protrusive force in migrating cells (DesMarais et al., 2004; Ghosh et al., 2004). To further confirm that Rac2 regulates cofilin activation and subsequent FBE generation at the leading edge of migrating cells, we investigated the subcellular localization of total cofilin and phosphorylated cofilin (P-cofilin) in WT, Rac1N, and Rac2N neutrophils. Epifluorescence microscopy images show that WT and Rac1N neutrophils display abundant cofilin and complete absence of P-cofilin in the F-actin rich leading edge during fMLP-mediated chemotaxis (Fig. 4). However, Rac2N cells show no loss of P-cofilin from the same leading edge of fMLP-activated neutrophils (Fig. 4). These results confirm that Rac2 is required for mediating cofilin dephosphorylation and subsequent FBE generation at the leading edge of chemotaxing neutrophils. As we have described previously in Glogauer et al. (2003) and reflected in Fig. 4, Rac knockouts demonstrate distinct morphological changes when compared with WT cells. Although Rac1N neutrophils display an abnormally elongated morphology caused by impaired Rho activation (Pestonjamasp et al., 2006), Rac2N neutrophils polarize and orient toward the source of chemoattractant but fail to migrate efficiently, resulting in a poorly defined uropod (Sun et al., 2004).
Figure 4.
Rac2 regulates the subcellular localization of active/dephosphorylated cofilin. (A) P-cofilin is absent in the F-actin–rich leading edge in activated WT and Rac1N cells only. Neutrophils were stimulated with fMLP for 60 s, fixed in PFA, and immunostained with anti–P-cofilin (Alexa 488) and TRITC phalloidin (F-actin). After fMLP stimulation, P-cofilin is not found at the F-actin–rich leading edge in WT and Rac1N neutrophils (arrows) but is still present in the leading edge of the Rac2N cells. (B) Rac2 regulates dephosphorylation of cofilin at the leading edge. Neutrophils were stimulated with fMLP for 60 s, fixed, and immunostained for total cofilin (anti-cofilin/Alexa 488) and F-actin (TRITC phalloidin). Dephosphorylated cofilin levels increased at the F-actin–rich leading edge in fMLP-stimulated WT and Rac1N neutrophils (arrows) but not in the leading edge of the Rac2N cells. Results are representative of 40 cells per group. Bar, 12 μm.
FBE generation: relative roles of cofilin, uncapping, and Arp2/3
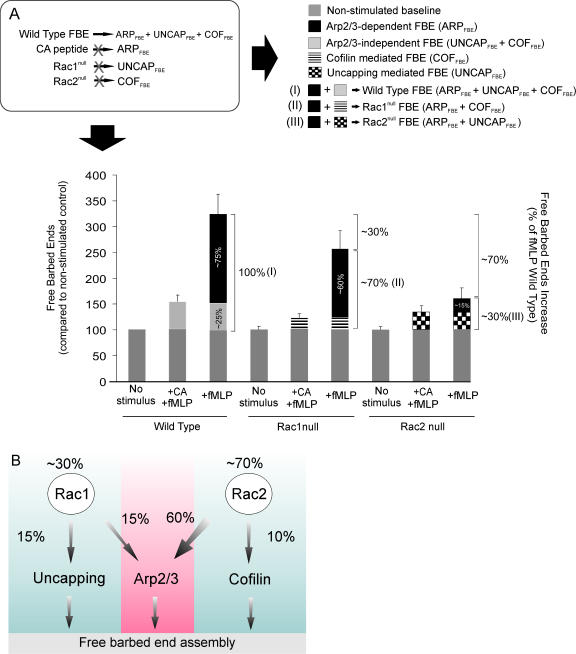
Although we recognize that there is likely to be interplay and interdependence between the three FBE generation mechanisms, we have used our findings and observations to model and approximate the relative roles of uncapping, cofilin severing, and Arp2/3 nucleation in FBE generation downstream of the fMLP receptor. At present, three primary mechanisms have been identified as being responsible for generating FBE in rapidly migrating cells such as neutrophils and cancer cells (Glogauer et al., 2000; Condeelis, 2001). In WT neutrophils, FBEs are generated through either Arp2/3 (ARPFBE), uncapping (UNCAPFBE), and/or cofilin severing (COFFBE): WTFBE = ARPFBE + UNCAPFBE + COFFBE. To determine the proportion (%) that each of these mechanisms contributes to total fMLP-mediated FBEs we used the five following observations to solve the above equation (Fig. 5 A, top left).
Figure 5.
Relative roles of Arp2/3, uncapping, and cofilin in fMLP-mediated FBE formation. (A) Rac2 is the primary regulator of fMLP-mediated FBE formation. Rac1N and Rac2N were treated with 1 μM CA peptide (ARP2/3 inhibitor) before OG permeabilization and 1 μM fMLP activation (60 s). Quantification of FBE formation was determined using a pyrene-actin nucleation assay. In each group, the ARPFBE formation was calculated by subtracting the residual FBE after treatment with the CA peptide from the fMLP-stimulated control samples (FBEfMLP − FBEfMLP+CA). Data were normalized to the nonactivated WT control. Results shown are based on nine independent experiments. P < 0.0001. The relative roles of ARPFBE, UNCAPFBE, and COFFBE were evaluated based on the assumptions drawn from the described observations (legend). Error bars represent ± SEM. (B) The relative roles of Arp2/3, uncapping, and cofilin severing in fMLP-mediated FBE generation. As described, Rac1 and Rac2 regulate FBE formation through uncapping and cofilin activation, respectively. In addition, Rac2 has a key role in the regulation of Arp2/3 leading to FBE. This is consistent with our previous demonstration that Rac2 activates Cdc42 (Sun et al., 2004).
First, neutrophils without both Rac1 and Rac2 (double null) show an uncapping and a cofilin dephosphorylation–severing defect. Double null neutrophils treated with the CA domain peptide from WASP, which inhibits Arp2/3-mediated nucleation and was previously described in Barkalow et al. (1996) and Glogauer et al. (2000), do not show any detectable increase in FBEs downstream of the fMLP receptor (unpublished data), confirming the validity of the equation WTFBE = ARPFBE + UNCAPFBE + COFFBE.
Second, because we know that the CA peptide inhibits de novo nucleation through the Arp2/3 complex, we could determine the relative role played by Arp2/3 in the FBE formation downstream of fMLP stimulation by comparing the CA-pretreated and nontreated neutrophils after fMLP stimulation (ARPFBE = FBEfMLP − FBEfMLP+CA).
Third, because we know that Rac1 is responsible for uncapping (UNCAPFBE; Fig. 2), then Rac1null FBE = ARPFBE + COFFBE. We are now able to eliminate ARPFBE from the equation by using the CA peptide. As demonstrated in Fig. 5 A, this allows us to conclude that Rac1null FBE + CA = COFFBE = 10% of total FBEs.
Fourth, based on our findings that Rac2 is responsible for COFFBE (Figs. 3 and 4), FBE formation in Rac2N cells is attributed to filament uncapping and Arp2/3 (Rac2null FBE = ARPFBE + UNCAPFBE). We are able to eliminate ARPFBE from the equation by using the CA domain peptide. As demonstrated in Fig. 5 A, this allows us to conclude that Rac2null FBE + CA = UNCAPFBE = 15% of total FBEs. In addition to this observation, we also show that Rac2 is the primary regulator of Arp2/3, responsible for ∼60% of total ARPFBE.
Fifth, we are now able to determine that Arp2/3 is responsible for 75% of FBEs through two separate approaches: (a) fMLP-stimulated WT cells treated with the CA peptide show a 75% reduction in FBE generation; (b) combining the second and third findings allows us to calculate that 75% of FBEs are dependent on Arp2/3 because we have accounted for uncapping (15%) and cofilin (10%).
Rac1 and Rac2 differential regulation of FBEs
In the present investigation, we analyzed the specific roles of Rac1 and Rac2 on FBE formation downstream of the fMLP receptor in neutrophils. We describe for the first time that Rac1 and Rac2 differentially regulate FBE formation in neutrophils through uncapping and Arp2/3/cofilin, respectively (Fig. 5 B). Here we show that Rac1 is responsible for UNCAPFBE formation (∼15%). We also show that Rac2 is the key regulator of FBE formation in murine neutrophils (∼70%) by regulating both actin depolymerizing factor/cofilin (COFFBE, ∼10%) and the Arp2/3 complex (ARPFBE, ∼60%). Previous studies have demonstrated that cofilin is a major generator of FBEs in migrating cancer cells (Zebda et al., 2000; Condeelis, 2001). Recently, DesMarais et al. (2004) also showed that cofilin directly initiates FBE formation and works synergistically with Arp2/3 to create a burst of actin nucleation. Using a different approach, Kiuchi et al. (2007) demonstrated that cofilin is also required to generate the necessary supply of actin monomers, which add on to FBEs during actin assembly at the leading edge of migrating cells. Thus, cofilin appears to have dual roles in the leading edge of migrating cells through its generation of FBEs and its critical role in supplying free monomers required for actin assembly. Our experimental approach measuring FBE generation, using incorporation of pyrene actin in Rac knockouts treated with CA peptide, has allowed us to confirm and quantify the relative role of cofilin in FBE generation. From our data we are able to demonstrate that cofilin does make a small but considerable contribution to FBEs (COFFBE, ∼10%) in neutrophils downstream of fMLP activation, independent of its critical role in regulating the availability of free actin monomers. Importantly, our observation that Rac2 is the primary regulator of the Arp2/3 complex is consistent with previous work showing that Cdc42 activation in these murine neutrophils is downstream of Rac2 (Sun et al., 2004). Future studies will focus on the mechanisms through which Rac2 regulates cofilin activity.
Materials and methods
Animal models and neutrophil preparations
All procedures described were performed in accordance with the Guide for the Humane Use and Care of Laboratory Animals and were approved by the University of Toronto Animal Care Committee. Rac1-conditional null and Rac2−/− mice were generated according to a protocol described in Glogauer et al. (2003). In brief, Rac1 was selectively disrupted in granulocytes/neutrophils by using a conditional Rac1c/−LysMcre in which the Cre recombinase is expressed under the control of the murine lysozyme M gene regulatory region. This approach generated Rac1 deletion in neutrophils at birth (Glogauer et al., 2003). Rac1 conditionals were bred with Rac2N mice (Roberts et al., 1999) and the resulting offspring enabled the generation of mice with neutrophils deficient in either Rac1, Rac2, or both. Genotyping for Rac1, Rac2, and LysM alleles was performed as described in Glogauer et al. (2003). Neutrophil isolation was performed as described in Sun et al. (2004). More than 85% of cells isolated were neutrophils as assessed by Wright-Giemsa staining. Viability as determined by trypan blue exclusion was >90%.
Phosphocofilin immunoblot and immunostaining
Isolated bone marrow neutrophils were exposed to 1 μM fMLP at 37°C for 60 s and immediately subjected to 15% SDS-PAGE. Nitrocellulose membranes were incubated overnight with 1:1,000 phosphocofilin (Ser3) and cofilin antibodies (Cell Signaling Technology) in TBS-Tween 20 solution and 5% fat-free milk. Membranes were incubated with goat anti–rabbit IgG peroxidase conjugates (GE Healthcare) followed by chemiluminescence visualization (ECL; PerkinElmer). Immunoblots were scanned (300 dpi; Perfection 1250; Epson) and analyzed by densitometry (ImageJ 1.35s; National Institutes of Health). For cofilin and P-cofilin immunostaining, 106 neutrophils were cultured on BSA-coated slides for 10 min at 37°C and stimulated with fMLP for 1 min followed by fixation with 4% PFA. Fixed cells were washed in PBS, permeabilized with 0.1% Triton X-100 in PBS for 4 min, and blocked with 1% BSA for 30 min. Cells were incubated with anti-phosphocofilin or anti-cofilin (Cell Signaling Technology), diluted 1:50, and detected with Alexa Flour 488 goat anti–rabbit IgG (Invitrogen). Cells were also stained with 1:400 rhodamine phalloidin at room temperature for 30 min followed by epifluorescence microscopy analysis. Images were visualized using an Eclipse E100 (Nikon), a 40/0.95 Plan Apo objective (Nikon), and a digital camera (C4742-80; Hamamatsu). Images were acquired using Simple PCI software version 5.3 (Compix). All figures were created using CorelDRAW 12.0.
FBE assay
To analyze actin nucleation activity, we determined the ability of permeabilized neutrophils to accelerate spontaneous actin assembly measured as enhancement of pyrene-actin fluorescence with polymerization (Glogauer et al., 2000). We permeabilized neutrophils (5 × 106/ml) for 10 s using 0.2% OG (PHEM buffer containing 10 μM phallacidin, 42 nM leupeptin, 10 mM benzamidine, and 0.123 mM aprotinin). We stopped the permeabilization process by diluting the detergent with 3 vol of buffer B (1 mM Tris, 1 mM EGTA, 2 mM MgCl2, 10 mM KCl, 5 mM β-mercaptoethanol, and 5 mM ATP; pH 7.4) and then stimulated the cells with 1 μM fMLP for 60 s. We then assayed for FBEs by adding pyrene-labeled rabbit skeletal muscle actin (Glogauer et al., 2000) to a final concentration of 1 μM and followed the fluorescence increase using a microplate reader (FLUOstar optima; BMG Labtech) with fluorescence excitation and emission wavelengths of 355 and 405 nm, respectively. For some experiments cells were stimulated with fMLP for 60 s after cell permeabilization. There were no differences in FBE numbers whether the cells were stimulated before or after permeabilization, as this technique maintains receptor signaling to actin assembly after the brief OG permeabilization (Glogauer et al., 2000). To confirm that we were measuring FBEs and not pointed ends, 2 μM cytochalasin D (CD) was added to block barbed ends. In all experiments >97% of all fluorescence increase was inhibited by the CD (Glogauer et al., 2000). The CA peptide was used to block Arp2/3 complex–mediated nucleation (Rohatgi et al., 2000). As described in Glogauer et al. (2000), 1 μM CA peptide was present in the media at the time of cell permeabilization to allow for peptide entry into the permeabilized cells, and this was then followed by fMLP stimulation and pyrene incorporation quantification as described in this paragraph.
Gelsolin, adducin, and CapZ quantification
Gelsolin and CapZ were measured in the supernatant of OG-permeabilized cells and on the respective cell lysates. One million murine neutrophils were permeabilized with 0.2% OG buffer and then stimulated with 1 μM fMLP for up to 60 s. The supernatants were collected at 10, 30, and 60 s and subjected to SDS-PAGE electrophoresis. The remaining cells were lysed and collected separately, followed by SDS-PAGE electrophoresis. Proteins were transferred to a membrane and blotted with 1:2,000 anti-gelsolin, 1:2,000 anti–adducin α (Santa Cruz Biotechnology, Inc.), or 1:5,000 anti-CapZ primary antibodies (BD Biosciences). The anti-gelsolin antibody was provided by C.A. McCulloch (University of Toronto, Toronto, Canada). The release of capping proteins was evaluated by the amount of capping proteins found in the supernatant of permeabilized cells. Results were analyzed with ImageJ 1.35s.
CIN transfection
Primary neutrophils were transfected using a previously described protocol (Magalhaes et al., 2007). In brief, murine neutrophils were suspended in 100 μl of Nucleofector Solution V (Amaxa Biosystems) and supplemented with 6 μg of vector DNA of WT CIN tagged with GFP (GFP-CIN-WT) or GFP control vector (Gohla et al., 2005). The GFP-CIN-WT construct was supplied by G.M. Bokoch (The Scripps Research Institute, La Jolla, CA). The pmaxGFP DNA construct (Amaxa Biosystems) was used as the control. The cells were transfected using the program Y-001 (Amaxa Biosystems). The cells were carefully recovered with 1,000 μl of 37°C Iscove's Modified Dulbecco's medium and transferred to 1 ml of Iscove's Modified Dulbecco's medium with 10% FBS, followed by a 2-h recovery time in a humidified 37°C 5% CO2 incubator. Cells were allowed to recover on nontissue culture 12-well plates. In some experiments, the cells were subjected to SDS-PAGE electrophoresis and immunoblotted for P-cofilin to monitor cofilin activation (dephosphorylation). A >45% transfection efficiency was achieved using this protocol.
Statistical analysis
Statistical analysis was performed using the 14.0 software (SPSS Inc.). Multiple comparisons were performed by analysis of variance associated with the Bonferroni and Tukey's honestly significant difference tests for post hoc testing. All results represent at least three independent experiments. Statistical significance was defined as P < 0.05. Data are expressed as mean ± SEM.
Acknowledgments
Thanks to Laurent Sabbagh (University of Toronto).
This work is supported by a Canadian Institute for Health Research (CIHR) operating grant (MOP-53136) and a CIHR New Investigator Award to M. Glogauer. M.A.O. Magalhães is a CIHR strategic training fellow (STP-53877) and recipient of a University of Toronto Open Scholarship.
C.X. Sun and M.A.O. Magalhães contributed equally to this paper.
Abbreviations used in this paper: ARPFBE, ARP2/3-dependent FBE; CD, cytochalasin D; CIN, chronophin; COFFBE, cofilin-mediated FBE; FBE, free barbed end; fMLP, formyl-methionyl-leucyl-phenylalanine; P-cofilin, phosphorylated cofilin; Rac1N, Rac1 null neutrophil; UNCAPFBE, uncapping-mediated FBE; WT, wild-type.
References
- Barkalow, K., W. Witke, D.J. Kwiatkowski, and J.H. Hartwig. 1996. Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J. Cell Biol. 134:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch, G.M. 1995. a. Chemoattractant signaling and leukocyte activation. Blood. 86:1649–1660. [PubMed] [Google Scholar]
- Bokoch, G.M. 1995. b. Regulation of the phagocyte respiratory burst by small GTP-binding proteins. Trends Cell Biol. 5:109–113. [DOI] [PubMed] [Google Scholar]
- Bokoch, G.M. 2000. Regulation of cell function by Rho family GTPases. Immunol. Res. 21:139–148. [DOI] [PubMed] [Google Scholar]
- Bokoch, G.M. 2005. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 15:163–171. [DOI] [PubMed] [Google Scholar]
- Condeelis, J. 2001. How is actin polymerization nucleated in vivo? Trends Cell Biol. 11:288–293. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., F. Macaluso, J. Condeelis, and M. Bailly. 2004. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 117:3499–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, D.C., L.C. Sanders, G.M. Bokoch, and G.N. Gill. 1999. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1:253–259. [DOI] [PubMed] [Google Scholar]
- Falet, H., G. Chang, B. Brohard-Bohn, F. Rendu, and J.H. Hartwig. 2005. Integrin alpha(IIb)beta3 signals lead cofilin to accelerate platelet actin dynamics. Am. J. Physiol. Cell Physiol. 289:C819–C825. [DOI] [PubMed] [Google Scholar]
- Ghosh, M., X. Song, G. Mouneimne, M. Sidani, D.S. Lawrence, and J.S. Condeelis. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746. [DOI] [PubMed] [Google Scholar]
- Glogauer, M., J. Hartwig, and T. Stossel. 2000. Two pathways through Cdc42 couple the N-formyl receptor to actin nucleation in permeabilized human neutrophils. J. Cell Biol. 150:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glogauer, M., C.C. Marchal, F. Zhu, A. Worku, B.E. Clausen, I. Foerster, P. Marks, G.P. Downey, M. Dinauer, and D.J. Kwiatkowski. 2003. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J. Immunol. 170:5652–5657. [DOI] [PubMed] [Google Scholar]
- Gohla, A., J. Birkenfeld, and G.M. Bokoch. 2005. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7:21–29. [DOI] [PubMed] [Google Scholar]
- Hall, A. 1992. Ras-related GTPases and the cytoskeleton. Mol. Biol. Cell. 3:475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T.Y., C. DerMardirossian, and G.M. Bokoch. 2006. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18:26–31. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- Kiuchi, T., K. Ohashi, S. Kurita, and K. Mizuno. 2007. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177:465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes, M.A., F. Zhu, H. Sarantis, S.D. Gray-Owen, R.P. Ellen, and M. Glogauer. 2007. Expression and translocation of fluorescent-tagged p21-activated kinase-binding domain and PH domain of protein kinase B during murine neutrophil chemotaxis. J. Leukoc. Biol. 82:559–566. [DOI] [PubMed] [Google Scholar]
- Mouneimne, G., V. DesMarais, M. Sidani, E. Scemes, W. Wang, X. Song, R. Eddy, and J. Condeelis. 2006. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16:2193–2205. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp, K.N., C. Forster, C. Sun, E.M. Gardiner, B. Bohl, O. Weiner, G.M. Bokoch, and M. Glogauer. 2006. Rac1 links leading edge and uropod events through Rho and myosin activation during chemotaxis. Blood. 108:2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., L. Blanchoin, and R.D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- Roberts, A.W., C. Kim, L. Zhen, J.B. Lowe, R. Kapur, B. Petryniak, A. Spaetti, J.D. Pollock, J.B. Borneo, G.B. Bradford, et al. 1999. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 10:183–196. [DOI] [PubMed] [Google Scholar]
- Rohatgi, R., H.Y. Ho, and M.W. Kirschner. 2000. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150:1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C.X., G.P. Downey, F. Zhu, A.L. Koh, H. Thang, and M. Glogauer. 2004. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood. 104:3758–3765. [DOI] [PubMed] [Google Scholar]
- Zebda, N., O. Bernard, M. Bailly, S. Welti, D.S. Lawrence, and J.S. Condeelis. 2000. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]