Abstract
Anumber of mammalian genes are expressed from only one of the two homologous chromosomes, selected at random in each cell. These include genes subject to X-inactivation, olfactory receptor (OR) genes, and several classes of immune system genes. The means by which monoallelic expression is established are only beginning to be understood. Using a cytological assay, we show that the two homologous alleles of autosomal random monoallelic loci differ from each other in embryonic stem (ES) cells, before establishment of monoallelic expression. The Polycomb Group gene Eed is required to establish this distinctive behavior. In addition, we found that when Eed mutant ES cells are differentiated, they fail to establish asynchronous replication timing at OR loci. These results suggest a common mechanism for random monoallelic expression on autosomes and the X chromosome, and implicate Eed in establishing differences between homologous OR loci before and after differentiation.
Introduction
Gene expression in diploid cells is generally biallelic: RNA is transcribed from both alleles of a gene in each cell. However, it is becoming clear that a substantial subset of genes is expressed monoallelically, despite having identical DNA sequences. Monoallelically expressed genes fall into two major classes: imprinted genes and random monoallelic genes.
Imprinted genes are expressed exclusively from either the maternally or paternally inherited chromosome. For example, only the maternal allele of the mouse Cdkn1c locus (Hatada and Mukai, 1995) and the paternal allele of the Igf2 gene (DeChiara et al., 1991) are expressed. For imprinted genes, the identity of the expressed allele is predetermined, often by differential DNA methylation established in the male and female gametes (Razin and Cedar, 1994).
Random monoallelic genes, on the other hand, can be expressed from either the maternal or paternal chromosome. A dramatic example of random monoallelic expression is mammalian X inactivation, in which one of the two X chromosomes in a female cell is transcriptionally silenced (Lyon, 1961). The choice of which X chromosome to silence is made early in embryonic development. Subsequently, the inactive X is clonally inherited, resulting in adult females with mosaic expression of X-linked genes from the maternally and paternally inherited X chromosomes. In addition to X-linked genes, an increasing number of random monoallelic genes are being identified on autosomes. These genes include olfactory receptors (ORs) (Chess et al., 1994), several immune-system genes including natural killer cell receptors and interleukins (Pernis et al., 1965; Cebra et al., 1966; Held et al., 1995; Hollander et al., 1998), and the cell adhesion molecule p120 catenin (Gimelbrant et al., 2005). OR gene choice has an added layer of complexity in that only one allele of one of the ∼1,000 olfactory loci located in tandem arrays across the genome is expressed in each olfactory neuron (Serizawa et al., 2004).
The mechanism by which one and only one allele of a gene is chosen at random to be expressed remains mysterious. We have recently shown that the homologous X chromosomes in female mouse embryonic stem (ES) cells adopt different, mutually exclusive states even before X inactivation is initiated (Mlynarczyk-Evans et al., 2006). Furthermore, these two states correlate with the fate of the chromosome upon differentiation of ES cells carrying mutations that predetermine the fates of the active and inactive X chromosomes. In wild-type ES cells, the maternal and paternal X chromosomes can switch back and forth between these states, but upon X inactivation, the states appear to be fixed as the active and inactive X chromosomes. The two states are detected as a tendency for replicated loci on one chromosome to appear as single pinpoints by FISH in paraformaldehyde (PFA)-fixed cells, while loci on the homologous chromosome tend to appear as doublet signals. We refer to this phenomenon as singlet/doublet signals independent of asynchronous DNA replication, or SIAR.
In this paper, we find that SIAR is a general characteristic of random monoallelic genes in ES cells. Establishment of SIAR is dependent on the Polycomb Group protein Eed, an essential component of the histone H3 lysine 27 methyltransferase complex. Furthermore, Eed is also required for asynchronous replication of random monoallelic genes in differentiated cells. Together, these results suggest that a common mechanism, involving chromatin modifications, underlies both X inactivation and autosomal random monoallelic expression.
Results
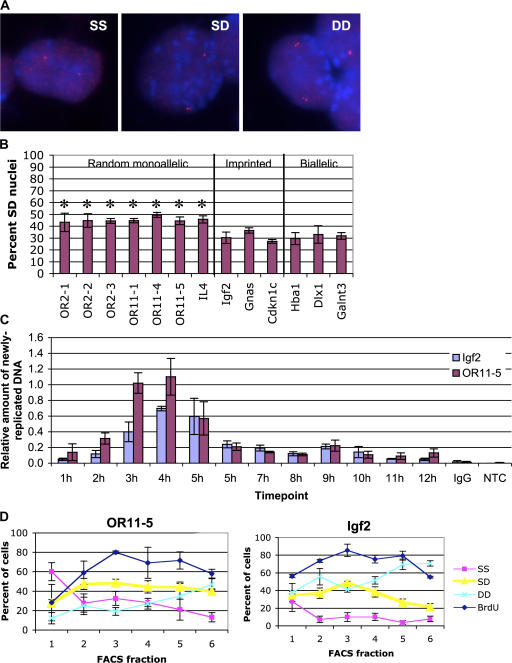
We wished to examine whether SIAR was peculiar to the X chromosome, or whether it was a more general characteristic of monoallelically expressed genes. To determine whether future monoallelic genes on autosomes behave similarly to X-linked genes, FISH was performed on PFA-fixed mouse ES cells using probes to random monoallelic genes, imprinted genes, and biallelically expressed controls (Table I). Genes destined to be randomly monoallelically expressed in differentiated cells displayed a singlet FISH signal on one allele and a doublet signal on the other allele in a high percentage of S-phase ES cells (Fig. 1, A and B). Imprinted and biallelically expressed genes displayed a lower frequency of singlet/doublet (SD) cells than the future random monoallelic genes (Fig. 1 B).
Table I.
BACs used as FISH probes
| BAC | Probe name | Location |
|---|---|---|
| RP23-52P17 | OR2-1 | 36.4 Mb, Chr. 2 |
| RP24-342H16 | Galnt3 | 65.9 Mb, Chr. 2 |
| RP23-419H3 | Dlx1 | 71.3 Mb, Chr. 2 |
| RP23-318N4 | OR2-2 | 88.8 Mb, Chr. 2 |
| RP23-63C10 | OR2-3 | 112 Mb, Chr. 2 |
| RP23-147E9 | Gnas | 174 Mb, Chr. 2 |
| RP23-17N3 | Igf2 | 137 Mb, Chr. 7 |
| RP23-124B2 | Cdkn1c | 138 Mb, Chr. 7 |
| RP23-71G18 | Hba1 | 31.9 Mb, Chr. 11 |
| RP24-212H23 | OR11-1 | 49.3 Mb, Chr. 11 |
| RP23-226J16 | IL-4 | 53 Mb, Chr. 11 |
| RP24-260A8 | OR11-4 | 73.3 Mb, Chr. 11 |
| RP24-317D24 | OR11-5 | 87.7 Mb, Chr. 11 |
BACs located within odorant receptor arrays are referred to by the name of the array (Zhang and Firestein, 2002), with the addition of OR11-5, which is located on chromosome 11 distal to OR11-4.
Figure 1.
Autosomal random monoallelic genes display SIAR. (A) Representative FISH images of cells displaying SS, SD, or DD signals. DNA-FISH was performed on wild-type ES cells using the OR probe OR11-5 (Table I) labeled with Cy3 (red). DNA was stained with DAPI (blue). OR probes tended to show a relatively high nonspecific background signal by FISH, probably due to cross-hybridization with other OR genes. (B) Quantitation of the percentage of nuclei displaying SD FISH signals for the random monoallelic OR and IL-4 loci; the imprinted genes Igf2, Gnas, and Cdkn1c; and the biallellically expressed genes Hba1, Dlx1, and Galnt3. Asterisks indicate samples with a significantly greater % SD than Hba1 using an unpaired t test (P ≤ 0.001). Error bars represent one standard deviation in each direction. See Fig. S1 for complete scoring of SS, SD, and DD signals (available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1). (C) Replication timing assay. Cells were arrested in G1 with mimosine, and released. At 1-h intervals, cells were BrdU labeled, DNA was isolated, and BrdU-containing DNA was immunoprecipitated. Sequences from OR11-5 (specifically, the OR gene Olfr464), Igf2, and a loading control, consisting of BrdU-labeled human DNA added before immunoprecipitation, were analyzed by quantitative PCR amplification. Water (no template control, NTC) and DNA immunoprecipitated with nonspecific serum (IgG) were amplified as negative controls. PCR primer sequences are given in Table II. (D) Percentage of nuclei with SD FISH signals in FACS fractions. DNA was stained with Hoeschst, and cells were sorted (Fig. S1) by DNA content onto slides. FISH was performed on PFA-fixed cells using probes to the OR array OR11-5 and the imprinted gene Igf2. Percent BrdU positive (blue diamonds), singlet/singlet (SS; pink squares), singlet/doublet (SD; yellow triangles), and doublet/doublet (DD; aqua crosses) cells are shown. Both OR11-5 and Igf2 showed the expected decrease in the percentage of SS cells in the early fractions and the expected increase in DD signals in later fractions. Error bars represent one standard deviation in each direction. For Igf2, the fraction of SD cells in fraction 3 was significantly higher than in fractions 1, 5, and 6 (P ≤ 0.001) and somewhat higher than in fractions 2 and 4 (P < 0.05) based on a t test. For OR11-5, the frequency of SD cells was significantly lower in fraction 1 (P < 0.005) than in the remainder of the fractions.
The high frequency of SD FISH signals for X-linked genes in female ES cells (Mlynarczyk-Evans et al., 2006) does not reflect asynchronous replication of these loci. Imprinted genes, in contrast, have been shown to replicate asynchronously in ES cells (Gribnau et al., 2003). We asked whether the elevated frequency of SD FISH signals for autosomal random monoallelic loci could be attributed to asynchronous replication. First, we assayed the replication timing of Olfr464, an OR in the OR11-5 array, and the imprinted gene Igf2. ES cells were released from a late G1 block, DNA was isolated at 12 one-hour intervals, newly replicated BrdU-labeled DNA was immunoprecipitated, and Olfr464 or Igf2 sequences were detected by qPCR. Replication of both Olfr464 and Igf2 occurred around the 3–5-h window after release (Fig. 1 C).
We then assayed the cell cycle window in which cells with SD FISH signals occurred to determine whether the cells with SD signals peak at a single point. Such a result would be consistent both with our replication timing data and with the slight replication asynchrony that has been reported for these genes (Singh et al., 2003; Gimelbrant and Chess, 2006). ES cells were FACS sorted by DNA content and fixed with PFA, and the proportion of cells with SD FISH signals was determined in each of the six fractions (Fig. 1 D). For Igf2, the percentage of cells with SD signals peaked in the third fraction and then decreased, consistent with the documented asynchronous replication of this locus (Gribnau et al., 2003). For the OR array OR11-5, the percentage of cells with SD signals increased between the first two fractions and remained at a relatively constant level throughout the remaining fractions.
In combination, the analyses of replication timing and SD FISH signals across the cell cycle indicate that one of the two OR11-5 alleles exhibits a singlet FISH signal in a significant fraction of ES cells, even when both loci have replicated. Thus, a substantial fraction of the observed SD FISH signals for OR11-5 cannot be attributed to asynchronous replication.
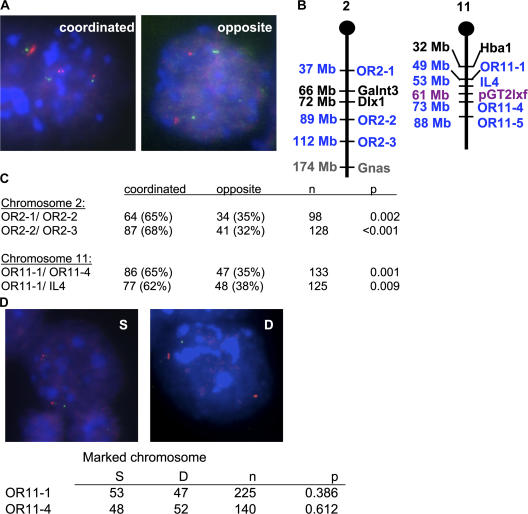
In differentiated cells, random monoallelic genes do display asynchronous replication (Chess et al., 1994; Mostoslavsky et al., 2001). When multiple random monoallelic genes occur on the same chromosome, they replicate early on one homologue and late on the other, despite the fact that intervening biallelically expressed genes do not exhibit asynchronous replication (Singh et al., 2003). We therefore tested whether loci on the same chromosome are coordinated in their behavior even before differentiation. To do this, we performed FISH in ES cells with pairs of probes to random monoallelic genes on the same chromosome (Fig. 2 A). Two pairs of OR arrays on chromosome 2, a pair of OR arrays on chromosome 11, and an OR array and interleukin-4 on chromosome 11 were tested pairwise (Fig. 2 B). All four pairs displayed singlet signals on one chromosome and doublet signals on the other in ∼65% of cells in which each locus exhibited one singlet and one doublet allele (Fig. 2 C). This is significantly different from the 50% of cells that are predicted to exhibit this pattern if behavior of loci on the same chromosome is not coordinated.
Figure 2.
SIAR of autosomal genes is coordinated and switchable. (A) Appearance of random monoallelic loci as SD FISH signals is coordinated along the length of a chromosome. Representative FISH images of cells displaying coordinated or opposite SD signals for pairs of OR probes on the same chromosome. DNA-FISH was performed on wild-type ES cells using the OR probes OR11-1 (left) or OR2-3 (right) labeled with Cy3 (red), and OR11-4 (left) or OR2-2 (right) labeled with biotin and detected with FITC-avidin (green). DNA was stained with DAPI (blue). (B) Locations of loci examined on chromosomes 2 and 11. OR arrays are named according to the system of Zhang and Firestein (2002). Random monoallelic genes are labeled in blue, biallelic genes in black, imprinted genes in gray, and the chromosome 11 transgene used as a marker in violet. (C) Quantitation of the percentage of cells displaying coordinated or opposite SD signals for four pairs of probes. A χ-square test was used to determine P values for coordination, with a null hypothesis of no coordination. (D) The identities of the singlet and doublet alleles are not fixed in ES cells. Representative FISH images of cells from a clonal population displaying a singlet or doublet signal on a marked chromosome. Combined RNA/DNA-FISH was performed on ES cells carrying a transgene on one copy of chromosome 11 (BayGenomics line RRR379) using the OR probes OR11-1 labeled with Cy3 (red), and plasmid pGT2lxf labeled with FITC (green). Because OR genes are not expressed in ES cells, these probes detected DNA only; the pGT2lxf probe detected both RNA and DNA. The frequency of SD FISH signals for each probe by itself in RRR379 ES cells was comparable to that in wild-type male (E14) or female (ES2-1) ES cells (not depicted). DNA was stained with DAPI (blue). Quantitation of the percentage of cells displaying a singlet or doublet signal for the OR11-1 or OR11-4 probe on the pGT2lxf-containing chromosome are given in the table below. A χ-square test was used to determine P values, with a null hypothesis of a locus on the marked chromosome being equally likely to appear as a singlet or a doublet.
The identity of the homologue that contains the early replicating random monoallelic genes is fixed in clonally derived mouse embryo fibroblasts (MEFs) (Singh et al., 2003), prompting us to examine whether the same chromosome always shows an elevated frequency of singlet FISH signals in clonally derived ES cells. FISH was performed in an allele-specific manner, using an ES cell line containing a transgene on one copy of chromosome 11. In ES cells derived from a single progenitor, the singlet signal for either of the two OR probes on chromosome 11 appeared on the transgene-containing chromosome in approximately half the cells (Fig. 2 D). Thus, the identity of the chromosome exhibiting the singlet FISH signal for autosomal random monoallelic loci switches in ES cells, in contrast to the fixed behavior of these loci in differentiated cells.
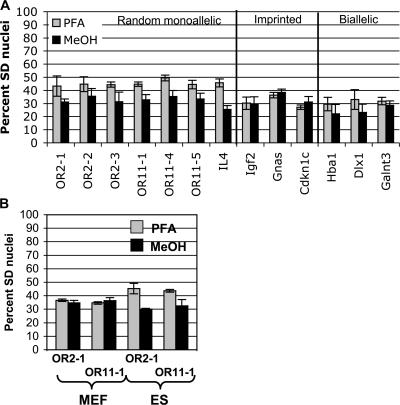
The appearance of an already-replicated allele as a singlet FISH signal suggests that the sister chromatids remain closely apposed such that the individual chromatids cannot be distinguished. To test whether intact chromatin structure is necessary to observe the high percentage of SD cells for autosomal monoallelic genes, we compared two fixation methods. PFA fixation, as used above, preserves nuclear organization, while fixation with methanol/acetic acid (MeOH) removes proteins from DNA and disrupts chromatin organization (Hendzel and Bazett-Jones, 1997). FISH for autosomal random monoallelic genes in MeOH-fixed samples revealed a lower percentage of SD cells than in PFA-fixed samples; imprinted genes, on the other hand, displayed similar frequencies of SD cells in MeOH- and PFA-fixed samples (Fig. 3 A). These results confirm that SIAR requires relatively intact chromatin structure (Mlynarczyk-Evans et al., 2006).
Figure 3.
SIAR is dependent on intact nuclear structure and precedes establishment of monoallelic expression. (A) SD FISH signals for random monoallelic genes, but not imprinted genes, are reduced in methanol-fixed samples. FISH was performed on MeOH-fixed wild-type ES cells using probes for random monoallelic OR genes and IL-4; the imprinted genes Igf2, Gnas, and Cdkn1c; and the biallellically expressed genes Dlx1, Hba1, and Galnt3 (data for PFA-fixed cells is provided for comparison, and is the same as in Fig. 1 B). The frequency of SD FISH signals in PFA-fixed (gray) and MeOH-fixed (black) samples is shown. Error bars represent one standard deviation in each direction. The observed frequency of SD FISH signals for Hba1 and Igf2 in MeOH-fixed ES cells was consistent with previously published work (Simon et al., 1999; Gribnau et al., 2003). See Fig. S2 for complete scoring of SS, SD, and DD signals in MeOH-fixed cells and a statistical analysis of the difference between PFA- and MeOH-fixed cells (available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1). (B) Differentiated cells do not display SIAR. DNA-FISH was performed on PFA- (gray) and MeOH-fixed (black) wild-type MEFs using OR array probes OR11-1 and OR11-4. Data for ES cells (Fig. 1 B and Fig. 2 B) is shown for comparison. Error bars represent one standard deviation in each direction. The observed frequency of SD FISH signals for OR arrays in MeOH-fixed MEFs was consistent with previously published work (Chess et al., 1994; Simon et al., 1999). See Fig. S3 for complete scoring of SS, SD, and DD signals in PFA- and MeOH-fixed MEFs (available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1).
SIAR at X-linked loci is restricted to undifferentiated ES cells (Mlynarczyk-Evans et al., 2006). To determine whether SIAR at autosomal loci is also limited to undifferentiated cells, before the establishment of monoallelic expression, we performed FISH for two OR arrays, OR2-1 and OR11-1, in MEFs. Both probes displayed a lower frequency of SD cells in MEFs compared with ES cells, and there was no significant difference in the percentage of SD cells in PFA- and MeOH-fixed MEFs (Fig. 3 B). The SD cells observed in MEFs presumably reflect the asynchronous replication of OR arrays in differentiated cells (Chess et al., 1994).
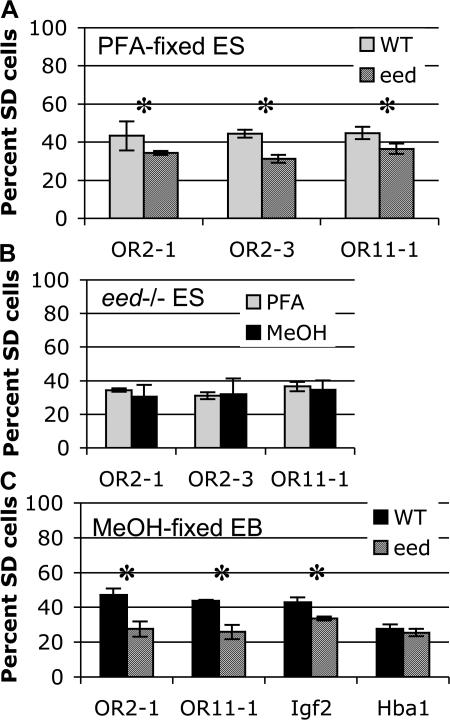
The chromatin difference that underlies the difference in appearance of future random monoallelic loci by FISH remains unknown. The loss of SIAR in MeOH-fixed cells implies that intact chromatin structure is required; for this reason, possible candidates for the cause of SIAR include proteins that play a role in chromatin modification. We performed FISH on ES cells mutant for either Eed (Montgomery et al., 2005), which is required for histone H3 methylation on lysine 27, or the maintenance DNA methyltransferase Dnmt1 (Gribnau et al., 2003) to see if they are required for SIAR. ES cells mutant for Dnmt1 did not display a significant difference in the frequency of SD cells for OR2-1 or OR11-1 probes compared with wild-type cells (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1). Mutation of Eed, in contrast, did result in a significant reduction in the frequency of SD signals for OR arrays (Fig. 4 A). In addition, the frequency of SD signals was the same for PFA-fixed and MeOH-fixed Eed mutant ES cells (Fig. 4 B), further suggesting that these cells no longer display SIAR, because disruption of nuclear organization no longer affects SD signal frequency. Together these data indicate that Eed is necessary for SIAR at OR2-1 and OR11-1.
Figure 4.
Eed is required for SIAR. (A) Mutation of Eed reduces SIAR in ES cells. The frequency of SD FISH signals for three OR arrays in PFA-fixed wild-type (gray) and Eed knockout (striped) ES cells is shown. Asterisks indicate a significant difference between wild-type and Eed as determined by an unpaired t test (P < 0.005). Error bars represent one standard deviation in each direction. See Fig. S4 for complete scoring of SS, SD, and DD signals (available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1). (B) PFA-fixed and MeOH-fixed Eed −/− ES cells display a similar frequency of SD signals. The frequency of SD FISH signals for three OR arrays in PFA-fixed (gray) and MeOH-fixed (black) Eed knockout ES cells is shown. Error bars represent one standard deviation in each direction. See Fig. S4 for complete scoring of SS, SD, and DD signals. (C) Asynchronous replication of random monoallelic loci is lost in differentiated Eed −/− cells. The frequency of SD FISH signals for two OR arrays, Igf2, and Hba1 in MeOH-fixed wild-type (black) and Eed knockout (striped) differentiated cells is shown. EBs were differentiated for 17 d, and differentiation was tested using an alkaline phosphatase assay. 97.5% of wild-type cells and 95% of Eed −/− cells were negative for phosphatase activity. Asterisks indicate a significant difference between wild-type and Eed as determined by an unpaired t test (P < 0.001). Error bars represent one standard deviation in each direction. See Fig. S4 for complete scoring of SS, SD, and DD signals.
We examined whether Eed was also necessary to establish asynchronous replication timing of OR genes in differentiated cells. Wild-type and Eed −/− ES cells were differentiated as embryoid bodies for 17 d. FISH for OR2-1, OR11-1, Igf2, and Hba1 was performed on MeOH-fixed differentiated cells. Differentiated wild-type cells showed a high frequency of SD FISH signals at monoallelically expressed loci (Fig. 4 C), consistent with previously published results in MEFs (Singh et al., 2003). In differentiated Eed −/− cells, the frequency of SD signals for the OR arrays was reduced to the level seen for the biallelically expressed Hba1. This suggests that asynchronous replication of ORs was lost in differentiated Eed −/− cells (Fig. 4 C). In combination, our results show that Eed plays dual roles in regulation of random monoallelic autosomal genes: it is necessary for SIAR before differentiation and asynchronous replication timing of these loci after differentiation.
Discussion
The similar behavior of autosomal and X-linked genes before random monoallelic expression lends support to the idea that the mechanism of random choice is at least partially conserved between autosomes and X chromosomes (Singh et al., 2003). On both the X chromosome (Mlynarczyk-Evans et al., 2006) and autosomes, alleles of future random monoallelic genes differ from each other in a switchable fashion that is dependent on intact nuclear structure and coordinated among loci on the same chromosome. Our current model (Mlynarczyk-Evans et al., 2006) is that the observed differences in frequency of singlet and doublet FISH signals reflect an underlying difference in chromatin structure between the chromosomes that affects the likelihood that the replicated loci on each chromosome will separate enough to appear as a doublet signal by FISH.
As there are no known cell lines in which OR choice is predetermined, it is not possible to correlate autosomal SIAR with future expression states, as was done with X-linked loci (Mlynarczyk-Evans et al., 2006). However, autosomal random monoallelic loci display asynchronous replication in differentiated cells, even in cell types in which they are not expressed, and this asynchronous replication has been proposed to underlie the choice of which allele will be expressed in specific differentiated cell types (Mostoslavsky et al., 2001). Eed mutant ES cells that lack SIAR also lose asynchronous replication after differentiation (Fig. 4 C), suggesting that the chromatin difference underlying SIAR of autosomal random monoallelic loci may be required for the later asynchronous replication of the loci.
Although it remains possible that Eed affects SIAR in ES cells and asynchronous replication in differentiated cells through two independent pathways, our results are consistent with the hypothesis that SIAR is a precursor to both random monoallelic expression and asynchronous replication. Eed is not required at the time of random X chromosome inactivation in order for that process to occur normally, based on studies of Eed −/− embryos (Kalantry and Magnuson, 2006). However, maternal stores of Eed present earlier in embryogenesis may have already established the underlying chromatin differences that are visualized as SIAR. Alternatively, it is conceivable that SIAR of X-linked and autosomal loci is mediated by different genes, or that SIAR is important for asynchronous replication, but not monoallelic expression. Because no female Eed −/− ES cell lines currently exist, it remains to be seen whether SIAR of X-linked loci is affected by loss of Eed.
The behavior of the Eed −/− ES and differentiated cells suggests that Eed-mediated histone methylation is involved in asynchronous replication in differentiated cells as well as in the differences between homologous alleles of future random monoallelic genes in ES cells. Although it is well-established that histone methylation, expression status, and replication timing are closely correlated, it has been less clear whether particular histone modifications are a cause or effect of replication timing (Wu et al., 2006). The histone methyltransferase Suv39h1, which methylates histone H3 lysine 9, has recently been shown to affect the timing of replication of pericentric heterochromatin in the mouse (Wu et al., 2006). Together with our results, this suggests that a combination of histone modifications may play a causal role in establishing replication timing of particular loci.
Materials and methods
Cell lines and culture
Mouse cell lines used in this study included: ES2-1, wild-type female ES cells (Marahrens et al., 1997); E14, wild-type male ES cells (Hooper et al., 1987); RRR379, male ES cells carrying an insertion of pGT0Lxf at the epsin2 locus on chromosome 11 (BayGenomics); Eed −/− male ES cells (Montgomery et al., 2005), Dnmt −/− male ES cells (Gribnau et al., 2003), and wild-type female MEFs.
MEFs and EBs were cultured in Diff medium: Knockout DME (Invitrogen) with 10% fetal bovine serum (FBS), 1× nonessential amino acids (UCSF Cell Culture facility), 1× l-glutamine, 1× penicillin/streptomycin (UCSF Cell Culture facility), and b-mercaptoethanol. ES cells were cultured in ES medium: same as Diff medium, plus 1,000 U/ml leukemia inhibitory factor (LIF), following standard protocols. Differentiation of EBs was tested using the ELF Phosphatase Detection kit (American Type Culture Collection).
FACS
Wild-type female ES cells were labeled with BrdU (GE Healthcare) and stained with 40 ug/ml Hoechst 33342 (Molecular Probes) for 45 min before harvesting for flow cytometry. The cells were resuspended in ES medium containing 40 ug/ml Hoechst 33342, 7% Cell Dissociation Buffer (Invitrogen), and 10 mM EDTA, and sorted using a FACSDiVa Cell Sorter (Becton Dickinson). DNA content was measured based on the intensity of Hoechst emission using a HQ445/50 bandpass filter. Cells were sorted into six fractions containing similar numbers of cells onto multiwell slides pretreated with 1 mg/ml poly-l-lysine and allowed to settle and adhere.
FISH
All cells were labeled with BrdU for 30 min before fixation. Cells to be PFA fixed were cytospun onto slides, washed 30 s with ice-cold CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 10 mM Pipes, pH 6.8), 30 s with cold CSK + 0.5% Triton X-100, and 30 s with cold CSK, then fixed for 10 min at room temperature in 4% PFA, 1× PBS (Marahrens et al., 1998). For MeOH fixation, trypinized cells were treated with 0.075 M KCl for 10 min on ice, washed four times with a 3:1 MeOH/acetic acid solution, and then dropped onto slides (Gribnau et al., 2003).
FISH for genomic DNA was performed essentially as previously described (Gribnau et al., 2003). PFA-fixed samples were pretreated with 0.01% pepsin in 0.01 M HCl for 4 min at 37°C, fixed for 5 min in 4% PFA/1× PBS at room temperature, and treated for 30 min in 0.1 mg/ml RNaseA at 37°C. After dehydration through an ethanol series, samples were denatured for 3–8 min on an 80°C heat block. For combined RNA/DNA-FISH, the RNaseA treatment was omitted. MeOH-fixed slides were not pretreated, and were denatured for 30 s on the heat block. BACs (Table I) were directly labeled with Cy3-dCTP by random priming for use as probes. The frequency of nuclei displaying SS, SD, and DD signals for single probes were scored in S-phase (BrdU positive) cells. To ensure that singlet and doublet signals were being scored consistently, three of the authors independently scored several blinded slides for SIAR, obtaining comparable results. Linked sequences can be reliably scored as being on the same chromosome over distances of up to 50 Mb (Ensminger and Chess, 2004); all pairwise DNA-FISH experiments in this study were performed within this distance range.
Microscopy
All microscopy was performed at room temperature, on slides mounted with Vectashield (Vector Laboratories). FISH results were analyzed using a fluorescent microscope (BX60; Olympus) with a 100× oil immersion objective, NA 1.30. Images were captured with a digital camera (ORCA-ER; Hamamatsu) and Openlab 4.0.1 software. Grayscale images were combined into an RGB image using Photoshop, with Cy3, FITC, and DAPI images pasted into the red, green, and blue channels, respectively. The Photoshop Levels tool was used to adjust the upper and lower input levels for each channel to match the upper and lower boundaries of the image histogram. No gamma adjustments were made.
Replication timing assay
To assay replication timing, cells were arrested in late G1 with mimosine, then released into the cell cycle. At 12 one-hour time points after release, cells were labeled with BrdU and DNA was isolated. 0.5 ug of BrdU- labeled human DNA was mixed with 10 ug of DNA from each time point as a control for immunoprecipitation efficiency. BrdU-labeled DNA was immunoprecipitated with either a monoclonal α-BrdU antibody (Becton Dickinson) or mouse IgG and Protein G–Sepharose 4 Fast Flow beads (GE Healthcare) and resuspended in 1 mM Tris, 0.1 mM EDTA, pH 8.0, in a final volume of 1 ml.
Immunoprecipitated DNA from each time point and a dilution series of genomic DNA were used as templates for qPCR amplification of sequences from OR11-5, Igf2, and the human DNA control. PCR primer sequences are given in Table II. The results were quantitated using the relative standard curve method as described in ABI User Bulletin #2, normalizing to the human DNA control.
Table II.
PCR primer sequences
| Primer name | Primer sequence |
|---|---|
| MOR240-qf (Olfr464) | CACCATCATGAACACCAAGC |
| MOR240-qr (Olfr464) | GAGAACCAGCATCCCACTGT |
| Igf2-qf | GAGTTCAGAGAGGCCAAACG |
| Igf2-qr | TAGTGTGGGACGTGATGGAA |
| hORF-qf (loading control) | TGTGTGCTCCTGGATCTCTG |
| hORF-qr (loading control) | AACACCTGGTTCCACCTCTG |
Statistics
An unpaired t test was used to examine which genes displayed a significantly greater percentage of SD signals than the biallelic control Hba1, with P ≤ 0.001. A t test was also used to compare the frequency of SD FISH signals in PFA- and MeOH-fixed cells (Fig. S2 B). All P values to determine statistical significance of coordination or switching were calculated using a χ–square test, with a null hypothesis of a 50:50 (random) distribution.
Online supplemental material
Figure S1 shows full scoring of FISH signal appearance in PFA-fixed ES cells and ES cell FACs profile. Figure S2 shows FISH signal appearance in MeOH-fixed ES cells. Figure S3 shows full scoring of FISH signal appearance in MEFs. Figure S4 shows FISH signal appearance in Dnmt1 −/− and Eed −/− mutant cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1.
Supplementary Material
Acknowledgments
We thank T. Fazzio and S. Lomvardas for critical reading of the manuscript and M. Bigos for assistance with FACS.
MKA was supported by a National Institutes of Health (NIH) National Research Service Award. S. Mlynarczyk-Evans was supported by a National Science Foundation Graduate Research Fellowship and a Julius R. and Patricia A. Krevans Fellowship. B. Panning is a Pew Scholar. This work was supported by grants from the NIH and the Sandler Foundation.
S. Mlynarczyk-Evans' present address is Department of Developmental Biology, Stanford University, Stanford, CA 94305.
Abbreviations used in this paper: ES cell, embryonic stem cell; MEF, mouse embryonic fibroblast; MeOH, methanol/acetic acid; OR, olfactory receptor; PFA, paraformaldehyde; SD, singlet/doublet; SIAR, singlet-doublet independent of asynchronous replication.
References
- Cebra, J.J., J.E. Colberg, and S. Dray. 1966. Rabbit lymphoid cells differentiated with respect to alpha-, gamma-, and mu-heavy polypeptide chains and to allotypic markers Aa1 and Aa2. J. Exp. Med. 123:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess, A., I. Simon, H. Cedar, and R. Axel. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. [DOI] [PubMed] [Google Scholar]
- DeChiara, T.M., E.J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 64:849–859. [DOI] [PubMed] [Google Scholar]
- Ensminger, A.W., and A. Chess. 2004. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum. Mol. Genet. 13:651–658. [DOI] [PubMed] [Google Scholar]
- Gimelbrant, A.A., and A. Chess. 2006. An epigenetic state associated with areas of gene duplication. Genome Res. 16:723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant, A.A., A.W. Ensminger, P. Qi, J. Zucker, and A. Chess. 2005. Monoallelic expression and asynchronous replication of p120 catenin in mouse and human cells. J. Biol. Chem. 280:1354–1359. [DOI] [PubMed] [Google Scholar]
- Gribnau, J., K. Hochedlinger, K. Hata, E. Li, and R. Jaenisch. 2003. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 17:759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada, I., and T. Mukai. 1995. Genomic imprinting of p57KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat. Genet. 11:204–206. [DOI] [PubMed] [Google Scholar]
- Held, W., J. Roland, and D.H. Raulet. 1995. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 376:355–358. [DOI] [PubMed] [Google Scholar]
- Hendzel, M.J., and D.P. Bazett-Jones. 1997. Fixation-dependent organization of core histones following DNA fluorescent in situ hybridization. Chromosoma. 106:114–123. [DOI] [PubMed] [Google Scholar]
- Hollander, G.A., S. Zuklys, C. Morel, E. Mizoguchi, K. Mobisson, S. Simpson, C. Terhorst, W. Wishart, D.E. Golan, A.K. Bhan, and S.J. Burakoff. 1998. Monoallelic expression of the interleukin-2 locus. Science. 279:2118–2121. [DOI] [PubMed] [Google Scholar]
- Hooper, M., K. Hardy, A. Handyside, S. Hunter, and M. Monk. 1987. HPRT- deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 326:292–295. [DOI] [PubMed] [Google Scholar]
- Kalantry, S., and T. Magnuson. 2006. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon, M.F. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 190:372–373. [DOI] [PubMed] [Google Scholar]
- Marahrens, Y., B. Panning, J. Dausman, W. Strauss, and R. Jaenisch. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11:156–166. [DOI] [PubMed] [Google Scholar]
- Marahrens, Y., J. Loring, and R. Jaenisch. 1998. Role of the Xist gene in X chromosome choosing. Cell. 92:657–664. [DOI] [PubMed] [Google Scholar]
- Mlynarczyk-Evans, S., M. Royce-Tolland, M.K. Alexander, A.A. Andersen, S. Kalantry, J. Gribnau, and B. Panning. 2006. X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 4:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, N.D., D. Yee, A. Chen, S. Kalantry, S.J. Chamberlain, A.P. Otte, and T. Magnuson. 2005. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 15:942–947. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky, R., N. Singh, T. Tenzen, M. Goldmit, C. Gabay, S. Elizur, P. Qi, B.E. Reubinoff, A. Chess, H. Cedar, and Y. Bergman. 2001. Asynchronous replication and allelic exclusion in the immune system. Nature. 414:221–225. [DOI] [PubMed] [Google Scholar]
- Pernis, B., G. Chiappino, A.S. Kelus, and P.G. Gell. 1965. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med. 122:853–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin, A., and H. Cedar. 1994. DNA methylation and genomic imprinting. Cell. 77:473–476. [DOI] [PubMed] [Google Scholar]
- Serizawa, S., K. Miyamichi, and H. Sakano. 2004. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 20:648–653. [DOI] [PubMed] [Google Scholar]
- Simon, I., T. Tenzen, B.E. Reubinoff, D. Hillman, J.R. McCarrey, and H. Cedar. 1999. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 401:929–932. [DOI] [PubMed] [Google Scholar]
- Singh, N., F.A. Ebrahimi, A.A. Gimelbrant, A.W. Ensminger, M.R. Tackett, P. Qi, J. Gribnau, and A. Chess. 2003. Coordination of the random asynchronous replication of autosomal loci. Nat. Genet. 33:339–341. [DOI] [PubMed] [Google Scholar]
- Wu, R., P.B. Singh, and D.M. Gilbert. 2006. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 174:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., and S. Firestein. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5:124–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.