Abstract
We have used oligonucleotides containing appropriately placed fluorophores and quenchers to measure the stability of 15mer intermolecular triplexes with third strands consisting of repeats of TTT, TTC, TCC and TCTC. In the presence of 200 mM sodium (pH 5.0) triplexes that contain only T·AT triplets are unstable and melt below 30°C. In contrast, triplets with repeats of TTC, TCC and CTCT melt at 67, 72 and 76°C, respectively. The most stable complex is generated by the sequence containing alternating C+·GC and T·AT triplets. All four triplexes are stabilised by increasing the ionic strength or by the addition of magnesium, although triplexes with a higher proportion of C+·GC triplets are much less sensitive to changes in the ionic conditions. The enthalpies of formation of these triplexes were estimated by examining the concentration dependence of the melting profiles and show that, in the presence of 200 mM sodium at pH 5.0, each C+·GC triplet contributes about 30 kJ mol–1, while each T·AT contributes only 11 kJ mol–1. Kinetic experiments with these oligonucleotides show that in 200 mM sodium (pH 5.0) repeats of TCC and TTC have half-lives of ∼20 min, while the triplex with alternating C+·GC and T·AT triplets has a half-life of ∼3 days. In contrast, the dissociation kinetics of the triplex containing only T·AT are too fast to measure.
INTRODUCTION
The formation of intermolecular triplexes offers a means for targeting unique DNA sequences and has potential for use in antigene therapy (1–4). DNA triple helices are formed by the binding of synthetic oligonucleotides within the major groove of duplex DNA, where they make specific hydrogen bond interactions with groups on the exposed faces of the purine bases (5–8). Two main structural motifs have been defined in which the third strand runs parallel or antiparallel to the duplex purine strand. Antiparallel triplexes contain G·GC, A·AT and T·AT triplets (9) while parallel triplexes contain C+·GC and T·AT triplets (10).
Although triple helices form with considerable sequence specificity, they are generally much less stable than their duplex counterparts. This is mainly because they require the assembly of three polyanionic strands and their formation is critically dependent on the ionic strength (11–14). High concentrations of monovalent ions or lower concentrations of divalent ions, especially magnesium, promote parallel triplex formation (9,12,15,16). Although monovalent cations stabilise triplexes, they can compete with magnesium and thereby reduce its stabilising effect (12,17). This inhibition can be reversed by raising the magnesium concentration (11,17). For solutions that contain more than one type of cation the effect of each ion will depend on its valency (11). It has been suggested that the cation requirement of triplex formation depends on the T·AT and C+·GC content and that the effect of ionic strength decreases with increasing C+·GC content (18). Triplexes with third strands rich in thymines tend to be more sensitive to salt concentrations than those rich in cytosines (19,20). In supercoiled plasmids, which form intramolecular triplexes (H-DNA), magnesium promotes a switch between the conformation containing C+·GC triplets to the one containing G·GC triplets (21).
Parallel triplexes require conditions of low pH, which are necessary for protonation of the third strand cytosines. Runs of contiguous C+·GC triplets are destabilising as they further decrease the pK value of cytosine N3 (22). However, several studies have shown that, provided the pH is low enough to ensure formation of the C+·GC triplet, this triplet imparts a greater stability than T·AT (23–27). This effect may be due to the positive charge on C+, which partly overcomes the charge repulsion and the favourable disposition of this positive charge within the π-stack. There have been few studies on the effect of sequence on triplex stability, but it has been suggested that the most stable triplexes contain alternating C+·GC and T·AT triplets (27).
Kinetic studies on DNA triplexes have shown that these form very slowly, with association rate constants of ∼103 M–1 s–1, about three orders of magnitude slower than DNA duplexes (12,17,28–35). Some reports have suggested that the rate of triplex association decreases with temperature, probably because it occurs via a nucleation–zipper mechanism and lower temperatures stabilise the transient intermediates, which contain a few productive triplets (12). The dissociation rates of DNA triplexes are also slow. Values obtained by different techniques with a variety of different sequences have suggested half-lives of between 30 min and several days (12,17,26,33,35).
We have recently developed a novel technique for measuring the melting profiles of DNA triplexes (36). This technique uses synthetic oligonucleotides which contain appropriately positioned fluorophores and quenchers. These are in close proximity in the assembled triplex and the fluorescence is quenched. When the triplex melts the fluorophore and quencher are separated and there is a large increase in fluorescence. These experiments are performed in a Roche LightCycler, allowing the simultaneous determination of 32 melting profiles, and only require small quantities of oligonucleotide (20 µl of a 0.25 µM solution). In our previous study we demonstrated the feasibility of this approach using short intramolecular triplexes. In this paper we have used this technique to compare the stability of four 15mer intermolecular triplexes under a variety of conditions and have developed this assay to measure the kinetics of triplex dissociation.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
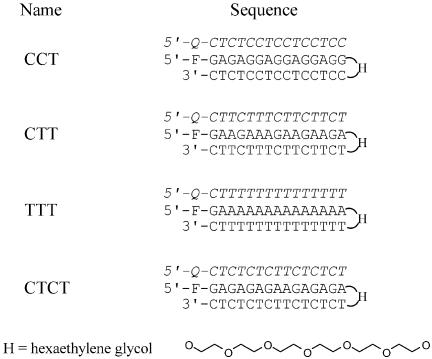
Oligodeoxyribonucleotides were purchased from Oswel Research Products Ltd (Southampton, UK) and were purified by HPLC. These were synthesised on an Applied Biosystems 394 DNA/RNA synthesiser on the 0.2 µmol scale. Fluorescein or methyl red was incorporated at various positions using Fam-cap-dU and MeRed-dR as previously described (36). In order to avoid any potential problems with misannealing we used intramolecular duplexes in which the two strands were connected by a single hexaethylene glycol moiety (H) (Fig. 1). The fluorophore (fluorescein) was incorporated at the 5′-end of the duplex DNA and the quencher (methyl red) was attached at the 5′-end of the third strand oligonucleotide. The intramolecular duplexes and their respective third strand oligonucleotides are shown in Figure 1. This arrangement of fluorophore and quencher allows us to increase the amount of third strand without affecting the total fluorescence signal. The 15mer sequences were chosen so as to generate triplexes with different arrangements of C+·GC and T·AT triplets and are based around repeats of (CCT)n, (CT)n, (CTT)n and Tn. The symmetrical repeating structure of each complex was deliberately broken so as to prevent strand slippage. In each case the fluorophore was located next to the same base (G) so as to minimise any fluorescence emission differences.
Figure 1.
Sequences of the intramolecular duplexes and third strand oligonucleotides used in this work. Fluorescein (F, Fam-cap-dU) was incorporated at the 5′-end of the duplexes and methyl red (Q, MeRed-dR) at the 5′-end of each third strand. H indicates hexaethylene glycol, which joins the two duplex strands. The third strands are shown in italic.
It is theoretically possible for these self-complementary duplexes to dimerise, forming intermolecular complexes instead of the intended intramolecular hairpin structures. Although triplex formation will still occur on these intermolecular duplexes, their formation will affect the quantitative analysis. We therefore performed fluorescence melting studies on these hairpins as a function of strand concentration. There is a small increase in the fluorescence signal when these duplexes melt, which is due to changes in the local environment of the terminal fluorescein and which is much smaller than the changes observed on melting of the triplexes, which involves separation of the fluorophore and quencher moieties. Nonetheless, this was sufficient to enable us to compare the melting profiles of the duplexes at concentrations between 0.25 and 10 µM. We find that there are only small changes in the Tm of each duplex over this 40-fold change in concentration range (2.0°C for CCT, 0.6°C for CTT and 2.1°C for CTCT). We were unable to measure this transition for TTT as, for unknown reasons, melting of this duplex is not accompanied by a fluorescence change. As a result of these small concentration-dependent changes in Tm, together with the single melting transition observed with these duplexes, it appears that the intramolecular hairpin structure is the predominant form in these studies.
Fluorescence melting curves of intermolecular triplexes
Fluorescence melting profiles were determined using a Roche LightCycler as previously described (36). The principle of these experiments is that when a triplex is formed, the fluorophore and quencher are in close proximity and the fluorescence is quenched. On denaturing the complex, the fluorophore and quencher are separated and there is a large increase in fluorescence. Triplexes were prepared in 50 mM sodium acetate pH 5.0, containing 150 mM NaCl. Each sample (20 µl) contained 0.25 µM duplex DNA and 0.25–10 µM triplex-forming oligonucleotide. The complexes were denatured by heating to 95°C at a rate of 0.1°C s–1 and maintained at this temperature for 5 min before cooling to 30°C at 0.1°C s–1. Samples were then held at 30°C for 5 min before melting again by heating to 95°C at 0.1°C s–1. The fluorescence was recorded during both melting and annealing phases. The LightCycler excites the samples at 488 nm and the emission was measured at 520 nm. Since this technique measures the changes in fluorescence that accompany separation of the fluorophore and quencher, the signal will be most strongly affected by opening of the terminal Hoogsteen pairs, rather than dissociation of the entire third strand. However, simple fraying of the termini will still leave the fluorescent groups in close proximity and dissociation of the entire third strand will be a highly cooperative process. With this proviso, the fluorescence profiles provide a good approximation for the dissociation of the third strand. Tm values were determined from the first derivatives of the melting profiles using the Roche LightCycler software and were reproducible to within 0.5°C. Unless otherwise stated, the Tm values quoted refer to the second melting transition. For most complexes there was little or no hysteresis between the melting and annealing curves. However, with the CTCT triplex the Tm determined from the melting profiles was ∼2–3°C higher than that determined from the annealing phase. This hysteresis arises when the complex is not at thermodynamic equilibrium during the temperature changes and is due to slow rates of dissociation and/or association. In these cases the Tm was determined by increasing the temperature in 1°C steps, leaving the samples to equilibrate for 2 min after each temperature rise before recording the fluorescence.
Determination of kinetic parameters by temperature jump experiments
The kinetics of triplex dissociation were determined by measuring the rate of change of fluorescence after rapidly increasing the temperature, in a manner similar to that of temperature jump relaxation kinetics. In order to ensure proper assembly of each triplex the mixtures were first melted by heating to 95°C and reannealed by cooling to 30°C at 0.1°C s–1. The complex was then equilibrated in the LightCycler for 5 min at a temperature 15°C below its Tm. The temperature was then rapidly increased by 10°C at the fastest rate on the LightCycler (20°C s–1) and the time-dependent change in fluorescence was recorded over the next minute. This increase in temperature caused some of the triplex to dissociate, producing an increase in fluorescence. Although the theoretical dead time under these conditions is only 0.5 s, we ignored all fluorescence changes that occurred in the first 2.5 s during equilibration to the new temperature. Successive temperature jumps were then recorded on the same sample. Before performing a new temperature jump on the same sample we ensured that it was still properly annealed by heating it again to 95°C and slowly cooling to 30°C. This was then equilibrated at the new starting temperature for 5 min before performing another temperature jump. In this way each experiment consisted of six cycles with the same sample starting at 15, 13, 11, 9, 7 and 5°C below the Tm value. Appropriate control experiments confirmed that the kinetics were not affected by longer equilibration times or the order in which the various temperature changes were measured. The time-dependent changes in fluorescence were fitted by an exponential function Ft = Ff × (1 – e–kt) + F0, where Ft is fluorescence at time t, Ff is final fluorescence and F0 is the initial fluorescence. The temperature jump experiments each contained 0.25 µM intramolecular duplexes and between 0.5 and 10 µM triplex-forming oligonucleotide. Each experiment was repeated at least three times. The observed relaxation rate constant (k) should be a function of the oligonucleotide concentration and the dissociation (k–1) and association (k1) rate constants, according to the equation k = k–1 + k1[DNA], where [DNA] is the total oligonucleotide concentration (third strand plus duplex). Values of k1 and k–1 at each temperature were determined from the slope of plots of k against [DNA]. Arrhenius plots for the dissociation constants (k–1) showed the expected linear relationship between ln(k–1) and 1/T, from which activation energies were derived and values at 37°C were obtained by extrapolation. Similar plots for k1 gave unusual non-linear Arrhenius plots as explained in Results.
RESULTS
Melting profiles
There have been many examples in which the stability of DNA triple helices has been determined by measuring their thermal stabilities. This is usually achieved by measuring the increase in absorbance (at 260 or 280 nm) as the complex dissociates. These UV melting studies suffer from a number of limitations. Firstly, the melting transition for the third strand may overlap with that of the duplex and it often cannot be properly resolved. Secondly, the absorbance changes are small (typically 25%) and, thirdly, it is not feasible to add an excess of the third strand as this increases the absorbance of the sample. We have previously shown that fluorescently labelled oligonucleotides can be used for determining the thermal stability of intramolecular DNA triplexes in a high throughput assay using the Roche LightCycler. When the complex is folded, the fluorophore and quencher are in close proximity and the fluorescence is quenched. When the complex melts, the fluorophore and quencher are separated and there is a large increase in the fluorescence signal. In the present study we have extended this technique and used it to compare the properties of several intermolecular triplexes. For these studies we have used intramolecular duplexes that contain a fluorescent group at the 5′-end, while the quencher is attached to the 5′-end of the triplex-forming oligonucleotide. Since the third strand is not fluorescent its concentration can be varied over a large range without affecting the fluorescence signal. This can therefore be used to examine some relatively unstable triplexes for which stoichiometric quantities of duplex and third strand do not give complete complex formation. In these experiments we assume that the terminal fluorescent groups do not affect triplex stability. Since the Tm values that we report below are similar to those reported in other studies, these groups only have a small effect on stability. In addition, any effects will be constant for all the triplexes since the fluorophore and quencher are placed in the same position for all the complexes.
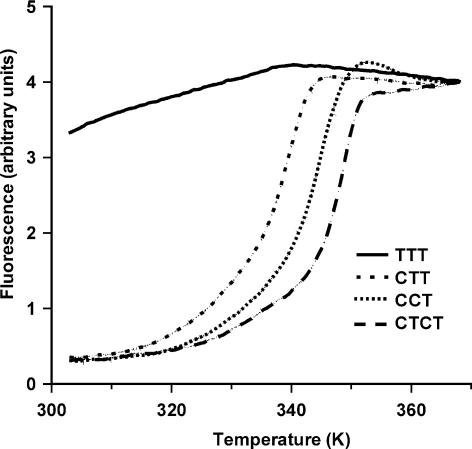
Figure 2 shows representative fluorescence melting profiles for the four triplexes, which contain different arrangements of C+·GC and T·AT triplets. In these experiments the fluorescently labelled duplex concentration was 0.25 µM and the third strand was added in a 16-fold excess (4 µM). It can be seen that, under these conditions (50 mM sodium acetate pH 5.0, containing 150 mM NaCl), triplexes CCT, CTT and CTCT produce clear fluorescent melting curves which are consistent with formation of the expected complexes. These triplexes have Tm values of 71.9, 66.7 and 75.7°C, respectively. There was no significant difference between the melting and annealing curves for CTT and CCT, while for CTCT melting occurred ∼2°C higher than annealing. In contrast it can be seen that the TTT triplex does not produce a melting transition and maintains a high fluorescence throughout. This demonstrates that this triplex, which contains mainly T·AT triplets, is much less stable than the ones that also contain C+·GC triplets. However, the stability of these triplexes is not a simple function of the C+·GC content, but is maximal for the triplex containing alternating C+·GC and T·AT triplets.
Figure 2.
Fluorescence melting curves for the intermolecular triplexes measured in 50 mM sodium acetate pH 5.0, containing 150 mM NaCl. For each curve the concentration of the third strand oligonucleotide (containing the methyl red quencher) was 4 µM and the concentration of the fluorescently labelled intramolecular duplex was 0.25 µM.
Since triplex formation requires the interaction between three polyanions, it is sensitive to the ionic conditions. We therefore determined the effects of NaCl and MgCl2 on these melting profiles. The results with TTT are shown in Figure 3, and the Tm values for all four oligonucleotides are summarised in Table 1. It can be seen that the TTT triplex is very sensitive to the ionic strength. In the absence of magnesium this complex cannot be detected at sodium ion concentrations below 400 mM. Addition of magnesium at concentrations >5 mM also stabilises this complex in the presence of 200 mM sodium. As expected, the other triplexes are also sensitive to the ionic conditions, but to a lesser extent. The addition of 1 M NaCl increases the melting temperatures of CCT, CTT and CTCT by 6.2, 21.0 and 10.6°C, respectively, while addition of 50 mM MgCl2 stabilises these by 2.2, 11.3 and 5.4°C. The sensitivity to ionic strength appears to depend on the proportion of T·AT triplets; sequences with a greater number of C+·GC triplets are less sensitive to changes in the ionic conditions. As a result of the different sensitivities of CCT and CTCT to ionic strength, CCT is the most stable triplex at the lowest ionic strengths. We analysed these salt-dependent effects by plotting 1/Tm against ln[Na+] as described by Plum and Breslauer (19). The slopes of these graphs are presented in Table 2. The number of ions (ΔNa+) released upon dissociation of these triplexes is given by (slope × ΔH)/R, where R is the universal gas constant and ΔH was estimated as described below. These values are included in Table 2. It can be seen that all four triplexes are stabilised by sodium, although this effect is much smaller for those that contain a greater proportion of C+·GC triplets. When these values are plotted against the percentage of C+·GC triplets (67% of CCT, 50% for CTCT, 33% of CTT and 6.7% for TTT) then a simple least squares fit suggests that melting of a 15mer triplex composed entirely of T·AT triplets would be accompanied by the release of 4.8 ± 0.4 Na+; in contrast, a triplex consisting entirely of C+·GC triplets would melt with the uptake of 0.1 Na+. These results suggest that while T·AT triplets are stabilised by addition of sodium, this ion has a small destabilising effect on C+·GC triplets.
Figure 3.
Fluorescence melting curves for intermolecular triplex TTT in the presence of increasing concentrations of NaCl and MgCl2. For the left-hand panel, showing the effects of added NaCl, the samples were prepared in 50 mM sodium acetate pH 5.0, containing 0, 200, 350, 500, 700 or 1000 mM NaCl. For the right-hand panel (showing the effects of added MgCl2), the samples were prepared in 50 mM sodium acetate pH 5.0, containing 150 mM NaCl plus 0, 1, 5, 10, 20 or 50 mM MgCl2. For both panels the ionic strength increases from left to right.
Table 1. Effects of NaCl and MgCl2 on the melting temperatures of the four intermolecular triplexes.
| Triplex | NaCl (mM) | Tm (°C) | MgCl2 (mM) | Tm (°C) |
|---|---|---|---|---|
| CCT | 0 | 72.5 | 0 | 71.9 |
| 50 | 71.8 | 1 | 73.2 | |
| 100 | 71.8 | 5 | 74.7 | |
| 150 | 71.9 | 10 | 74.8 | |
| 200 | 72.5 | 20 | 74.8 | |
| 350 | 74.5 | 50 | 75.0 | |
| 500 | 75.3 | |||
| 700 | 75.8 | |||
| 1000 | 78.7 | |||
| TTT | 0 | nd | 0 | nd |
| 50 | nd | 1 | nd | |
| 100 | nd | 5 | 32.7 | |
| 150 | nd | 10 | 39.0 | |
| 200 | nd | 20 | 44.7 | |
| 350 | 33.0 | 50 | 50.5 | |
| 500 | 39.5 | |||
| 700 | 45.3 | |||
| 1000 | 51.5 | |||
| CTT | 0 | 57.8 | 0 | 66.7 |
| 50 | 62.0 | 1 | 68.5 | |
| 100 | 64.7 | 5 | 72.8 | |
| 150 | 66.7 | 10 | 74.7 | |
| 200 | 68.3 | 20 | 76.0 | |
| 350 | 71.8 | 50 | 77.8 | |
| 500 | 74.3 | |||
| 700 | 76.5 | |||
| 1000 | 78.8 | |||
| CTCT | 0 | 71.7 | 0 | 75.7 |
| 50 | 72.8 | 1 | 76.5 | |
| 100 | 74.2 | 5 | 78.5 | |
| 150 | 75.7 | 10 | 79.5 | |
| 200 | 76.0 | 20 | 80.0 | |
| 350 | 78.2 | 50 | 80.7 | |
| 500 | 79.7 | |||
| 700 | 80.7 | |||
| 1000 | 82.3 |
The experiments with different concentrations of NaCl were performed in 50 mM sodium acetate pH 5.0, while the experiments with different concentrations of MgCl2 were performed in 50 mM sodium acetate containing 150 mM NaCl. In each case the duplex concentration was 0.25 µM, with 4 µM triplex-forming oligonucleotide. nd, no triplex detected.
Table 2. Effects of [NaCl] and pH on the melting temperatures of the four intermolecular triplexes.
| Sequence | δ(1/Tm)/δln[Na+] | ΔNa+ | δ(1/Tm)/δpH | ΔH+ |
|---|---|---|---|---|
| TTT | 1.9 ± 0.1 × 10–4 | 4.7 ± 0.2 | 4.8 ± 0.4 × 10–5 | 0.5 ± 0.1 |
| CTT | 6.0 ± 0.1 × 10–5 | 2.3 ± 0.1 | 1.9 ± 0.2 × 10–4 | 3.2 ± 0.3 |
| CTCT | 3.0 ± 0.2 × 10–5 | 1.7 ± 0.1 | 2.3 ± 0.2 × 10–4 | 5.7 ± 0.5 |
| CCT | 1.7 × 0.4 × 10–5 | 0.7 ± 0.2 | 3.5 ± 0.7 × 10–4 | 6.9 ± 1.4 |
The experiments at different concentrations of sodium were performed in 50 mM sodium acetate pH 5.0, supplemented with sodium chloride, while the experiments at different pH were performed in 50 mM sodium acetate pH 5–6.5 or 50 mM HEPES, each containing 150 mM NaCl. ΔNa+ and ΔH+, the number of sodium ions and protons, respectively, that are released upon disruption of the triplex were calculated from these slopes and the ΔH values as previously described (19).
As expected, the melting profiles of these intermolecular triplexes are pH dependent and the Tm values shift to lower temperatures at high pH values. This effect is particularly pronounced for triplexes that contain a greater number of C+·GC triplets. Melting profiles could not be obtained for the CCT, CTT and CTCT triplexes above pH 6.5. The TTT triplex could still be detected at pH 7.5, although its Tm dropped by 13°C relative to pH 5.0, as this also contains a single C+·GC triplet. These pH effects were analysed by plotting 1/Tm against pH, as previously described (19), generating a straight line the slope of which is equal to R × ΔH+ × log10/ΔH, where ΔH+ is the number of protons released upon disruption of the triplex. These slopes, together with the values of ΔH+, are shown in Table 2.
Since these triplexes are intermolecular complexes, formed by the bimolecular interaction of a third strand with the duplex, their Tm values should increase with oligonucleotide concentration. One advantage of this fluorescent melting technique is that we can increase the concentration of the third strand without affecting the fluorescence signal. We have therefore examined the effect of third strand concentration on the Tm values of these triplexes. The results of these experiments are shown in Figure 4. The melting curves for the CCT, CTT and CTCT triplexes were obtained in 50 mM sodium acetate pH 5.0, 150 mM NaCl, while the TTT triplex was measured in the same buffer containing 50 mM MgCl2; in each case the duplex concentration was 0.25 µM and the third strand was varied between 0.5 and 5.0 µM. These data were obtained by increasing the temperature in steps of 1°C every 2 min, so as to avoid the hysteresis observed with CTCT at faster rates of heating. It can be seen that, as expected, the Tm of each complex increases with increasing third strand concentration, although the effects are greatest for TTT as this is the weakest complex. Thermodynamic parameters for the formation of these complexes can be obtained by plotting 1/Tm against ln(C), where C is the total nucleic acid concentration (duplex plus third strand), producing a graph with a slope of R/ΔH (shown in Supplementary Material, Figure S1). Although these plots produce straight lines, it should be noted that this analysis assumes that the enthalpy (ΔH) is independent of temperature (i.e. ΔCp = 0). Although this assumption is commonly made when analysing DNA melting curves, one study has shown that a triplex containing 5 C+·CG and 10 T·AT has a ΔCp of –1 kcal mol K–1 (37). By this means the ΔH values for the four triplexes were calculated to be –376 ± 23, –202 ± 16, –316 ± 16 and –476 ± 76 kJ mol–1 for CCT, TTT, CTT and CTCT, respectively. There is a good correlation between ΔH and the number of C+·CG triplets. Indeed, plotting ΔH against the number of C+·CG triplets gives a line with a slope of 30.3 ± 0.9 kJ mol–1 and an intercept of 168 ± 6 kJ mol–1. This simple analysis suggests that for a 15mer triplex at pH 5.0, each C+·CG triplet contributes 30 kJ mol–1 to the enthalpy, while each T·AT triplet contributes only 11 kJ mol–1.
Figure 4.
Effect of third strand oligonucleotide concentration on the fluorescence melting curves. In each case the intramolecular duplex concentration was 0.25 µM. The concentration of the third strands was 0.5, 1, 2, 3, 4 or 5 µM, increasing from left to right, except for CTCT, for which only curves for 1, 3 and 5 µM are shown. The TTC, TCC and CTCT experiments were performed in 50 mM sodium acetate pH 5.0, containing 150 mM NaCl, while those for TTT were performed in 50 mM sodium acetate pH 5.0, containing 150 mM NaCl and 50 mM MgCl2.
Dissociation rate constants (k–1)
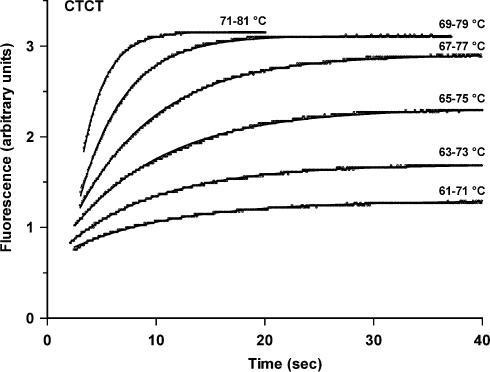
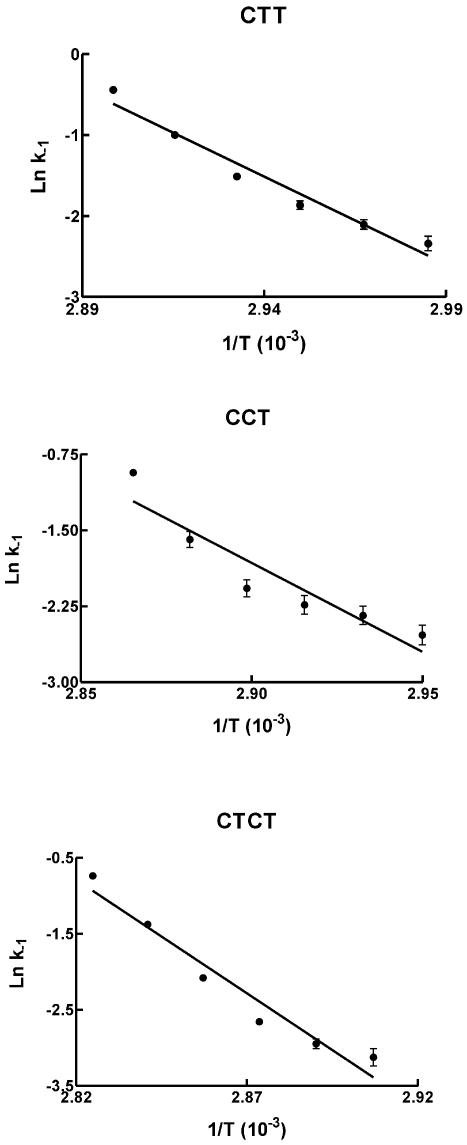
The dissociation kinetics of these triple helices were determined by temperature jump experiments as described in Materials and Methods. The rapid increase in temperature shifts the equilibrium towards the dissociated species, producing a time-dependent increase in fluorescence. These experiments were repeated over a range of different temperatures. Representative examples of these dissociation profiles are shown in Figure 5 for the CTCT triplex, using 0.25 µM duplex target and 4 µM third strand oligonucleotide. In these experiments we equilibrated the sample for 5 min at each new temperature before performing the temperature jump. Longer incubation times (up to 60 min) had no effect on the reaction profiles. We also showed that the temperature jump experiment could be repeated up to four times with the same sample without any effect on dissociation profiles, confirming that triplexes reform after cooling and that the DNA is not damaged or degraded during the experiment. These profiles show time-dependent increases in fluorescence and demonstrate slow triplex dissociation, even at these elevated temperatures. The changes in fluorescence were fitted to simple exponential curves, providing the apparent dissociation rate constant (kapp). For these bimolecular reactions the pseudo-first order rate constant (kapp) will be a function of the total oligonucleotide concentration (D = third strand plus duplex) and will be described by kapp = k–1 + k1[D], where k–1 and k1 are the dissociation and association rate constants. Each of these experiments was therefore repeated with a range of third strand oligonucleotide concentrations (0.25–10 µM) and the individual rate constants at each temperature were determined from plots of kapp against D. Examples of the linear relationship between kapp and [D] are shown in Figure 6. The association (k1) and dissociation (k–1) rate constants for the CCT, CTT and CTCT triplexes at different temperatures are shown in Table 3. Similar experiments with the TTT triplex produced profiles that were too fast to measure, even under ionic conditions which promoted triplex formation. As expected, the dissociation rate constants increase with temperature and Arrhenius plots constructed from these data are shown in Figure 7, from which the activation energies (Ea) and dissociation rate constants at 37°C were estimated. It can be seen that the dissociation rate constants for the CTT and CCT triplexes are similar, with t½ values at 37°C estimated to be ∼20 min. In contrast, the CTCT triplex is much more stable and has a dissociation half-life of >3 days at 37°C. The association constants showed a complex temperature dependence, from which it is not possible to estimate thermodynamic parameters.
Figure 5.
Temperature jump kinetic profiles for triplex CTCT. The intramolecular duplex concentration was 0.25 µM and the third strand oligonucleotide was 4 µM. The reactions were performed in 50 mM sodium acetate pH 5.0, containing 150 mM NaCl.
Figure 6.
Dependence of the pseudo-first order rate constants (kapp) on total oligonucleotide concentration. The data shown are for triplex CCT for a temperature jump from 62 to 72°C and for CTCT with a temperature jump from 65 to 75°C.
Table 3. Kinetic parameters for formation of the CCT, CTT and CTCT triplexes.
| Triplex | Temperature jump (°C) | k–1 (s–1) | k1 (µM–1 s–1) | k–1 37°C (s–1) | t½ 37°C (min) | Ea off (kJ mol–1) |
|---|---|---|---|---|---|---|
| CCT | 56–66 | 0.08 ± 0.01 | 0.020 ± 0.003 | 5.4 × 10–4 | 21.4 | 145 ± 27 |
| 58–68 | 0.10 ± 0.01 | 0.019 ± 0.006 | ||||
| 60–70 | 0.12 ± 0.02 | 0.025 ± 0.004 | ||||
| 62–72 | 0.13 ± 0.02 | 0.042 ± 0.004 | ||||
| 64–74 | 0.21 ± 0.03 | 0.043 ± 0.006 | ||||
| 66–76 | 0.40 ± 0.02 | 0.030 ± 0.004 | ||||
| CTT | 52–62 | 0.10 ± 0.01 | 0.016 ± 0.003 | 4.5 × 10–4 | 25.4 | 180 ± 18 |
| 54–64 | 0.12 ± 0.01 | 0.023 ± 0.003 | ||||
| 56–66 | 0.16 ± 0.01 | 0.023 ± 0.003 | ||||
| 58–68 | 0.22 ± 0.02 | 0.019 ± 0.003 | ||||
| 60–70 | 0.37 ± 0.01 | 0.010 ± 0.002 | ||||
| 62–72 | 0.66 ± 0.03 | 0.001 ± 0.005 | ||||
| CTCT | 61–71 | 0.044 ± 0.009 | 0.012 ± 0.001 | 2.5 × 10–6 | 4560 | 248 ± 29 |
| 63–73 | 0.052 ± 0.006 | 0.010 ± 0.001 | ||||
| 65–75 | 0.070 ± 0.004 | 0.0093 ± 0.0006 | ||||
| 67–77 | 0.125 ± 0.004 | 0.0055 ± 0.0006 | ||||
| 69–79 | 0.25 ± 0.01 | 0.0024 ± 0.0013 | ||||
| 71–81 | 0.48 ± 0.02 | 0.004 ± 0.003 |
The association (k1) and dissociation (k–1) rate constants were determined from exponential fits to kinetic profiles at different third strand oligonucleotide concentrations. The activation energy for dissociation (Ea off) and the k–1 at t½ at 37°C were estimated from the Arrhenius plots shown in Figure 7.
Figure 7.
Arrhenius plots of the dissociation constants (k–1) determined for triplexes CTT, CCT and CTCT.
DISCUSSION
Our previous studies have shown that fluorescently labelled oligonucleotides can be used for determining the stability of intramolecular triplexes (36). The present study demonstrates that this technique can be used to examine the thermodynamic and kinetic properties of intermolecular complexes. This has several advantages over conventional UV melting methods. Firstly, the fluorescence technique is much more sensitive and requires small amounts of material. Secondly, we can observe melting of the third strand without any interference from melting of the underlying duplex. Thirdly, by incorporating the fluorophore on the duplex target with the quencher on the third strand, it is possible to add an excess of the third strand without affecting the total fluorescence. Fourthly, we have used a variation of this technique to determine the kinetic properties of these DNA triplexes.
Comparison of C+·GC and T·AT
Several studies have now shown that at low pH C+·GC triplets impart a greater triplex stability than T·AT (23–27). The data presented in this paper confirm this observation and demonstrate that the most stable triplexes contain alternating C+·GC and T·AT triplets, as predicted by Roberts and Crothers (27). By examining the ΔH values for the formation of the different triplexes we suggest that at pH 5.0 and in the presence of 200 mM sodium the enthalpy of formation of an isolated C+·GC triplet is about three times that of T·AT. This contrasts with a previous study, which showed that the enthalpic contributions of T·AT/T·AT, T·AT/C+·GC and C+·GC/C+·GC base triplet stacks are very similar (26). However, it should be noted that this includes contributions for the different effects of protons and sodium ions on the two triplexes. Since these two triplets are isomorphous, we assume that the increased stability of C+·GC arises from its positive charge, which either assists in overcoming the charge repulsion from the polyanionic backbone or makes favourable interactions with the π-stacked bases.
Effect of metal ions
The stability of DNA triplexes is known to be sensitive to the ionic conditions, as their formation involves an interaction between three polyanions. The present study demonstrates that the T·AT triplet is much more sensitive to the ionic strength than C+·GC. Triplexes that contain only T·AT triplets are especially sensitive to the concentration of both sodium and magnesium and the TTT triplet melts below 30°C even in the presence of 250 mM sodium. In contrast to the overall stability, which is optimal for triplexes that contain alternating C+·GC and T·AT, the effects of sodium and magnesium decrease with increasing numbers of C+·GC triplets. Triplexes that contain predominantly C+·GC are least affected by changes in the concentrations of these cations.
Dissociation kinetics
We have also been able to use the fluorescently labelled oligonucleotides to assess the kinetic stability of these triplexes. Previous studies have shown that triplex association and dissociation are both very slow processes (12,17,28–35). Triplex association rate constants are typically ∼103 M–1 s–2, about three orders of magnitude slower than the formation of duplex DNA. Published values for triplex dissociation rate constants suggest half-lives of between 30 min and several days at 37°C. This fluorescent technique cannot be used directly to measure dissociation rates at 37°C as it is only applicable to temperatures for which there are fluorescence changes in the melting profile. However, the dissociation rate at physiological temperature can be obtained by extrapolation of the Arrhenius plots. Inspection of the Arrhenius plots presented in Figure 7 reveals that these curves may be non-linear, suggesting that other processes may be occurring in addition to the simple dissociation process. This curvature is especially pronounced for the CCT triplex and may indicate an effect due to the larger number of charged cytosines, which may also vary in a temperature-dependent fashion. By using this method we have shown that the dissociation rate is dependent on the sequence. Triplexes CCT and CTT have half-lives of ∼20 min at 37°C, while the dissociation of CTCT is about 200 times slower, with a t½ of ∼3 days. In contrast, the kinetic profiles with TTT were too fast to measure under any of the conditions tested.
In theory this method should allow determination of the association rate constants for triplex formation (k1). Although values for k1 were determined from the slopes of the apparent rate constants against DNA concentration, these did not yield useful Arrhenius plots. We can envision several reasons for this failure. Firstly, the measured rates do not show a strong variation with temperature and there is considerable inaccuracy in the estimated slopes. Secondly, and more importantly, it has been suggested that triplex association rates may decrease with increasing temperature (12). This is especially evident for the k1 values estimated for CTCT (Table 3). It is thought that this arises because triplex formation occurs via a nucleation–zipper mechanism; lower temperatures stabilise the transient intermediates and thereby facilitate association.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from Cancer Research UK and the European Union. The Roche LightCycler was partly funded by the BBSRC (JREI).
REFERENCES
- 1.Praseuth D., Guieysse,A.L. and Hélène,C. (1999) Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta, 1489, 181–206. [DOI] [PubMed] [Google Scholar]
- 2.Neidle S. (1997) Recent developments in triple-helix regulation of gene expression. Anticancer Drug Des., 12, 433–442. [PubMed] [Google Scholar]
- 3.Knauert M.P. and Glazer,P.M. (2001) Triplex-forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet., 10, 2243–2251. [DOI] [PubMed] [Google Scholar]
- 4.Vasquez K.M. and Wilson,J.H. (1998) Triplex-directed modification of genes and gene activity. Trends Biochem. Sci., 23, 4–9. [DOI] [PubMed] [Google Scholar]
- 5.Sun J.S., Garestier,T. and Hélène,C. (1996) Oligonucleotide directed triple helix formation. Curr. Opin. Struct. Biol., 6, 327–333. [DOI] [PubMed] [Google Scholar]
- 6.Thuong N.T. and Hélène,C. (1993) Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angew. Chem. Int. Ed. Engl., 32, 666–690. [Google Scholar]
- 7.Soyfer V.N. and Potoman,V.N. (1996) Triple-helical Nucleic Acids. Springer-Verlag, New York, NY. [Google Scholar]
- 8.Fox K.R. (2000) Targeting DNA with triplexes. Curr. Med. Chem., 7, 17–37. [DOI] [PubMed] [Google Scholar]
- 9.Moser H.E. and Dervan,P.B. (1987) Sequence-specific cleavage of double-helical DNA by triple helix formation. Science, 238, 645–650. [DOI] [PubMed] [Google Scholar]
- 10.Beal P.A. and Dervan,P.B. (1991) Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science, 251, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 11.Singleton S.F. and Dervan,P.B. (1993) Equilibrium association constants for oligonucleotide-directed triple helix formation at single sites: linkage to cation valence and concentration. Biochemistry, 32, 13171–13179. [DOI] [PubMed] [Google Scholar]
- 12.Rougée M., Faucon,B., Mergny,J.L., Barcelo,F., Giovannangeli,C., Garestier,T. and Hélène,C. (1992) Kinetics and thermodynamics of triple-helix formation: effects of ionic strength and mismatches. Biochemistry, 31, 9269–9278. [DOI] [PubMed] [Google Scholar]
- 13.Wu P., Kawamoto,Y., Hara,H. and Sugimoto,N. (2002) Effect of divalent cations and cytosine protonation on thermodynamic properties of intermolecular DNA double and triplex helices. J. Inorg. Biochem., 91, 277–285. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto N., Wu,P., Hara,H. and Kawamoto,Y. (2001) pH and cation effects on the properties of parallel pyrimidine motif DNA triplexes. Biochemistry, 40, 9396–9405. [DOI] [PubMed] [Google Scholar]
- 15.Plum G.E., Park,Y.-W., Singleton,S.F. and Dervan,P.B. (1990) Thermodynamic characterization of the stability and the melting behaviour of a DNA triplex: a spectroscopic and calorimetric study. Proc. Natl Acad. Sci. USA, 87, 9437–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilch D.S, Levenson,C. and Shafer,R.H. (1990) Structural analysis of the (dA)10·2(dT)10 triple helix. Proc. Natl Acad. Sci. USA, 87, 1942–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maher L.J., Dervan,P.B. and Wold,B. (1990) Kinetic analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry, 29, 8820–8826. [DOI] [PubMed] [Google Scholar]
- 18.Latimer L.J.P., Hampel,K. and Lee,J.S. (1989) Synthetic repeating sequence DNAs containing phosphorothioates: nuclease sensitivity and triplex formation. Nucleic Acids Res., 17, 1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plum G.E. and Breslauer,K.J. (1995) Thermodynamics of an intramolecular DNA triple helix: a calorimetric and spectroscopic study of the pH and salt dependence of thermally induced structural transitions. J. Mol. Biol., 248, 679–695. [DOI] [PubMed] [Google Scholar]
- 20.Wilson W.D., Hopkins,H.P., Mizan,S., Hamilton,D.D. and Zon,G. (1994) Thermodynamics of DNA triplex formation in oligomers with and without cytosine base. J. Am. Chem. Soc., 116, 3607–3608. [Google Scholar]
- 21.Kohwi Y. and Kohwi-Shigematsu,T. (1988) Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc. Natl Acad. Sci. USA, 85, 3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiessling L.L, Griffin,L.C. and Dervan,P.B. (1992) Flanking sequence effects within the pyrimidine triple-helix motif characterized by affinity cleavage. Biochemistry, 31, 2829–2834. [DOI] [PubMed] [Google Scholar]
- 23.Asensio J.L., Lane,A.N., Dhesi,J., Bergvist,S. and Brown,T. (1998) The contribution of cytosine protonation to the stability of parallel DNA triple helices. J. Mol. Biol., 275, 811–822. [DOI] [PubMed] [Google Scholar]
- 24.Völker J. and Klump,H.K. (1994) Electrostatic effects in DNA triple helices. Biochemistry, 33, 13502–13508. [DOI] [PubMed] [Google Scholar]
- 25.Keppler M.D. and Fox,K.R. (1997) Relative stability of triplexes containing different numbers of T.AT and C+.GC triplets. Nucleic Acids Res., 25, 4464–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soto A.M., Loo,J. and Marky,L.A. (2002) Energetic contributions for the formation of TAT/TAT, TAT/CGC+ and CGC+/CGC+ base triplet stacks. J. Am. Chem. Soc., 124, 14355–14363. [DOI] [PubMed] [Google Scholar]
- 27.Roberts R.W. and Crothers,D.M. (1996) Prediction of the stability of DNA triplexes. Proc. Natl Acad. Sci. USA, 93, 4320–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blake R.D., Massoulié,J. and Fresco,J.R. (1967) A spectral approach to the equilibria between polyriboadenylate and polyribouridylate and their complexes. J. Mol. Biol., 20, 291–308. [PubMed] [Google Scholar]
- 29.Xodo L.E. (1995) Kinetic analysis of triple-helix formation by pyrimidine oligodeoxynucleotides and duplex DNA. Eur. J. Biochem., 228, 918–926. [DOI] [PubMed] [Google Scholar]
- 30.Paes H.M. and Fox,K.R. (1997) Kinetic studies on the formation of intermolecular triple helices. Nucleic Acids Res., 25, 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox K.R. (1995) Kinetic studies on the formation of acridine-linked DNA triple helices. FEBS Lett., 357, 312–316. [DOI] [PubMed] [Google Scholar]
- 32.Anthony T. and Subramanian,V. (2002) A molecular beacon strategy for real-time monitoring of triplex DNA formation kinetics. Antisense Nucleic Acid Drug. Dev., 12, 145–154. [DOI] [PubMed] [Google Scholar]
- 33.Protozanova E. and Macgregor,R.B. (1996) Kinetic footprinting of DNA triplex formation. Anal. Biochem., 243, 92–99. [DOI] [PubMed] [Google Scholar]
- 34.Sarai A., Sugiura,S., Torigoe,H. and Shindo,H. (1993) Thermodynamic and kinetic analyses of DNA triplex formation: application of filter-binding assay. J. Biomol. Struct. Dyn., 11, 245–252. [DOI] [PubMed] [Google Scholar]
- 35.Ellouze C., Piot,F. and Takajashi,M. (1997) Use of fluorescein-labelled oligonucleotide for analysis of formation and dissocation kinetics of T:A:T triple-stranded DNA: effect of divalent cations. J. Biochem., 121, 521–526. [DOI] [PubMed] [Google Scholar]
- 36.Darby R.A.J., Sollogoub,M., McKeen,C., Brown,L., Risitano,A., Brown,L., Barton,C., Brown,T. and Fox,K.R. (2002) High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a Light Cycler. Nucleic Acids Res., 30, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamiya M., Torigoe,H., Shindo,H. and Sarai,A. (1996) Temperature dependence and sequence specificity of DNA triplex formation: an analysis using isothermal titration calorimetry. J. Am. Chem. Soc., 118, 4532–4538. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.