Abstract
The identification of caspases as major regulators of apoptotic cell death in animals initiated a quest for homologous peptidases in other kingdoms. With the discovery of metacaspases in plants, fungi, and protozoa, this search had apparently reached its goal. However, there is compelling evidence that metacaspases lack caspase activity and that they are not responsible for the caspaselike activities detected during plant and fungal cell death. In this paper, we attempt to broaden the discussion of these peptidases to biological functions beyond apoptosis and cell death. We further suggest that metacaspases and paracaspases, although sharing structural and mechanistic features with the metazoan caspases, form a distinct family of clan CD cysteine peptidases.
Cell death and caspases
Cellular fate is predominantly determined by the processes of division, differentiation, and death. A cell is considered dead when the plasma membrane has lost its integrity or when it is fragmented into so-called apoptotic bodies (Kroemer et al., 2005), but a plethora of definitions based on morphological parameters tries to capture the manifold types of mammalian cell death and the routes toward it. Originally, apoptosis was described as the type of cell death characterized by rounding and shrinking of the cell, chromatin condensation (pyknosis), nuclear fragmentation (karyorrhexis), and budding of discrete plasma membrane–lined portions of cytoplasm (blebbing; Kerr et al., 1972). During apoptosis in mammalian tissues, the plasma membrane remains intact until late stages, thereby preventing an unwanted inflammatory response (Krysko et al., 2006). Autophagy is defined by a vacuolization of the cytoplasm. Autophagic vacuoles have two membranes and contain degenerating organelles and cytosolic content (Gozuacik and Kimchi, 2007). The third main type of cell death, necrosis, is characterized by cell swelling (oncosis), organelle dilation, and subsequent rupture of the plasma membrane (Festjens et al., 2006).
The acquisition of apoptotic morphology is, in most cases, associated with and depends on the activation of Cys-dependent Asp-specific peptidases (caspases; Alnemri et al., 1996; Leist and Jäättelä, 2001). Caspases (clan CD, family C14) cleave their substrates after Asp, are synthesized as inactive zymogens, and can be divided into two types based on their overall structure and activation modes. Effectors or executioner caspases are activated by proteolytic separation of the large (p20) and small (p10) subunits, resulting in active (p20)2(p10)2 heterotetramers. Initiator caspases have an N-terminal extension, the prodomain, that is needed to recruit them into protein complexes that function as activation platforms, called apoptosomes (Riedl and Salvesen, 2007). Their activation does not require proteolytic cleavage but relies on conformational changes after oligomerization (Fuentes-Prior and Salvesen, 2004). Once triggered, initiator caspases can ignite a cascade by the proteolytic activation of effector caspase zymogens. The effector caspases ultimately cleave numerous substrates, thereby causing the typical morphological features of apoptosis (Kumar, 2007; Timmer and Salvesen, 2007). Members of the CD clan of proteases are characterized by their specificity for the residue at the N-terminal side of the scissile bond, the P1 residue, in their substrates. For caspases, substrate recognition additionally requires three or more residues N terminal to P1-Asp. Based on the optimal substrate oligopeptide sequence, caspase activity can be specifically measured by synthetic peptides C-terminally coupled to a fluorogenic moiety, such as 7-amido-4-methylcoumarin (AMC). Upon cleavage by caspases, an increase of fluorescence is proportional to caspase activity. Despite their omnipresence during apoptosis, caspases are also involved in nonapoptotic events, including inflammation, cell proliferation, and cell differentiation. Therefore, the reciprocal conclusion that caspase activities are strictly correlated with apoptosis is invalid (Lamkanfi et al., 2007).
As in animals, cell death is an essential part of the life cycle of plants. From seed germination until seed production, developmental cell death is manifested. A few well known examples are cell death during terminal differentiation of the vascular tracheary elements, leaf and flower senescence, elimination of reproductive organs in unisexual flowers, pollen rejection in the self-incompatibility response, fruit dehiscence, or pod shattering (van Doorn and Woltering, 2005). In addition, plants attempt to block the invasion of biotrophic pathogens via the hypersensitive response, leading to localized cell death at the site of infection (Jones and Dangl, 2006). Typical animal apoptotic features such as pyknosis, karyorrhexis, internucleosomal DNA cleavage, cell shrinkage, and the formation of apoptotic bodies have been observed in dying plant cells (van Doorn and Woltering, 2005). Importantly, during cell death, caspaselike activities are easily detected by synthetic fluorogenic oligopeptide substrates, and cell death can often be attenuated by synthetic caspase-specific inhibitors (Woltering, 2004). With the sequencing of the complete genome of the model plant Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000), these caspaselike activities have steered an intensive but frustrating search for caspase genes within plants.
The discovery of metacaspases and paracaspases
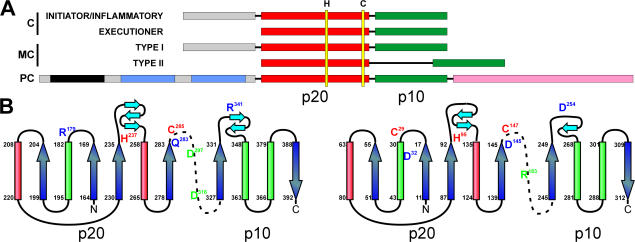
At the end of 2000, distant caspase relatives were discovered in silico in plants, fungi, and protozoa and were designated metacaspases (Uren et al., 2000). The sequences of previously found caspaselike proteins (paracaspases) in metazoans and in the slime mold Dictyostelium discoideum had been used in an iterative PSI-BLAST search of plant-expressed sequence tags (Aravind et al., 1999). Paracaspases contain a prodomain consisting of a death domain and one or two Ig domains, whereas two types of metacaspases can be distinguished (Fig. 1 A). Type I metacaspases have an N-terminal extension reminiscent of the prodomain in initiator and inflammatory caspases. Type II metacaspases lack such a prodomain but harbor a linker region between the putative large and small subunits (Uren et al., 2000; Vercammen et al., 2004). Both meta- and paracaspases contain a conserved catalytic His/Cys dyad, and structure predictions show that they bear the core of the caspase/hemoglobinase fold (Fig. 1 B), which is the determining structural feature of all clan CD Cys proteases (Rawlings and Barrett, 1993; Aravind and Koonin, 2002).
Figure 1.
Structural properties of caspases, paracaspases, and metacaspases. (A) Schematic representation of the domains of caspases (C), metacaspases (MC), and paracaspases (PC). The catalytic domains consist of a large p20 (red) and small p10 subunit (green). Positions of the catalytic His and Cys residues are indicated by yellow bars. Prodomains of inflammatory and proapoptotic initiator caspases and type I metacaspases are in gray. The N-terminal domain of paracaspases contains a death domain (black) and one or two Ig domains (blue). The C-terminal region of paracaspases, which is involved in ubiquitination, is shown in pink. (B) Topological diagram of the structure of human caspase 8 and metacaspase 9 of Arabidopsis thaliana. Catalytic His and Cys residues are labeled in red, (putative) S1 pocket–forming residues are in blue, and maturation sites are in green. Figure layout is adapted from the diagram for human caspase 8 in Fuentes-Prior and Salvesen (2004). The secondary structure of AtMC9 was predicted using the Protein Structure Prediction Server (McGuffin et al., 2000).
Phylogeny of the caspases, metacaspases, and paracaspases
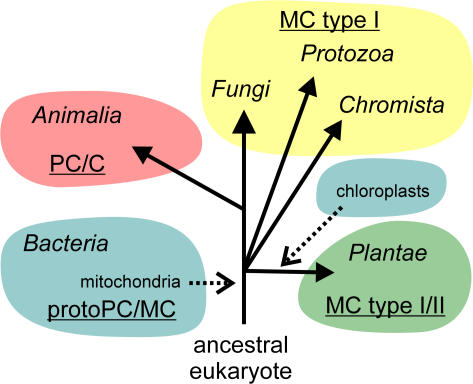
In Eukaryota, paracaspases and caspases are restricted to animal genomes (kingdom Animalia), and metacaspases are present in the kingdoms Protozoa, Fungi, Plantae, and Chromista, whereas in Prokaryota, meta-/paracaspase-like proteins are found in both Archaea and Eubacteria (Fig. 2). Previous phylogenetic analysis of eukaryotic caspases, metacaspases, and paracaspases has suggested that these groups are about equally distant from each other. These findings have led to the hypothesis that eukaryotic metacaspases originate from a horizontal gene transfer (HGT) between the mitochondrial endosymbionts, α-proteobacteria, and the early eukaryotes (Koonin and Aravind, 2002). Still, the origin of caspases and paracaspases remains elusive in such a scenario. Furthermore, meta-/paracaspase-like proteins can be found not only in α-proteobacteria but also in all groups of Bacteria, including cyanobacteria, the ancestors of chloroplasts in plants. Also, it is striking that only type I metacaspases can be found in Protozoa, Fungi, and Chromista, whereas both type I and II are present in Plantae, including green plants, glaucophyta, and rhodophyta. Therefore, an alternative hypothesis would be that caspases, paracaspases, and type I metacaspases have a common ancestor originating from HGT between mitochondrial endosymbionts and host eukaryotic cells. Type II metacaspases might possibly be derived from a second HGT event during the establishment of plastids from endosymbiotic cyanobacteria.
Figure 2.
Phylogenic distribution of caspases, metacaspases, paracaspases, and bacterial para-/metacaspase-like proteins. C, caspase; MC, metacaspase; PC, paracaspase; protoPC/MC, bacterial para-/metacaspase-like proteins. Classification of cellular life into one prokaryotic (Bacteria, including Archaebacteria) and five eukaryotic kingdoms (Fungi, Protozoa, Chromista, Animalia, and Plantae) as described in Cavalier-Smith (2004). HGT events are indicated by dotted arrows.
The alleged paracaspase of D. discoideum is a surprising element in this phylogenetic distribution because slime molds belong to the Protozoa kingdom. Phylogenetic analysis of the sequence of its putative catalytic p20 subunit reveals that it is almost equally related to that of caspases, metacaspases, paracaspases, and their bacterial homologues, making its classification as a separate paracaspase not well founded (our unpublished data). Also, its prodomain lacks a death domain and Ig domains, which is typical of animal paracaspases. Therefore, it is tempting to classify the D. discoideum protease as a metacaspase rather than a paracaspase.
Catalytic properties of metacaspases
In the Arabidopsis genome, nine metacaspase genes are present: three of type I (Arabidopsis thaliana metacaspase 1 [AtMC1] to AtMC3) and six of type II (AtMC4 to AtMC9; Vercammen et al., 2004). Upon overproduction in Escherichia coli, type II metacaspases autoprocess and display a Cys-dependent proteolytic activity against synthetic P1-Arg substrates, whereas AtMC9 also cleaves P1-Lys substrates, albeit with low efficiency (Vercammen et al., 2004, 2006; Watanabe and Lam, 2005). Type I metacaspases from Arabidopsis do not autoprocess upon recombinant overproduction and, like mammalian initiator caspases, possibly require induced oligomerization within an activation platform (Fuentes-Prior and Salvesen, 2004). A positional scanning synthetic combinatorial library screening with purified recombinant AtMC9 confirmed the preference for P1-Arg. The optimized tetrapeptide substrate Ac-Val-Arg-Pro-Arg-AMC had a kcat/KM of 4.6 × 105 M−1 s−1 and, thus, can be considered a very efficient substrate for AtMC9 (Vercammen et al., 2006).
The P1 preference of clan CD proteases is dictated by conserved amino acids distributed throughout the mature protease that together form the S1 pocket (Fuentes-Prior and Salvesen, 2004). In caspases, Arg179, Gln283, and Arg341 (according to caspase-1 residue numbering) form a basic S1 pocket for optimal binding of the acidic P1-Asp within their substrates (Fuentes-Prior and Salvesen, 2004). When the available sequences of eukaryotic metacaspases, paracaspases, and bacterial meta-/paracaspase homologues are aligned to those of animal caspases, Gln283 is replaced by an Asp, and Arg341 is replaced by Asp or Glu in both para- and metacaspases. Arg179 of caspases aligns to Leu. Six residues more C terminal, a highly conserved Asp is present that aligns with Asp163 of bacterial gingipain R, which is known to coordinate binding of the P1-Arg of substrates of this peptidase (Eichinger et al., 1999). Together, these residues are ideally positioned to create a highly acidic S1 pocket that is perfectly suited to accept the basic P1 residues Arg and Lys (Fig. 1 B). As the predicted S1 pocket–forming residues are strictly conserved in all known sequences of para- and metacaspases, the Arg/Lys specificity is very probably shared by all of them. The determined P1 specificity of metacaspases of other plants and of yeast and protozoa confirmed this hypothesis (Bozhkov et al., 2005; Watanabe and Lam, 2005; González et al., 2007).
The fact that no close bacterial caspase homologues have been identified yet would reflect an animal-specific evolutionary process of gene duplications and progression of the caspases from Arg/Lys toward Asp specificity. Until now, attempts to detect the protease activity of paracaspases have been unsuccessful (Snipas et al., 2004), but paracaspases may have retained their preference for basic P1 residues in their substrates. The reason for the shift in caspase P1 specificity remains unclear.
A biochemical particularity of AtMC9 is the presence of a second catalytic Cys, Cys29. Mutation analysis revealed that the primary Cys147 is necessary for autocatalytic processing and concomitant activation of AtMC9. However, once activated either autocatalytically or by exogenous AtMC9, proteolytic activity almost completely depends on Cys29 because replacement of this residue by Ala reduces protease activity by 99%. Furthermore, Cys147 but not Cys29 can be inactivated by S-nitrosylation. Thus, in the presence of nitric oxide, AtMC9 remains inactive until S-nitrosylation is reversed or until upstream proteases convert pro-AtMC9 into its mature form (Belenghi et al., 2007).
Are metacaspases involved in cell death?
The identification of metacaspase genes prompted the assessment of their potential involvement in cell death events in fungi, protozoa, and plants. The first data came from studies in baker's yeast (Saccharomyces cerevisiae). Overproduction of the single metacaspase YCA1 resulted in autocatalytic processing and rendered cells more sensitive to exogenous or aging-related oxidative stress, as determined by reduced clonogenicity (Madeo et al., 2002). However, it may not be surprising that overproduction of an active protease, causing endogenous stress, resulted in a higher sensitivity to exogenous stress. A yeast strain with a disrupted YCA1 gene (Δyca1) was also shown to be threefold less sensitive to H2O2, and ∼5% of the cells escaped from aging-related cell death (Madeo et al., 2002). Whether this observation reflects a direct involvement of YCA1 in cell death or this desensitization is caused by indirect effects, such as an altered protein turnover disturbing the balance of pro– and anti–cell death mediators, remains unclear. Indeed, after treatment of Δyca1 cells with H2O2, levels of oxidized proteins were much higher than those of wild-type cells (Khan et al., 2005). Concomitantly, the proteasome activity of Δyca1 cells increased and apoptosis decreased upon H2O2 treatment, as measured by phosphatidylserine (PS) externalization and DNA fragmentation. The reduced capability of Δyca1 cells compared with wild-type cells to cope with damaged proteins might explain the considerable decrease in cell viability after extended culture (i.e., >30 d; Herker et al., 2004). Whereas PS exposure and DNA fragmentation are genuine apoptotic markers, clonogenicity assays might also reflect other cellular states such as cell cycle arrest or metabolic deficiencies. Therefore, the clonogenicity results themselves do not exclude functions of metacaspases other than cell death involvement. In animal cells, PS exposure by dying cells functions as an “eat-me” signal for phagocytotic cells. However, the physiological function of PS exposure by yeast and plant cells remains intriguing because they possess a rigid cell wall and, thus, are incapable of phagocytosis.
Extracts of H2O2-treated YCA1-overproducing yeast were highly active toward the synthetic caspase substrates Val-Glu-Ile-Asp-AMC and Ile-Glu-Thr-Asp-AMC, suggesting that the YCA1 metacaspase behaved as a bona fide caspase (Madeo et al., 2002). These results were later contradicted: lysates from bacteria and H2O2-stimulated yeast overproducing YCA1 were not active against synthetic caspase substrates but cleaved P1-Arg and, to a lesser extent, P1-Lys substrates similarly to plant metacaspases (Watanabe and Lam, 2005). Thus, YCA1 involvement cannot be determined by using synthetic caspase substrates or inhibitors. Because YCA1-independent cell death (Büttner et al., 2007; for review see Váchová and Palková, 2007) and YCA1-independent caspaselike activities (Váchová and Palková, 2005; Hauptmann et al., 2006) have been reported, the involvement of metacaspase activity in yeast cell death remains debatable (for review see Váchová and Palková, 2007). The identification of endogenous YCA1 substrates will be crucial in unraveling the signaling pathways regulated by this metacaspase.
The genome of the pathogenic filamentous fungus Aspergillus fumigatus contains two type I metacaspases, CasA and CasB. With double knockout mutants, neither of these metacaspases was found to be necessary for virulence. In addition, stress-induced cell death did not depend on metacaspases despite the abrogation of apoptosis-related membrane PS exposure in CasA and CasB double knockout stationary-phase cultures. Interestingly, both CasA and CasB were required for growth in the presence of agents inducing endoplasmic reticulum stress, suggesting a prosurvival role for metacaspases rather than an involvement in cell death processes (Richie et al., 2007).
Of the five type I metacaspases of Trypanosoma brucei, only TbMCA4 caused retardation in growth, loss of respiratory competence, and subsequent decrease in clonogenicity when overproduced in baker's yeast (Szallies et al., 2002). Surprisingly, TbMCA4, like TbMCA1, lacks a catalytic Cys at the canonical location, although an adjacent Cys is present. Nevertheless, using different synthetic tetrapeptides with Asn, Asp, Arg, or Lys at the P1 position, no proteolytic activity in lysates of E. coli or yeast overproducing TbMCA4 could be demonstrated. Triple-null trypanosomes for TbMCA2, TbMCA3, and TbMCA5 had no altered cell death or enhanced susceptibility to stresses, but the rapid down-regulation of all three genes with induced RNAi resulted in an in vitro growth arrest (Helms et al., 2006). In Trypanosoma cruzi, two metacaspase genes, TcMCA3 and TcMCA5, have been reported. In untreated epimastigotes, the encoded proteins were distributed in the whole cell, but, after exposure to fresh human serum, which induces rapid apoptosis-like cell death, relocalization to the nucleus was observed. Upon overproduction of TcMCA5, epimastigotes of T. cruzi were more sensitive to fresh human serum–induced cell death (Kosec et al., 2006). Overproduction in yeast of the single type I metacaspase from Leishmania major, LmjMCA, slightly enhanced sensitivity toward H2O2, as measured by PS exposure. Interestingly, extracts of LmjMCA-overproducing yeast cells were proteolytically active toward P1-Arg synthetic substrates, demonstrating the probably universal preference for basic P1 residues of metacaspases (González et al., 2007). Plasmodium berghei has three type I metacaspases, of which PbMC2 and PbMC3 lack one or both of the catalytic site residues, but knockout mutants of the PbMC1 gene did not display any obvious phenotype (Le Chat et al., 2007).
As discussed in the section Phylogeny of the caspases, metacaspases, and paracaspases, we propose to classify the slime mold paracaspase as a metacaspase. Upon starvation, D. discoideum differentiates into multicellular fruiting bodies consisting of a spore mass supported by a stalk. During this process, stalk cells die in a caspase-independent autophagic cell death (Cornillon et al., 1994; Olie et al., 1998). Both differentiation and cell death were demonstrated to be independent of meta-/paracaspase action (Roisin-Bouffay et al., 2004).
In Norway spruce (Picea abies), metacaspases were studied in the context of developmental cell death during in vitro somatic embryogenesis. In this process, dying embryo suspensor cells contained elevated activity against the synthetic fluorogenic caspase-6 substrate Val-Glu-Ile-Asp-AMC. Accordingly, treatment with the synthetic inhibitor Val-Glu-Ile-Asp-fmk prevented differentiation of the suspensor and subsequent suspensor cell death (Bozhkov et al., 2004). Disruption of the type II metacaspase gene mcII-Pa abrogated the terminal differentiation and death of the suspensor cells and drastically reduced caspaselike activity, suggesting that mcII-Pa had caspase activity and was involved in cell death (Suarez et al., 2004). Later in vitro experiments have shown that mcII-Pa had Arg but not Asp specificity (Bozhkov et al., 2005). Because knocking down mcII-Pa not only disrupted cell death but also blocked embryonic differentiation, we speculate that mcII-Pa might be primarily involved in suspensor differentiation rather than in suspensor cell death. Possibly, mcII-Pa regulates the actin reorganization observed during suspensor differentiation (Smertenko et al., 2003), like mammalian caspases do in the cytoskeletal rearrangements during apoptosis (Mashima et al., 1999).
In Arabidopsis, mere constitutive overexpression or disruption of metacaspase genes does not lead to an obvious phenotype (Vercammen et al., 2006; Belenghi et al., 2007; our unpublished data), and, thus, a role for metacaspases in cell death or other processes has not been identified yet. Redundancy may exist between the various members of this family, or additional factors may be necessary to activate ectopically expressed metacaspases. A large amount of microarray data is available (http://www.arabidopsis.org/info/expression/ATGenExpress.jsp) describing the expression of >20,000 Arabidopsis genes (Zimmermann et al., 2004). Analysis of these data for the nine metacaspase genes could at least give a hint to their functional roles. Several metacaspase genes are strongly induced in senescing flowers, in response to various pathogens and elicitors, and during various abiotic stresses (Sanmartín et al., 2005; our unpublished data). As a prominent role for cell death has been demonstrated in responses to biotic and abiotic stresses, it might be tempting to deduce from these expression profiles that metacaspases play a role in cell death signaling. Alternatively, plants might first try to cope with stresses by rapid adaptation before sacrifice. In vivo reporter systems will be necessary to specifically pinpoint those cells expressing particular metacaspases. Also, metacaspase activity can be regulated on multiple posttranslational levels. For example, the activity of AtMC9 is regulated by autoprocessing, pH, a protease inhibitor (AtSerpin1), and S-nitrosylation (Vercammen et al., 2004, 2006; Belenghi et al., 2007). Likewise, AtMC4 and AtMC5 activity depends on calcium (Vercammen et al., 2004; Watanabe and Lam, 2005). Therefore, specific in vivo activity assays would strongly contribute to our understanding of the role of individual metacaspases in plant development and stress response.
Conclusions and perspectives
To date, metacaspases of plants, fungi, and protozoa have been shown to have Arg/Lys-specific activity (Vercammen et al., 2004, 2006; Bozhkov et al., 2005; Watanabe and Lam, 2005; González et al., 2007). Based on the available sequences, we speculate that all metacaspases and possibly also paracaspases share this specificity. As a consequence, the caspaselike activities reported to be involved in plant and fungal cell death most probably differ from the metacaspases. Other plant proteases exhibiting caspaselike activity and suggested to be involved in cell death include the legumains (also called vacuolar processing enzymes) and some subtilisins (Coffeen and Wolpert, 2004; Hara-Nishimura et al., 2005; Hatsugai et al., 2006).
Until now, the role of metacaspases in cell death still remained enigmatic, and both up- and down-regulation of metacaspases have yielded conflicting data. However, such approaches bear the risk that a constitutive perturbation of genes that are essential for normal cellular homeostasis leads to overinterpretation. Alternative routes toward unraveling the function of metacaspases could involve the identification of their substrates by using technologies that allow direct characterization of in vivo protein processing on a proteome-wide scale (Gevaert et al., 2006). Knowing the degradome specificity of metacaspases could reveal their role in cellular and developmental processes, including cell death. Overproduction of the cleavage fragments and/or of uncleavable mutant proteins would help elucidate the functional consequences of substrate cleavage by metacaspases.
We conclude that although metacaspases, paracaspases, and caspases contain a caspase fold and probably originated from a common ancestor gene, metacaspases and paracaspases are clearly distinct from caspases for the following reasons. First, the fact that these different proteases contain a caspase fold might not be a valid argument to group them together in the caspase family (clan CD, family C14; http://merops.sanger.ac.uk/). Legumains (clan CD, family C13) and gingipains (clan CD, family C25) constitute separate families that also contain a caspase fold (Chen et al., 1998; Eichinger et al., 1999). Second, the P1 preference of metacaspases is basic, whereas that of caspases is acidic. If metacaspases and caspases shared similar functions, we assume that both the proteases and their specific degradome would have coevolved. In view of the crucial functions of many of their substrates, this hypothesis is unlikely. Third, previous phylogenetic analyses of clan CD peptidases have shown that caspases constitute a separate group distinct from other clan CD peptidases, including metacaspases and paracaspases (Koonin and Aravind, 2002). Therefore, we believe that it might be expedient to regroup metacaspases and paracaspases into a separate family in the CD clan of Cys peptidases.
Acknowledgments
We thank Dr. Martine De Cock for assistance in preparing this manuscript.
This work was supported, in part, by grants from the Research Fund of Ghent University (Geconcerteerde Onderzoekstacties no. 12.0514.03 and 12.0505.02), the Interuniversity Poles of Attraction Programme-Belgian Science Policy (P6/18), the European Union (Epistem LSHB-CT-2005-019067; DeathTrain MRTN-CT-035624), and the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (2G.0218.06 and G.0133.05). D. Vercammen was a postdoctoral fellow of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen.
Abbreviations used in this paper: AMC, 7-amido-4-methylcoumarin; AtMC, Arabidopsis thaliana metacaspase; HGT, horizontal gene transfer; PS, phosphatidylserine.
References
- Alnemri, E.S., D.J. Livingston, D.W. Nicholson, G. Salvesen, N.A. Thornberry, W.W. Wong, and J. Yuan. 1996. Human ICE/CED-3 protease nomenclature. Cell. 87:171. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408:796–815. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and L.V. Koonin. 2002. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins. 46:355–367. [DOI] [PubMed] [Google Scholar]
- Aravind, L., V.M. Dixit, and E.V. Koonin. 1999. The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci. 24:47–53. [DOI] [PubMed] [Google Scholar]
- Belenghi, B., M.C. Romero-Puertas, D. Vercammen, A. Brackenier, D. Inzé, M. Delledonne, and F. Van Breusegem. 2007. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 282:1352–1358. [DOI] [PubMed] [Google Scholar]
- Bozhkov, P.V., L.H. Filonova, M.F. Suarez, A. Helmersson, A.P. Smertenko, B. Zhivotovsky, and S. von Arnold. 2004. VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ. 11:175–182. [DOI] [PubMed] [Google Scholar]
- Bozhkov, P.V., M.F. Suarez, L.H. Filonova, G. Daniel, A.A. Zamyatnin, S. Rodriguez-Nieto Jr., B. Zhivotovsky, and A. Smertenko. 2005. Cysteine protease mcII-Pa executes programmed cell death during plant organogenesis. Proc. Natl. Acad. Sci. USA. 102:14463–14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, S., T. Eisenberg, D. Carmona-Gutierrez, D. Ruli, H. Knauer, C. Ruckenstuhl, C. Sigrist, S. Wissing, M. Kollroser, K.-U. Fröhlich, et al. 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell. 25:233–246. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 2004. Only six kingdoms of life. Proc. Biol. Sci. 271:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.-M., N.D. Rawlings, R.A.E. Stevens, and A.J. Barrett. 1998. Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett. 441:361–365. [DOI] [PubMed] [Google Scholar]
- Coffeen, W.C., and T.J. Wolpert. 2004. Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell. 16:857–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillon, S., C. Foa, J. Davoust, N. Buonavista, J.D. Gross, and P. Golstein. 1994. Programmed cell death in Dictyostelium. J. Cell Sci. 107:2691–2704. [DOI] [PubMed] [Google Scholar]
- Eichinger, A., H.-G. Beisel, U. Jacob, R. Huber, F.-J. Medrano, A. Banbula, J. Potempa, J. Travis, and W. Bode. 1999. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18:5453–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festjens, N., T. Vanden Berghe, and P. Vandenabeele. 2006. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim. Biophys. Acta. 1757:1371–1387. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior, P., and G.S. Salvesen. 2004. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384:201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert, K., P. Van Damme, B. Ghesquière, and J. Vandekerckhove. 2006. Protein processing and other modifications analyzed by diagonal peptide chromatography. Biochim. Biophys. Acta. 1764:1801–1810. [DOI] [PubMed] [Google Scholar]
- González, I.J., C. Desponds, C. Schaff, J.C. Mottram, and N. Fasel. 2007. Leishmania major metacaspase can replace yeast metacaspase in programmed cell death and has arginine-specific cysteine peptidase activity. Int. J. Parasitol. 37:161–172. [DOI] [PubMed] [Google Scholar]
- Gozuacik, D., and A. Kimchi. 2007. Autophagy and cell death. Curr. Top. Dev. Biol. 78:217–245. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., N. Hatsugai, S. Nakaune, M. Kuroyanagi, and M. Nishimura. 2005. Vacuolar processing enzyme: an executor of plant cell death. Curr. Opin. Plant Biol. 8:404–408. [DOI] [PubMed] [Google Scholar]
- Hatsugai, N., M. Kuroyanagi, M. Nishimura, and I. Hara-Nishimura. 2006. A cellular suicide strategy of plants: vacuole-mediated cell death. Apoptosis. 11:905–911. [DOI] [PubMed] [Google Scholar]
- Hauptmann, P., C. Riel, L.A. Kunz-Schughart, K.-U. Fröhlich, F. Madeo, and L. Lehle. 2006. Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 59:765–778. [DOI] [PubMed] [Google Scholar]
- Helms, M.J., A. Ambit, P. Appleton, L. Tetley, G.H. Coombs, and J.C. Mottram. 2006. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J. Cell Sci. 119:1105–1117. [DOI] [PubMed] [Google Scholar]
- Herker, E., H. Jungwirth, K.A. Lehmann, C. Maldener, K.-U. Fröhlich, S. Wissing, S. Büttner, M. Fehr, S. Sigrist, and F. Madeo. 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., and J.L. Dangl. 2006. The plant immune system. Nature. 444:323–329. [DOI] [PubMed] [Google Scholar]
- Kerr, J.F., A.H. Wyllie, and A.R. Currie. 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 26:239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.A.S., P.B. Chock, and E.R. Stadtman. 2005. Knockout of caspase-like gene, YCA1, abrogates apoptosis and elevates oxidized proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 102:17326–17331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V., and L. Aravind. 2002. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 9:394–404. [DOI] [PubMed] [Google Scholar]
- Kosec, G., V.E. Alvarez, F. Agüero, D. Sánchez, M. Dolinar, B. Turk, V. Turk, and J.J. Cazzulo. 2006. Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Mol. Biochem. Parasitol. 145:18–28. [DOI] [PubMed] [Google Scholar]
- Kroemer, G., W.S. El-Deiry, P. Golstein, M.E. Peter, D. Vaux, P. Vandenabeele, B. Zhivotovsky, M.V. Blagosklonny, W. Malorni, R.A. Knight, et al. 2005. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 12:1463–1467. [DOI] [PubMed] [Google Scholar]
- Krysko, D.V., K. D'Herde, and P. Vandenabeele. 2006. Clearance of apoptotic and necrotic cells and its immunological consequences. Apoptosis. 11:1709–1726. [DOI] [PubMed] [Google Scholar]
- Kumar, S. 2007. Caspase function in programmed cell death. Cell Death Differ. 14:32–43. [DOI] [PubMed] [Google Scholar]
- Lamkanfi, M., N. Festjens, W. Declercq, T. Vanden Berghe, and P. Vandenabeele. 2007. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 14:44–55. [DOI] [PubMed] [Google Scholar]
- Le Chat, L., R.E. Sinden, and J.T. Dessens. 2007. The role of metacaspase 1 in Plasmodium berghei development and apoptosis. Mol. Biochem. Parasitol. 153:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist, M., and M. Jäättelä. 2001. Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2:589–598. [DOI] [PubMed] [Google Scholar]
- Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lächelt, M. Herlan, M. Fehr, K. Lauber, S.J. Sigrist, S. Wesselborg, and K.-U. Fröhlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell. 9:911–917. [DOI] [PubMed] [Google Scholar]
- Mashima, T., M. Naito, and T. Tsuruo. 1999. Caspase-mediated cleavage of cytoskeletal actin plays a positive role in the process of morphological apoptosis. Oncogene. 18:2423–2430. [DOI] [PubMed] [Google Scholar]
- McGuffin, L.J., K. Bryson, and J.T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics. 16:404–405. [DOI] [PubMed] [Google Scholar]
- Olie, R.A., F. Durrieu, S. Cornillon, G. Loughran, J. Gross, W.C. Earnshaw, and P. Golstein. 1998. Apparent caspase independence of programmed cell death in Dictyostelium. Curr. Biol. 8:955–958. [DOI] [PubMed] [Google Scholar]
- Rawlings, N.D., and A.J. Barrett. 1993. Evolutionary families of peptidases. Biochem. J. 290:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie, D.L., M.D. Miley, R. Bhabhra, G.D. Robson, J.C. Rhodes, and D.S. Askew. 2007. The Aspergillus fumigatus metacaspases CasA and CasB facilitate growth under conditions of endoplasmic reticulum stress. Mol. Microbiol. 63:591–604. [DOI] [PubMed] [Google Scholar]
- Riedl, S.J., and G.S. Salvesen. 2007. The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8:405–413. [DOI] [PubMed] [Google Scholar]
- Roisin-Bouffay, C., M.-F. Luciani, G. Klein, J.-P. Levraud, M. Adam, and P. Golstein. 2004. Developmental cell death in Dictyostelium does not require paracaspase. J. Biol. Chem. 279:11489–11494. [DOI] [PubMed] [Google Scholar]
- Sanmartín, M., L. Jaroszewski, N.V. Raikhel, and E. Rojo. 2005. Caspases. Regulating death since the origin of life. Plant Physiol. 137:841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko, A.P., P.V. Bozhkov, L.H. Filonova, S. von Arnold, and P.J. Hussey. 2003. Re-organisation of the cytoskeleton during developmental programmed cell death in Picea abies embryos. Plant J. 33:813–824. [DOI] [PubMed] [Google Scholar]
- Snipas, S.J., E. Wildfang, T. Nazif, L. Christensen, K.M. Boatright, M. Bogyo, H.R. Stennicke, and G.S. Salvesen. 2004. Characteristics of the caspase-like catalytic domain of human paracaspase. Biol. Chem. 385:1093–1098. [DOI] [PubMed] [Google Scholar]
- Suarez, M.F., L.H. Filonova, A. Smertenko, E.I. Savenkov, D.H. Clapham, S. von Arnold, B. Zhivotovsky, and P.V. Bozhkov. 2004. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 14:R339–R340. [DOI] [PubMed] [Google Scholar]
- Szallies, A., B.K. Kubata, and M. Duszenko. 2002. A metacaspase of Trypanosoma brucei causes loss of respiration competence and clonal death in the yeast Saccharomyces cerevisiae. FEBS Lett. 517:144–150. [DOI] [PubMed] [Google Scholar]
- Timmer, J.C., and G.S. Salvesen. 2007. Caspase substrates. Cell Death Differ. 14:66–72. [DOI] [PubMed] [Google Scholar]
- Uren, A.G., K. O'Rourke, L. Aravind, M.T. Pisabarro, S. Seshagiri, E.V. Koonin, and V.M. Dixit. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 6:961–967. [DOI] [PubMed] [Google Scholar]
- Váchová, L., and Z. Palková. 2005. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 169:711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váchová, L., and Z. Palková. 2007. Caspases in yeast apoptosis-like death: facts and artefacts. FEMS Yeast Res. 7:12–21. [DOI] [PubMed] [Google Scholar]
- van Doorn, W.G., and E.J. Woltering. 2005. Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10:117–122. [DOI] [PubMed] [Google Scholar]
- Vercammen, D., B. van de Cotte, G. De Jaeger, D. Eeckhout, P. Casteels, K. Vandepoele, I. Vandenberghe, J. Van Beeumen, D. Inzé, and F. Van Breusegem. 2004. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 279:45329–45336. [DOI] [PubMed] [Google Scholar]
- Vercammen, D., B. Belenghi, B. van de Cotte, T. Beunens, J.-A. Gavigan, R. De Rycke, A. Brackenier, D. Inzé, J.L. Harris, and F. Van Breusegem. 2006. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J. Mol. Biol. 364:625–636. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., and E. Lam. 2005. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 280:14691–14699. [DOI] [PubMed] [Google Scholar]
- Woltering, E.J. 2004. Death proteases come alive. Trends Plant Sci. 9:469–472. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., M. Hirsch-Hoffmann, L. Hennig, and W. Gruissem. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136:2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]