Abstract
The bacterial world is full of varying cell shapes and sizes, and individual species perpetuate a defined morphology generation after generation. We review recent findings and ideas about how bacteria use the cytoskeleton and other strategies to regulate cell growth in time and space to produce different shapes and sizes.
Bacteria are exceedingly tiny compared with most eukaryotic organisms, yet, far from being amorphous bags of biomolecules, they display a breathtaking array of cell morphologies from spheres, rods, and helices to tapered, branched, and flat shapes. Both shape and size are important for cell function, particularly with respect to diffusion and nutrient uptake. A sphere may seem a simple shape to achieve, but its surface area to volume ratio rapidly shrinks with increased size. Meanwhile, a rod can maintain a viable ratio with greater volumes. Other bacteria develop one or more long, thin appendages that effectively increase the exposed surface area without substantially increasing volume. The shape of a bacterium is not dictated by diffusion considerations alone. For example, cyanobacteria are able to survive on exposed sandstone surfaces by forming long cell filaments that can insert and lodge in multiple pores (Kurtz and Netoff, 2001). With a variety of strategies available to a cell, it seems likely that the morphology of each species is uniquely tailored for fitness and survival (Young, 2006).
Remarkably, these defined bacterial cell morphologies are maintained and propagated from one generation to the next, underscoring the importance of cell shape and size. Bacteria can also make morphological transitions in response to changes in environmental conditions. For example, Escherichia coli increases its length and diameter slightly when its growth rate is increased (Woldringh et al., 1980); the rod-shaped plant symbiont Sinorhizobium meliloti differentiates into Y-shaped nitrogen-fixing cells in plant cells (VandenBosch et al., 1989); uropathogenic E. coli cells lengthen into long filaments as part of an immune evasion response (Justice et al., 2006); and the spiral-shaped pathogen Helicobacter pylori adopts a spherical (coccoid) shape both in extended culture (Benaissa et al., 1996) and in stomach infections (Chan et al., 1994). These morphological responses and the faithful maintenance and propagation of cell morphology indicate that sophisticated control systems must exist to regulate cell morphogenesis.
The cell morphogenesis triumvirate: cell wall, turgor pressure, and cytoskeleton
Bacteria, like other organisms with walled cells such as plants and fungi, must temporally and spatially control cell wall synthesis to regulate cell morphogenesis. An essential component of the bacterial cell wall is the peptidoglycan (PG), a meshwork of glycan strands cross-linked by peptide bridges that is synthesized and modified by enzymes collectively named penicillin-binding proteins (PBPs) because of their penicillin-binding property. Gram-negative bacteria mainly have one single layer of PG, whereas Gram-positive bacteria have multiple layers that are linked to each other via short peptides (Höltje, 1998). Either way, the mono- or multilayered PG forms one single, giant molecule that surrounds the cytoplasmic membrane and protects it from the turgor pressure exerted by the cytoplasm. The wall restrains the turgor pressure to avoid swelling and lysis, and the turgor pressure, in turn, is regarded as one of the primary forces that stretches the wall, favoring bond breaking and new PG insertion during cell growth (Koch, 1985; Harold, 2002). Thus, bacteria, like other walled cell organisms, face two obstacles. First, the turgor pressure exerts equal force in all directions, which is problematic for achieving any nonspherical shape. Second, the integrity of the stress-bearing PG wall must be maintained at all times to avoid cell lysis, yet bonds must be broken to allow wall enlargement during growth and division. The latter is particularly challenging for Gram-negative bacteria with their single-layered PG.
A proposed solution is to restrict cell wall insertion to specific locales and to coordinate or couple PG synthetic activity with bond hydrolysis, perhaps in a holoenzyme complex comprising both synthetic and lytic enzymes (Höltje, 1998). Not only is this idea compatible with current models of glycan strand insertion, but affinity chromatography and plasmon resonance experiments have also demonstrated interactions in vitro among E. coli PG synthetic and lytic enzymes (Romeis and Höltje, 1994; Schiffer and Höltje, 1999; Vollmer et al., 1999). How do bacteria control cell wall synthesis and hydrolysis in time and space to produce specific shapes and sizes? Remarkably, despite the wide divergence in cell wall composition, structure, and metabolic activities between bacteria and walled eukaryotic cells, the spatio-temporal control of cell growth can be accomplished in both using a similar tool: the cytoskeleton. All three classes of cytoskeletal elements corresponding to eukaryotic tubulin, actin, and intermediate filament proteins are represented in bacteria (Fig. 1), with each playing an important role in cell morphogenesis.
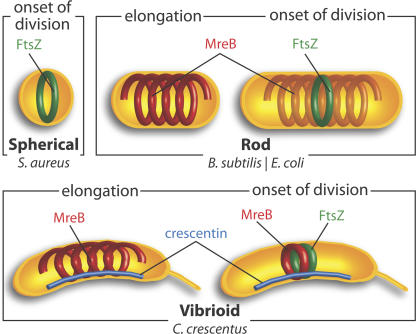
Figure 1.
The bacterial cytoskeleton. The only cytoskeletal element present in spherical bacteria such as S. aureus (top left) is the tubulin-like cell division protein FtsZ (green), which localizes in a ring at the onset of cell division, recruits other cell division proteins, and defines the division plane. Most rod-shaped bacteria (top right) also contain one or more actin-like MreB homologues (red), which exhibit helix-like localization patterns and are essential for cell width control. At the onset of cell division, the FtsZ ring forms and defines the division plane. C. crescentus, a vibrioid bacterium (bottom), contains a third cytoskeletal element, the intermediate filament-like crescentin (blue), which is required for cell curvature and localizes at the inner curvature of cells. At the onset of division in C. crescentus, MreB exhibits FtsZ-dependent relocalization from a helix-like to a ring-like pattern at the FtsZ ring location.
In this paper, we will primarily discuss the most common and best-studied cell shapes—the sphere, the rod, and the curved rod—and describe the main control mechanisms involved in cell shape and size regulation.
Spherical cells: not so simple after all
Because bacterial morphology is a result of where and when cell wall growth occurs, it is tempting to consider the humble sphere as the default shape, but the growth of spherical bacteria (cocci) is clearly not unregulated isotropic growth, which could only lead to continuous cell expansion. In fact, spatial restriction of cell wall growth and, thus, cell growth is required to curb such expansion and enable cell division. Spherical cells such as Staphylococcus aureus localize wall synthesis to a circular division septum that bisects the cell; cleavage and remodeling of this septum leads to the formation of two new hemispheres, one on each daughter cell (Fig. 2, top; Pinho and Errington, 2003). Thus, it is this flat septum-to-hemispherical modification that allows the expansion of cellular volume. Localization of growth at the division septum is dependent on the tubulin-like cytoskeletal protein FtsZ (Pinho and Errington, 2003), which forms a cytokinetic ring structure at the division plane (Fig. 1; Bi and Lutkenhaus, 1991) and is essential for cell division in bacteria. In the absence of FtsZ, cell wall growth in S. aureus occurs in an unregulated, dispersed manner, leading to cell expansion and lysis (Pinho and Errington, 2003). This is accompanied by failure of the PG synthesis enzyme PBP2 to localize at the division site, suggesting that FtsZ restricts cell wall synthesis activity to the division septum (Pinho and Errington, 2003).
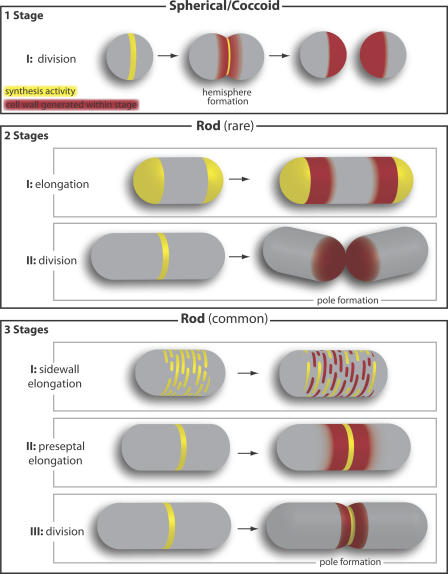
Figure 2.
Cell wall growth patterns. Yellow and red colors show where new cell wall synthesis is occurring and where cell wall material has been inserted within a particular stage, respectively. (top) Spherical (coccoid) bacteria synthesize a new cell wall at division only, when a division septum bisects the cell (left) and is remodeled to form two new hemispheres (middle). Each daughter cell has one new hemisphere produced during division (right). (middle) A few known rod-shaped bacteria (e.g., Corynebacterium) lack MreB and grow from their poles to elongate between rounds of division; thus, they have two stages of growth: elongation and division. During elongation, active wall synthesis at the cell poles leads to lengthening of the cell cylinder. During division, the division septum snaps in two to form two new poles. (bottom) Most rod-shaped bacteria have at least one MreB homologue and can have two elongation stages. The first is characterized by patchy or helical cell wall insertion along the sidewall of the cell cylinder between inert poles. Subsequently, before the onset of division, preseptal elongation occurs in an FtsZ ring–dependent fashion, during which wall synthesis is localized adjacent to the FtsZ ring. Finally, cell division occurs.
FtsZ and localized cell wall synthesis
Division of both cocci and rod-shaped bacteria requires that the FtsZ ring localize cell wall deposition at the site of division (Fig. 2). A growing body of evidence suggests that this occurs through the local control of substrate availability as well as through the indirect recruitment of PBPs. In S. aureus, PBP2 localization at the division site is dependent on substrate recognition, as either covalent modification of its active site or depletion of available substrates causes delocalization of PBP2 from the FtsZ ring (Pinho and Errington, 2005). Similarly, in Streptococcus pneumoniae, the localization of cell wall synthases at the division site depends on the availability of their substrate, which is limited to the midcell (Morlot et al., 2004). In Bacillus subtilis, use of fluorescent vancomycin, which labels both newly incorporated PG material and lipid-linked PG precursors in roughly equal proportions, shows strong septal staining that is dependent on both the FtsZ ring and the division-specific PBP2 (Daniel and Errington, 2003). In Caulobacter crescentus, FtsZ is required for the midcell localization of MurG, which catalyzes the last step in the synthesis of lipid-linked PG precursors (Aaron et al., 2007). Controlling localization of the PBP substrates is apparently not the whole story because a small, catalytically inactive region of E. coli PBP3 containing little more than a transmembrane helix can localize, albeit poorly, to the septum (Wang et al., 1998; Piette et al., 2004; Wissel et al., 2005). Collectively, these data suggest that FtsZ is an essential component of a system that integrates substrate availability and PBP recruitment to localize cell wall synthesis. This spatial control of cell growth by the tubulin homologue FtsZ is fascinatingly reminiscent of the interaction between microtubules and cell wall (cellulose) synthases in plants (Paredez et al., 2006).
The common rod shape
To achieve a rod shape, bacteria add an elongation phase between rounds of division. There are two main strategies that cells use to perform this elongation. One is to localize growth to the poles as in bacteria like Corynebacterium (Fig. 2, middle; Umeda and Amako, 1983; Daniel and Errington, 2003). This strategy may depend on polarly localized proteins such as the coiled coil–rich protein DivIVA (Ramos et al., 2003). The second strategy, which is much more widespread in bacteria, is to halt most PG synthesis after pole formation at the end of division and switch to growth along the cylindrical sidewalls of the cell, keeping the poles relatively inert (Fig. 2, bottom; de Pedro et al., 1997; Daniel and Errington, 2003). It should be noted that cell poles do not become inert immediately after division but appear to become progressively so (Mobley et al., 1984; Rafelski and Theriot, 2006; Tiyanont et al., 2006; Aaron et al., 2007).
The MreB family of bacterial actin homologues has multiple functions (Carballido-Lopez, 2006), including a critical role in controlling cell diameter during cell elongation. Depletion or drug-induced inactivation of MreB leads to a gradual cell widening during elongation, causing a growth-dependent rounding of the cell over time (Jones et al., 2001; Figge et al., 2004; Gitai et al., 2005; Kruse et al., 2005). MreB is only found in nonspherical bacteria (Jones et al., 2001), and some species have multiple MreB homologues (B. subtilis has three: MreB, Mbl, and MreBH), but most have only one (MreB). How MreB homologues regulate cell width during elongation is not well understood. It was initially thought that MreB acted like FtsZ, controlling the localization of cell wall synthesis, because the helix-like pattern of nascent PG in B. subtilis (Daniel and Errington, 2003) mimicked the helical localization of MreB homologues (Fig. 1; Jones et al., 2001; Shih et al., 2003; Figge et al., 2004). However, recent evidence suggests that the localization of nascent PG (Tiyanont et al., 2006) as well as PBP localization patterns (Scheffers et al., 2004; Divakaruni et al., 2005) are largely preserved in cells lacking MreB homologues, although there is the suggestion that MreB is required for the establishment but not the maintenance of PBP2 localization in C. crescentus (Dye et al., 2005).
One intriguing study demonstrated that MreBH of B. subtilis is required for the sidewall-specific localization of a PG hydrolase, LytE (Carballido-Lopez et al., 2006), raising the possibility that the bacterial actin-like cytoskeleton may regulate PG insertion by controlling the availability of hydrolase-generated insertion sites in the existing wall. In C. crescentus, the PG precursor–synthesizing enzyme MurG, which accumulates at the midcell region during preseptal and septal growth (Aaron et al., 2007; Mohammadi et al., 2007), is also found in a patchy pattern along the cell length, and the latter organization appears MreB dependent (Divakaruni et al., 2007). There is also evidence that MreB is in a complex with MurG in E. coli (Mohammadi et al., 2007). The actin-like cytoskeleton may be connected to cell wall enzymes through the membrane proteins MreC, MreD, and RodA (Kruse et al., 2005; Leaver and Errington, 2005), although this coupling may be species specific (Dye et al., 2005). In C. crescentus, MreC is required for the normal patchy pattern of a PG hydrolase, MltA (Divakaruni et al., 2007); collectively, these data hint at the existence of a morphogenetic network that coordinates PG precursor supply, synthesis, and hydrolysis.
Because rod-shaped bacteria depleted of tubulin-like FtsZ do not divide but continue to elongate into long filaments, FtsZ has been canonically viewed as being involved in division but not elongation. However, recent evidence shows that FtsZ can be responsible not only for division but also for a period of cell wall elongation from the central region of the cell (Fig. 2, bottom). In C. crescentus, the FtsZ ring localizes near the midcell well before the onset of division and, by recruiting MurG, spatially localizes the synthesis of PG precursors and directs a substantial portion of cell wall elongation before division (referred to as preseptal elongation; Aaron et al., 2007). Observations in E. coli also support the existence of localized FtsZ ring–dependent wall synthesis before septation (de Pedro et al., 1997). A recent E. coli study also suggests that FtsZ has an additional role in PG synthesis in the pole-proximal lateral sidewalls (Varma et al., 2007).
Adding curvature to the rod
There are many examples of bacteria that add another layer of morphological complexity to the common rod shape to produce a curved rod (vibrioid) or helical shape. There are a few known bacterial strategies for creating a curved shape. The spirochete Borrelia burgdorferi appears to generate a helical shape with periplasmic flagella through an obscure mechanism (Motaleb et al., 2000), and Spiroplasma has no cell wall but produces helical morphology with contractile cytoplasmic fibrils (Trachtenberg, 2004). In both cases, the fibers associated with helical shape are also involved with cellular motility. C. crescentus is the only walled bacterium with a curved rod-shaped morphology for which a cytoskeletal component of the curvature-generating mechanism has been identified thus far. This protein, crescentin, localizes specifically at the inner curvature of cells and shares striking similarities with eukaryotic intermediate filament proteins (Fig. 1, bottom; Ausmees et al., 2003). The consequence of crescentin loss is a straight rod cell shape (Ausmees et al., 2003).
There are little data about how a cytoskeletal structure might produce a curved or helical shape, and, thus, current models are largely theoretical. One model states that a cytoplasmic fiber attached to the cell envelope will produce a curved or helical shape if the filament either actively shortens or grows more slowly than the cell as a whole (Wolgemuth et al., 2005). This concept is borne out by physical modeling, and C. crescentus vibrioid cells, which elongate into helices after an extended time in stationary phase, obey one of the predictions of the model, namely that helical pitch will decrease as cells elongate (Wolgemuth et al., 2005). An alternative hypothesis derives from the popular view that bacterial cytoskeletal elements have little or no mechanical role but are passive scaffolds, or platforms for the localization of proteins that regulate cell wall synthesis. In this model, a filamentous structure like that of crescentin might localize a negative regulator of PG synthesis along only one side of a rod-shaped cell, causing cell wall growth asymmetry between sidewalls and, thereby, curvature toward the shorter cell wall.
Surprisingly, little is known about the mechanical properties of whole cells, which is critical for a better understanding of cell morphogenesis. If mechanically deformed, would bacterial cells be elastic (returning to their original shape like a piece of rubber) or plastic (holding their new shape like a piece of clay)? Intriguingly, it has recently been demonstrated that simple mechanical force can change the growth patterns of bacteria so that they adopt specific morphologies. When E. coli cells are placed in agarose microchambers of defined shapes and cell division is inhibited, the elongating cells take the shape of the chamber in which they were confined (Takeuchi et al., 2005). Remarkably, when cells are released from the chambers, they retain the induced shape (Takeuchi et al., 2005), indicating that the cells act plastically; physical confinement alters the construction of the cell wall to produce a given shape. Continued growth after release leads to a gradual loss of the induced morphology (Takeuchi et al., 2005). This result raises the possibility that cytoskeletal elements may work in a similar manner, exerting local forces within the cell that lead to specific patterns of growth. Future experiments using micromanipulation techniques on single cells will surely be fruitful in exploring this possibility.
More complex shapes
There are many complex bacterial shapes beyond what we have discussed above that remain exciting avenues of research. For instance, Streptomyces coelicolor, which forms a funguslike mycelium of branched hyphae, grows only from the tips of branches (Flärdh, 2003b). Apical growth and morphogenesis seem to depend on localization of the S. coelicolor DivIVA protein at hyphal tips, but its precise function is not yet clear (Flärdh, 2003a). Interestingly, MreB is not essential for vegetative growth in S. coelicolor but is required for the development of aerial hyphae and spores (Mazza et al., 2006). Another instance of a complex shape is the prosthecate bacteria, which produce thin extensions of the cell envelope (referred to as stalks) to increase cell surface area and nutrient uptake. In C. crescentus, the stalk elongates specifically from the base of a cell pole (Schmidt and Stanier, 1966; Aaron et al., 2007). How this occurs is unknown.
The Mollicutes are different from other bacteria in that they have defined shapes while having no cell wall; instead, they rely entirely on internal structures. For instance, the spiral shape of Spiroplasma is produced by a helical cytoskeletal ribbon of protein fibrils that follows the shortest helical path within the cell (Trachtenberg, 2004). Unequal local contraction of the constituent fibrils produces changes in helicity that propel the cell. Similarly, a cytoplasmic structure in Mycoplasma pneumoniae produces the attachment organelle, a fingerlike projection that is important for cell motility (Henderson and Jensen, 2006), but the exact structure and function of the cytoskeleton in these cells is unknown.
Control of cell size
In addition to their geometry, bacteria need to carefully control their size. The placement and frequency of division can alternatively produce long filamentous cells or short rods, curved rods or spirals, and equally or unequally sized daughter cells. The position and timing of cell division must also be regulated to avoid chromosome guillotining or production of anucleate cells. In virtually all bacteria, division is initiated by the formation of a ring of the tubulin homologue FtsZ at the division site (Fig. 1), and, thus, regulation of the location and timing of FtsZ ring formation controls cell division.
There are multiple systems to control FtsZ assembly and ring formation that differ somewhat among species. The nucleoid occlusion system (Woldringh et al., 1990) prevents division at DNA-containing cellular locales. It uses the proteins SlmA in E. coli and Noc in B. subtilis, which bind to DNA and prevent FtsZ polymerization (Wu and Errington, 2004; Bernhardt and de Boer, 2005). The Min system uses an FtsZ assembly inhibitor, MinC, that, through binding to the MinD ATPase, either oscillates (in E. coli) or is tethered to the polar regions (in B. subtilis; Marston et al., 1998; Hu et al., 1999; Raskin and de Boer, 1999). Therefore, the concentration of MinC is lowest at the midcell, favoring FtsZ assembly there. C. crescentus lacks the Min system and instead uses an FtsZ assembly inhibitor called MipZ, which binds to the chromosome-partitioning protein ParB (Thanbichler and Shapiro, 2006). Thus, MipZ follows ParB as the chromosomes separate, clearing MipZ from the central cell region and allowing the FtsZ ring to form (Thanbichler and Shapiro, 2006). Another system was recently uncovered in B. subtilis that couples nutrient availability to cell division (Weart et al., 2007). It uses UgtP, a sugar transferase that acts both in the teichoic acid biosynthesis pathway and as an FtsZ assembly inhibitor, to ensure that fast-growing cells gain sufficient mass before division occurs (Weart et al., 2007). Common to all these systems is a release of FtsZ assembly inhibition at the future division site.
Once established, the FtsZ ring is highly dynamic, with fluorescence recovery half-times of 8–9 s (Anderson et al., 2004), and it recruits several cell division proteins in hierarchical fashion (Margolin, 2005). As the cell divides, the FtsZ ring constricts until cell division is complete. It is unknown whether FtsZ generates a constrictive force itself or is merely compressed as it directs the synthesis of new cell poles. However, FtsZ is also present in wall-less bacteria, suggesting that it can itself generate a force (Wang and Lutkenhaus, 1996). In vitro, FtsZ forms straight protofilaments when GTP is bound and curved filaments when GDP is bound, suggesting that GTP hydrolysis–induced conformational changes may drive cell constriction (Lu et al., 2000), although this remains unproven.
Conclusions and perspectives
When it comes to cell morphogenesis, bacteria must meet the same challenges as walled eukaryotic cells, controlling the timing and location of cell wall synthesis to achieve specific shapes and sizes. Remarkably, similar to eukaryotes, bacteria use a highly regulated internal cytoskeleton for this spatio-temporal control to generate cell shapes ranging from the simple to the complex.
Still, many questions remain to be answered about the particulars of bacterial growth control. The cytoskeletal MreB family proteins are clearly important for cell width control and rod morphology, but how they achieve this function remains poorly understood. The chemical composition and bonding patterns of PG are well known, but the structure of PG has so far been inaccessible to observation techniques. How is the cell wall constructed at the molecular level in cocci, rods, and vibrioid or helical cells? Although many studies have focused on synthesis of the cell wall, few have considered membrane synthesis, which is also required for cell expansion and might be spatially or temporally regulated. A study of B. subtilis suggests that new membranes are primarily synthesized at the division septum (Nishibori et al., 2005), opening the door for further experiments. It is remarkable that cells can grow into various shapes in response to external forces (Takeuchi et al., 2005). How can force alter cell wall growth? Although there is evidence that cytoskeletal structures (e.g., the FtsZ ring) serve as scaffolds for guiding cell wall deposition, can they produce force (and enough of it) to locally alter growth and, thus, shape?
The amazing ability of bacteria to generate, maintain, and transmit different shapes and sizes is intimately connected with a strikingly high degree of cytoplasmic organization, including distinct cytoskeletal elements. Many exciting discoveries surely lie ahead as the workings of bacterial morphology control systems are brought to light.
Acknowledgments
Because of space limitations, we were unable to cite much work that has contributed to this field, and we extend our sincerest apologies to the authors whose work was omitted. We thank the Jacobs-Wagner laboratory for useful comments on the manuscript.
Research in our laboratory is supported by the National Institutes of Health and by the Pew Scholars Program in the Biological Sciences (sponsored by the Pew Charitable Trusts). M.T. Cabeen is also supported by the National Science Foundation Graduate Research Fellows Program.
Abbreviations used in this paper: PBP, penicillin-binding protein; PG, peptidoglycan.
References
- Aaron, M., G. Charbon, H. Lam, H. Schwarz, W. Vollmer, and C. Jacobs-Wagner. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 64:938–952. [DOI] [PubMed] [Google Scholar]
- Anderson, D.E., F.J. Gueiros-Filho, and H.P. Erickson. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees, N., J.R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 115:705–713. [DOI] [PubMed] [Google Scholar]
- Benaissa, M., P. Babin, N. Quellard, L. Pezennec, Y. Cenatiempo, and J.L. Fauchere. 1996. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect. Immun. 64:2331–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, T.G., and P.A. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell. 18:555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E.F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature. 354:161–164. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez, R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70:888–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez, R., A. Formstone, Y. Li, S.D. Ehrlich, P. Noirot, and J. Errington. 2006. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell. 11:399–409. [DOI] [PubMed] [Google Scholar]
- Chan, W.Y., P.K. Hui, K.M. Leung, J. Chow, F. Kwok, and C.S. Ng. 1994. Coccoid forms of Helicobacter pylori in the human stomach. Am. J. Clin. Pathol. 102:503–507. [DOI] [PubMed] [Google Scholar]
- Daniel, R.A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 113:767–776. [DOI] [PubMed] [Google Scholar]
- de Pedro, M.A., J.C. Quintela, J.V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni, A.V., R.R. Loo, Y. Xie, J.A. Loo, and J.W. Gober. 2005. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 102:18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni, A.V., C. Baida, C.L. White, and J.W. Gober. 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66:174–188. [DOI] [PubMed] [Google Scholar]
- Dye, N.A., Z. Pincus, J.A. Theriot, L. Shapiro, and Z. Gitai. 2005. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. USA. 102:18608–18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge, R.M., A.V. Divakaruni, and J.W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332. [DOI] [PubMed] [Google Scholar]
- Flärdh, K. 2003. a. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523–1536. [DOI] [PubMed] [Google Scholar]
- Flärdh, K. 2003. b. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564–571. [DOI] [PubMed] [Google Scholar]
- Gitai, Z., N.A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin- mediated segregation of a specific region of a bacterial chromosome. Cell. 120:329–341. [DOI] [PubMed] [Google Scholar]
- Harold, F.M. 2002. Force and compliance: rethinking morphogenesis in walled cells. Fungal Genet. Biol. 37:271–282. [DOI] [PubMed] [Google Scholar]
- Henderson, G.P., and G.J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60:376–385. [DOI] [PubMed] [Google Scholar]
- Höltje, J.V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA. 96:14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L.J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 104:913–922. [DOI] [PubMed] [Google Scholar]
- Justice, S.S., D.A. Hunstad, P.C. Seed, and S.J. Hultgren. 2006. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. USA. 103:19884–19889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A.L. 1985. How bacteria grow and divide in spite of internal hydrostatic pressure. Can. J. Microbiol. 31:1071–1084. [DOI] [PubMed] [Google Scholar]
- Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78–89. [DOI] [PubMed] [Google Scholar]
- Kurtz, H.D., and D.I. Netoff. 2001. Stabilization of friable sandstone surfaces in a desiccating, wind-abraded environment of south-central Utah by rock surface microorganisms. Journal of Arid Environments. 48:89–100. [Google Scholar]
- Leaver, M., and J. Errington. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 57:1196–1209. [DOI] [PubMed] [Google Scholar]
- Lu, C., M. Reedy, and H.P. Erickson. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182:164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6:862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A.L., H.B. Thomaides, D.H. Edwards, M.E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza, P., E.E. Noens, K. Schirner, N. Grantcharova, A.M. Mommaas, H.K. Koerten, G. Muth, K. Flärdh, G.P. van Wezel, and W. Wohlleben. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60:838–852. [DOI] [PubMed] [Google Scholar]
- Mobley, H.L., A.L. Koch, R.J. Doyle, and U.N. Streips. 1984. Insertion and fate of the cell wall in Bacillus subtilis. J. Bacteriol. 158:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, T., A. Karczmarek, M. Crouvoisier, A. Bouhss, D. Mengin-Lecreulx, and T. den Blaauwen. 2007. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 65:1106–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot, C., M. Noirclerc-Savoye, A. Zapun, O. Dideberg, and T. Vernet. 2004. The D,D-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51:1641–1648. [DOI] [PubMed] [Google Scholar]
- Motaleb, M.A., L. Corum, J.L. Bono, A.F. Elias, P. Rosa, D.S. Samuels, and N.W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA. 97:10899–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibori, A., J. Kusaka, H. Hara, M. Umeda, and K. Matsumoto. 2005. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 187:2163–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez, A.R., C.R. Somerville, and D.W. Ehrhardt. 2006. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 312:1491–1495. [DOI] [PubMed] [Google Scholar]
- Piette, A., C. Fraipont, T. Den Blaauwen, M.E. Aarsman, S. Pastoret, and M. Nguyen-Disteche. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J. Bacteriol. 186:6110–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho, M.G., and J. Errington. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871–881. [DOI] [PubMed] [Google Scholar]
- Pinho, M.G., and J. Errington. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799–807. [DOI] [PubMed] [Google Scholar]
- Rafelski, S.M., and J.A. Theriot. 2006. Mechanism of polarization of Listeria monocytogenes surface protein ActA. Mol. Microbiol. 59:1262–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, A., M.P. Honrubia, N. Valbuena, J. Vaquera, L.M. Mateos, and J.A. Gil. 2003. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology. 149:3531–3542. [DOI] [PubMed] [Google Scholar]
- Raskin, D.M., and P.A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA. 96:4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., and J.V. Höltje. 1994. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J. Biol. Chem. 269:21603–21607. [PubMed] [Google Scholar]
- Scheffers, D.J., L.J. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51:749–764. [DOI] [PubMed] [Google Scholar]
- Schiffer, G., and J.V. Höltje. 1999. Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031–32039. [DOI] [PubMed] [Google Scholar]
- Schmidt, J.M., and R.Y. Stanier. 1966. The development of cellular stalks in bacteria. J. Cell Biol. 28:423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, Y.L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA. 100:7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, S., W.R. DiLuzio, D.B. Weibel, and G.M. Whitesides. 2005. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 5:1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler, M., and L. Shapiro. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 126:147–162. [DOI] [PubMed] [Google Scholar]
- Tiyanont, K., T. Doan, M.B. Lazarus, X. Fang, D.Z. Rudner, and S. Walker. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. USA. 103:11033–11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg, S. 2004. Shaping and moving a spiroplasma. J. Mol. Microbiol. Biotechnol. 7:78–87. [DOI] [PubMed] [Google Scholar]
- Umeda, A., and K. Amako. 1983. Growth of the surface of Corynebacterium diphtheriae. Microbiol. Immunol. 27:663–671. [DOI] [PubMed] [Google Scholar]
- VandenBosch, K.A., N.J. Brewin, and E.L. Kannenberg. 1989. Developmental regulation of a Rhizobium cell surface antigen during growth of pea root nodules. J. Bacteriol. 171:4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, A., M. de Pedro, and K.D. Young. 2007. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 189:5692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, W., M. von Rechenberg, and J.V. Höltje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726–6734. [DOI] [PubMed] [Google Scholar]
- Wang, L., M.K. Khattar, W.D. Donachie, and J. Lutkenhaus. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180:2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and J. Lutkenhaus. 1996. Characterization of the ftsZ gene from Mycoplasma pulmonis, an organism lacking a cell wall. J. Bacteriol. 178:2314–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart, R.B., A.H. Lee, A.C. Chien, D.P. Haeusser, N.S. Hill, and P.A. Levin. 2007. A metabolic sensor governing cell size in bacteria. Cell. 130:335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissel, M.C., J.L. Wendt, C.J. Mitchell, and D.S. Weiss. 2005. The transmembrane helix of the Escherichia coli division protein FtsI localizes to the septal ring. J. Bacteriol. 187:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh, C.L., N.B. Grover, R.F. Rosenberger, and A. Zaritsky. 1980. Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. II. Experiments with Escherichia coli B/r. J. Theor. Biol. 86:441–454. [DOI] [PubMed] [Google Scholar]
- Woldringh, C.L., E. Mulder, J.A. Valkenburg, F.B. Wientjes, A. Zaritsky, and N. Nanninga. 1990. Role of the nucleoid in the toporegulation of division. Res. Microbiol. 141:39–49. [DOI] [PubMed] [Google Scholar]
- Wolgemuth, C.W., Y.F. Inclan, J. Quan, S. Mukherjee, G. Oster, and M.A. Koehl. 2005. How to make a spiral bacterium. Phys. Biol. 2:189–199. [DOI] [PubMed] [Google Scholar]
- Wu, L.J., and J. Errington. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 117:915–925. [DOI] [PubMed] [Google Scholar]
- Young, K.D. 2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70:660–703. [DOI] [PMC free article] [PubMed] [Google Scholar]