Abstract
Pericentric heterochromatin transcription has been implicated in Schizosaccharomyces pombe heterochromatin assembly and maintenance. However, in mammalian systems, evidence for such transcription is inconsistent. We identify two populations of RNA polymerase II–dependent mouse γ satellite repeat sequence–derived transcripts from pericentric heterochromatin that accumulate at different times during the cell cycle. A small RNA species was synthesized exclusively during mitosis and rapidly eliminated during mitotic exit. A more abundant population of large, heterogeneous transcripts was induced late in G1 phase and their synthesis decreased during mid S phase, which is coincident with pericentric heterochromatin replication. In cells that lack the Suv39h1,2 methyltransferases responsible for H3K9 trimethylation, transcription occurs from more sites but is still cell cycle regulated. Transcription is not detected in quiescent cells and induction during G1 phase is sensitive to serum deprivation or the cyclin-dependent kinase inhibitor roscovatine. We demonstrate that mammalian pericentric heterochromatin transcription is linked to cellular proliferation. Our data also provide an explanation for inconsistencies in the detection of such transcripts in different systems.
Introduction
The centromeres of eukaryotic chromosomes are flanked by pericentric heterochromatin that is highly variable between species in size and repetitive DNA sequence composition but remarkably conserved in chromatin protein composition and structure from fission yeast to humans (Huisinga et al., 2006). Pericentric heterochromatin structure is essential for accurate chromosome segregation during mitosis (Peters et al., 2001; Pidoux and Allshire, 2004) and is similar in composition to constitutive heterochromatin found at other chromosome regions that also contain repetitive sequences and transposable elements, where it functions to silence transcription, reduce the frequency of recombination, and promote long-range chromatin interactions (Jia et al., 2004; Grewal and Elgin, 2007). Heterochromatin is composed of regular tightly packed arrays of hypoacetylated nucleosomes that are methylated at lysine 9 of histone H3 (MeK9H3), mediated by the Su(VAR)3-9 histone methyltransferases (Clr4 in fission yeast and Suv39h1,2 in mammals). MeK9H3 recruits the heterochromatin protein 1 (HP1) family of proteins (Swi6 in fission yeast), which in turn recruit Su(VAR)3-9 as part of a complex self-reinforcing network of proteins that are enriched at heterochromatic loci (Huisinga et al., 2006; Grewal and Elgin, 2007; Grewal and Jia, 2007). Although species-specific differences exist for some components of this network, the overall conservation of heterochromatin structure and function suggests that detailed mechanistic insights gained from experiments in fission yeast and flies will also apply to mammals.
Paradoxically, although constitutive heterochromatin functions to silence transcription, in fission yeast it has been shown that transcription from within pericentric heterochromatin is required for the formation and maintenance of heterochromatin and for sister chromatid cohesion (Kato et al., 2005; Grewal and Jia, 2007). Transcripts generated by RNA polymerase II are processed into siRNA that is in turn recognized by an RNAi-induced transcriptional silencing complex that is recruited to and required for heterochromatin assembly and gene silencing (Huisinga et al., 2006; Grewal and Elgin, 2007; Grewal and Jia, 2007). The RNAi pathway is also required for the formation of heterochromatin and silencing of repetitive sequences in Drosophila melanogaster (Grewal and Elgin, 2007). In mammalian cells, an unidentified RNA component is required for the association of HP1 with pericentric heterochromatin (Maison et al., 2002; Muchardt et al., 2002). However, mammalian homologues to certain key components of the fission yeast transcription–mediated gene silencing network have not been identified (Huisinga et al., 2006; Zaratiegui et al., 2007). Moreover, attempts to detect transcription from mammalian pericentric heterochromatin have met with varied levels of success, with discrepancies found both in the ability to detect such transcripts and the sizes of any transcripts detected (Harel et al., 1968; Flamm et al., 1969; Cohen et al., 1973; Maio and Kurnit, 1974; Gaubatz and Cutler, 1990; Rudert et al., 1995; Lehnertz et al., 2003; Rizzi et al., 2004; Cobb et al., 2005; Kanellopoulou et al., 2005; Martens et al., 2005; Murchison et al., 2005; Valgardsdottir et al., 2005).
One possible explanation for these inconsistencies is that the transcription of satellite DNA could be cell cycle regulated, making it difficult to detect in asynchronously growing cells or tissues in which most cells are not cycling. In fact, cell cycle regulation of heterochromatin transcription could provide a logical means to drive the reassembly of heterochromatin after the disruptive processes of DNA replication and mitosis, which might not be necessary in a quiescent cell. Here, we show that different types of RNA polymerase II–transcribed RNA species are synthesized from the AT-rich mouse γ (major) satellite repeat sequences at different times during the cell cycle: a small species induced specifically during mitosis and a large heterogeneous set of RNAs induced during late G1 and early S phase. Both were short lived and dependent on the passage of cells through the restriction point.
Results
Different RNA species corresponding to mouse γ satellite pericentric heterochromatin are detected at specific cell cycle stages
To examine satellite transcription during the cell cycle, mouse C127 cells were synchronized by selective detachment during mitosis and released into G1 phase for up to 7 h, at which time 5–10% of cells begin to enter S phase (Fig. 2 C; Gilbert and Cohen, 1987). To monitor S phase progression, a portion of mitotic cells were arrested at the G1/S boundary in the presence of the DNA polymerase inhibitor aphidicolin for 10–12 h and released into S phase for an additional 20 h. Total RNA from various time points was then isolated and analyzed by Northern blot hybridization using a mouse γ satellite DNA probe. As shown in Fig. 1 (A–C), molecules smaller than 200 nt were detected specifically in mitotic cells and were undetectable by 1 h after mitosis. These are smaller than the size of the γ satellite repeat (234 bp). When small RNAs were selectively enriched before Northern hybridization, hybridization signals were detected almost exclusively during mitosis (Fig. 1, D and E). Later in G1 phase, a more heterogeneous set of RNAs were detected that were mainly larger than 1 kb, which is consistent with previous papers (Gaubatz and Cutler, 1990; Rudert et al., 1995). These accumulated gradually during the course of G1, reaching a peak in late G1/early S phase, after which the amount of detectable RNA was substantially reduced but still higher than during early G1.
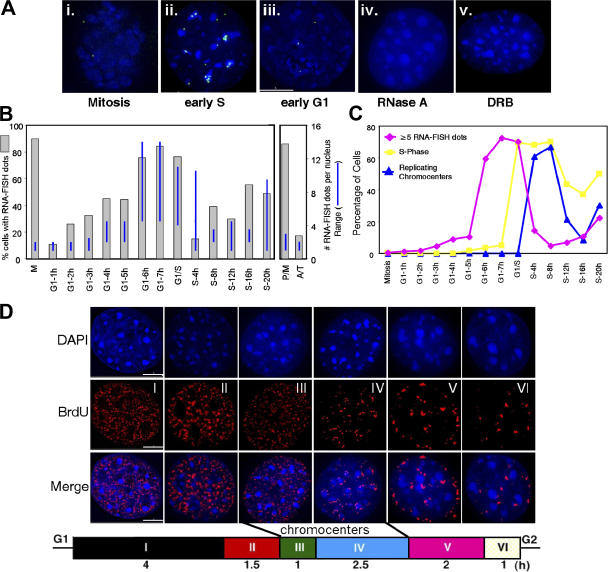
Figure 2.
RNA-FISH analysis of γ satellite transcription during the cell cycle. Aliquots of cells from Fig. 1 were subjected to RNA-FISH analysis with a γ satellite probe as described in Materials and methods. (A) Single z-section images of hybridized nuclei (spots that do not appear to overlap DAPI are above or below chromocenters in another plane). DNA (DAPI), blue; RNA-FISH, green. Control hybridizations were performed after treatment of early S phase nuclei with RNase A (iv) or treatment of cells for 1 h with DRB (v). No FISH signals were detected in any cells from either of these controls. (B) Quantification of the percentage of cells with any detectable RNA-FISH signal and the range (lowest to highest) in the number of transcription sites (RNA-FISH foci) per nucleus. Because M phase cells had been briefly treated (4 h) with nocodazole before mitotic shakeoff, we separately evaluated the presence of RNA-FISH foci during mitosis in asynchronously growing cell populations and quantified their presence in cells that were clearly in prophase or metaphase (P/M) versus anaphase or telophase (A/T). (C) The percentage of cells from B with at least five RNA-FISH foci per cell is plotted. Aliquots of these same cells were pulse labeled with BrdU and stained with anti-BrdU antibodies (and DAPI) to determine the percentage of cells in S phase (BrdU positive). Using the spatial patterns of BrdU labeling from these cells, as shown in D, we calculated the percentage of S phase (BrdU positive) cells that are engaged in replicating chromocenters, which contain γ satellite DNA. (D) Six spatial patterns of DNA synthesis can be distinguished in mouse fibroblasts representing different stages of S phase, as previously described in detail (Wu et al., 2005). DNA is stained with DAPI, and sites of DNA synthesis are visualized by indirect immunofluorescence with an antibody specific to BrdU-substituted DNA. Images have been deconvolved and a merge of the BrdU and DAPI staining patterns is shown to illustrate the two stages during which cells are engaged in the replication of chromocenters, used to score γ satellite replication in C and Fig. 3. A schematic of the length of time that C127 cells spend in each stage of S phase is given at the bottom (adapted from Wu et al., 2006a). Experiments were repeated for three independent synchronizations. At least 150 cells were counted for each time point. Bars, 5 μm.
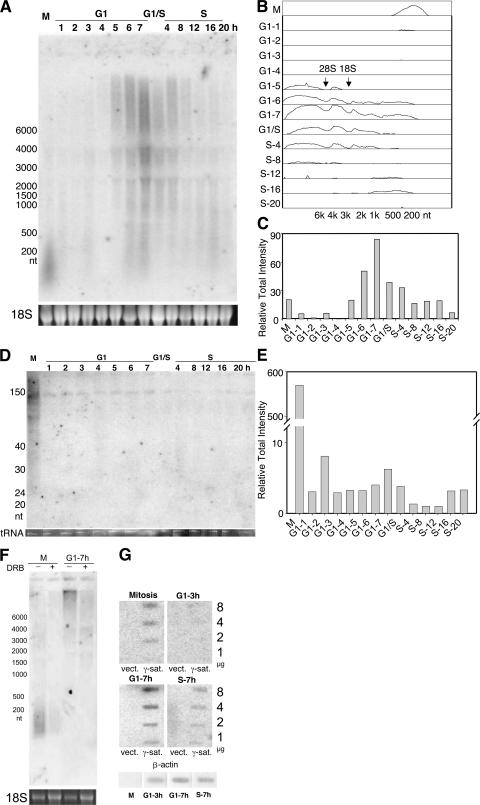
Figure 1.
Northern analysis of γ satellite transcripts during the cell cycle. (A) Mouse C127 cells were synchronized by mitotic selection and released into G1 phase. One group of cells was collected at hourly intervals thereafter, and a second group was treated with 10 μg/ml aphidicolin at G1-5 h for 10–12 h and released into S phase for 20 h. Cells were collected at the indicated time points. Total RNA was isolated and subjected to Northern hybridization using a γ satellite probe (see Materials and methods). 18S ribosomal RNA is shown at the bottom as loading control. (B) Densitometric traces of signal intensities for each lane in A after background subtraction. The positions of 18 and 28S ribosomal RNA are shown in the G1-5 lane. (C) Quantification of the total intensity of hybridization signal per lane for each sample in A. Similar results were obtained in three independent synchronization experiments. (D) Small RNA (<200 nt) was isolated from aliquots of the same cell populations and subjected to PAGE and Northern analysis using the same probe as in A. (E) Quantification of the total hybridization of the small RNA signals, as in C. Similar results were obtained in two independent experiments. (F) M phase (M) or G1-7 h cells were collected with or without prior DRB treatment (1 h), and Northern hybridization was performed as in A. (G) Nuclear run-on analysis of transcription. Labeled nascent RNA from nuclear run-on reactions with permeabilized cells synchronized at mitosis, G1-3 h, G1-7 h, and S-7 h were hybridized to the same major satellite plasmid (γ-sat.) used as a probe in A–F. The corresponding empty plasmid vector (vect.) was used as a negative control. Plasmids were immobilized on nylon filters using a slot blot at quantities of 1, 2, 4, and 8 μg per slot as indicated. As a positive control, 2 μg of a purified PCR product corresponding to the mouse β-actin gene was also hybridized with the same labeled nascent RNA. Similar results were obtained in two independent experiments.
To confirm the short half-life of these transcripts, we examined their sensitivity to the RNA polymerase II inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Fig. 1 F). DRB added for as little as 1 h strongly reduced levels of both the small transcripts during mitosis and the large transcripts at 7 h into G1 phase, confirming that both species have a relatively short half-life. To determine whether the relative abundance of these transcripts at different times during the cell cycle reflects their de novo transcription rates, we evaluated the synthesis of nascent transcripts from preengaged RNA polymerase complexes using nuclear run-on assays (Fig. 1 G). Transcription from γ satellite DNA was strongest during late G1 phase and was also detected in late S phase and mitosis, but was virtually undetectable during early G1 phase. In contrast, transcription from the β-actin gene could be detected at all times except mitosis. In fact, most transcription is silenced during mitosis by phosphorylation and the eviction of transcription factors (Prasanth et al., 2003). Together, these results demonstrate that small heterochromatic RNAs are synthesized de novo during mitosis and not processed from transcripts synthesized before mitosis, although they could be processed from larger transcripts synthesized during mitosis.
The short half-life for detection of both the mitotic and late G1/early S phase transcripts could be caused by the rapid degradation of the RNA or rapid modification of the transcripts in ways that prevent their detection by hybridization, such as RNA editing (Stuart and Panigrahi, 2002; Samuel, 2003). The adenosine-rich and potentially double-stranded (Kanellopoulou et al., 2005) transcripts produced from γ satellite DNA would make excellent substrates for hydrolytic deamination of adenosine residues to inosine residues by double-stranded RNA–specific adenosine deaminases. In fact, vigilin, a component of an adenosine deaminase acting on RNA complex, appears to colocalize with dense chromatin in monkey COS7 cells and, when overexpressed, associates with pericentric satellite sequences in human HEK293T cells (Wang et al., 2005). However, immunolocalization of vigilin with two independent antibodies revealed no colocalization of vigilin with DAPI-dense pericentric heterochromatin clusters (chromocenters) at any time during the cell cycle (unpublished data). Moreover, we sequenced RT-PCR products amplified with degenerate primers or primers designed against γ satellite regions that were unlikely to be affected by editing (Zhang and Carmichael, 2004). 10 different products from M phase, G1/S phase, and asynchronous cells were identical to the original γ satellite sequence (unpublished data). From these experiments, we conclude that A-to-I editing of γ satellite transcripts is not a major contributor to the rapid loss in detection of the mitotic transcripts.
Cell cycle regulation of the number of discrete transcription sites
To confirm these results using an alternative method, we used RNA-FISH. RNA-FISH detects nascent transcripts as they are produced at the site of transcription (Levsky et al., 2002; Osborne et al., 2004) and accurately reflects results obtained with the more laborious nuclear run-on method (Becker et al., 2002). RNA-FISH signals hybridizing to the mouse satellite probe were detected on the outer surface of chromocenters (Fig. 2 A, i–iii), which are easily visualized with a DAPI stain (Wu et al., 2006a). No sites were detected with a control probe that did not contain γ satellite sequences (unpublished data). Detection of these sites was completely abolished by treatment of nuclei with RNaseA (Fig. 2 A, iv), demonstrating that they did not result from unintentional DNA denaturation. Treatment of cells with DRB for 1 h before collection resulted in a complete inhibition of detectable RNA-FISH signals (Fig. 2 A, v). These controls demonstrate that the signals detected by RNA-FISH represent nascent RNA transcripts originating from γ satellite DNA within pericentric heterochromatin.
The number of transcription sites detected per cell was highly heterogeneous, ranging from 0 to >15. Hence, we quantified both the percentage of positive cells as well as the number of transcription sites per cell at each cell cycle stage (Fig. 2 B). During mitosis (Fig. 2 B; M, metaphase; P/M, prophase and metaphase), ∼90% of cells had one to three sites of transcription. This could be an underestimate because the signal intensity per site was weaker in mitotic cells (relative to later times in the cell cycle), possibly caused by the small size of the RNA during mitosis (Fig. 1). The percentage of positive cells dropped considerably during mitotic exit (Fig. 2 B; A/T, anaphase and telophase), and by early G1 phase, <10% of cells displayed one to four intermediate intensity transcription sites, which is consistent with the lack of detectable transcripts by Northern analysis (Fig. 1). The percentage of positive cells, the number of transcription sites per cell, and the intensity of each site all increased in late G1 and early S phase, followed by a dramatic drop by 4 h in S phase, with only ∼15% cells showing a strong FISH signal. As cells progressed toward the end of S phase, the number of positive cells began to increase again, but with fewer numbers of sites per cell, indicating that a low level of de novo transcription continues into late S phase. The variable increase in detectable sites per cell at 20 h may represent entry of cells into the subsequent cell cycle.
To simplify the distinction between high and low levels of transcription, we estimated the number of cells carrying out the late G1/early S phase mode of satellite DNA transcription by quantifying cells that have an early S phase number of detectable transcription foci (five or greater). This plot (Fig. 2 C, pink) resembles the Northern quantification shown in Fig. 1 C. To monitor the progression of these cells through S phase, cells were labeled with BrdU just before collection for RNA-FISH, and aliquots were stained with anti-BrdU antibodies (and DAPI). These results revealed that transcriptional induction clearly occurred before the onset of S phase and was down-regulated during mid S phase (Fig. 2 C, yellow).
We have previously shown that replication of mouse chromocenters takes place during mid S phase (Wu et al., 2006a), close to the time at which satellite DNA transcription decreases. Cells engaged in chromocenter replication can be easily scored because of the prominent intranuclear appearance of the DAPI-stained chromocenters (Fig. 2 D). Replication begins at the periphery of the chromocenters (Fig. 2 D, III) followed by a period during which virtually all DNA synthesis in the cell consists of chromocenter replication (IV; Guenatri et al., 2004; Wu et al., 2006a). When the percentage of BrdU-positive cells engaged in the replication of chromocenters (Fig. 2 D, III and IV) was quantified in the same cell populations used for Fig. 2 C (yellow), a sharp increase in their number was seen within the same 4-h period as the decrease in transcription of γ satellite DNA within the chromocenters (Fig. 2 C, blue).
Down-regulation of γ satellite transcription is coincident with replication of pericentric heterochromatin
The results in Fig. 2 C suggest that γ satellite transcription may be down-regulated upon chromocenter replication. To investigate this possibility, we repeated the experiments shown in Fig. 2 with more precise S phase time points, starting from the G1/S border through 7 h into S phase. These results (Fig. 3, A and B) revealed a sharp decrease in the percentage of cells positive for transcription between 3 and 4 h, which coincides with a sharp increase in cells replicating chromocenters. However, there were two concerns with these BrdU/RNA-FISH experiments. First, because the denaturation step necessary to reveal BrdU incorporation is incompatible with RNA-FISH detection, it was necessary to quantify each property in separate cell samples. Second, we wanted to rule out the possibility that the cell-synchronizing agent aphidicolin may have affected the results. Hence, to visualize replication of pericentric heterochromatin and transcription of satellite RNA simultaneously within individual asynchronously growing cells, we combined RNA-FISH with immunolocalization of the replication fork protein proliferating cell nuclear antigen (PCNA). After elimination of the soluble pool of PCNA that is not engaged in DNA synthesis (Dimitrova and Gilbert, 2000), PCNA staining patterns resembled BrdU patterns throughout S phase (Fig. 3 C), as was expected (Leonhardt et al., 2000). Hence, cells in G1 phase could be identified by their small, PCNA-negative nuclei, cells at different stages of S phase could be identified by their PCNA staining pattern (Fig. 3 C, I–VI), and cells in G2 phase could be identified as large PCNA-negative cells. PCNA and RNA-FISH signals did not colocalize throughout almost the entire duration of S phase (Fig. 3 C), with the exception of 16% of cells in very late S phase (VI), for reasons that are not understood.
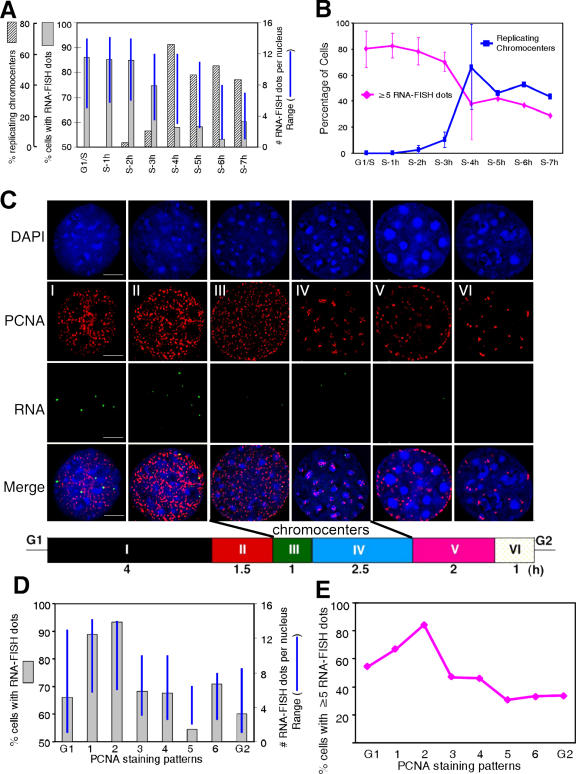
Figure 3.
Chromocenter replication coincides with down-regulation of γ satellite transcription. (A and B) C127 cells were synchronized at the G1/S boundary and released for the indicated time intervals. Cells were pulse labeled with BrdU for 30 min before collection and subjected to RNA-FISH and BrdU staining as in Fig. 2. (A) The percentage of cells replicating chromocenters, the percentage displaying any detectable RNA-FISH signals, and the range of RNA-FISH foci per cell were plotted as in Fig. 2 B. (B) The percentages of cells with at least five sites of transcription (RNA-FISH foci) and of replicating chromocenters (III and IV) were scored as in Fig. 2 C. Shown are the combined data from two independent experiments in which cells were collected at hourly intervals for either 4 or 7 h after release into S phase. More than 100 cells were counted for each time point in each experiment. The error bars represent the SD of two experiments. (C) Asynchronously growing cells were subjected to RNA-FISH with a γ satellite probe as in Fig. 2, and subsequently stained with fluorescent anti-PCNA antibodies. Shown here are deconvolved single z-section images. Simultaneous visualization of PCNA and RNA-FISH signals allows direct quantification of transcription during each stage of S phase without the need for synchronization. Cells in each stage of S phase were clearly identified, with PCNA patterns defined as BrdU patterns were in Fig. 2 D. G1 or G2 cells could be identified as smaller or larger PCNA-negative nuclei, respectively. A schematic of the length of time that C127 cells spend in each stage of S phase is shown at the bottom (adapted from Wu et al., 2006a). Bar, 5 μm. (D) The percentage of cells from C with any number of RNA-FISH signals and the range in number of signals per cell were scored and plotted as in Figs. 2 B and 3 A. Triton extraction removed most mitotic cells from the slide. (E) The percentage of cells with at least five RNA-FISH foci for each was scored as in Figs. 2 C and 3 B. At least 100 cells were counted for each stage, except PCNA patterns III and VI, which are the shortest periods, for which at least 50 cells were scored. Three independent repeats gave similar results.
As shown in Fig. 3 (D and E), transcription of γ satellite is considerably higher in early S phase and decreases starting with the onset of chromocenter replication (III). Moreover, the percentage of cells with more than five sites of γ satellite transcription increases from G1 to early S phase and then decreases at the time of chromocenter replication. These results confirm a general incompatibility between γ satellite transcription and replication during S phase, similar to what has been observed for individual sites of replication and transcription throughout S phase (Wei et al., 1998). It is possible that the reduction in transcription is exclusively caused by interference of replication with transcription. However, only a subset of pericentric regions are engaged in replication at any particular moment in time (Wu et al., 2006a), so it is unlikely that replication is simultaneously interfering with transcription of all pericentric regions.
Cell cycle regulation of γ satellite RNA is independent of Suv39h1,2-dependent epigenetic modifications
Suv39h1,2 is responsible for the trimethylation of lysine 9 of histone H3 (Me3K9H3) at pericentric heterochromatin in mice (Peters et al., 2001). In Suv39h1,2 double knockout mouse embryonic fibroblast (MEF) cells, Me3K9H3 is lost, HP1 dissociates, DNA methylation is drastically reduced, and the trimethylation of histone H4 lysine 20 (Me3K20H4) is lost (Peters et al., 2001; Lehnertz et al., 2003; Schotta et al., 2004; Kourmouli et al., 2005). These cells show karyotypic instability and elevated steady-state levels of γ satellite transcripts (Peters et al., 2001). Because these prior experiments were performed on asynchronously growing cells, the accumulation of γ satellite transcripts could have resulted either from elevated transcription rates or a disruption of cell cycle regulation resulting in transcription throughout the entire cell cycle.
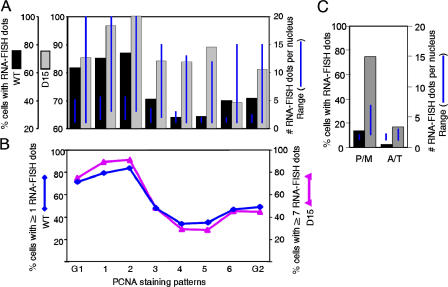
To distinguish between these possibilities, we performed PCNA/RNA-FISH staining in wild-type (WT) versus Suv39h1,2 double knockout (D15) MEFs, as described in Fig. 3. Although D15 had a substantially higher percentage of cells transcribing γ satellite DNA from considerably more sites than WT cells, both cell lines showed an increase in transcription transitioning from G1 to early S phase and a decline in transcription upon replication of chromocenters (Fig. 4 A), which is similar to C127 cells (Fig. 3 D). Mitotic transcription was also elevated in D15 (Fig. 4 C). To compare the percentage of cells transcribing γ satellite transcripts at late G1/early S phase levels, as was done for C127 cells in Fig. 3 E, we adjusted our criteria for the number of RNA-FISH foci per cell to reflect the relatively low level of transcription in WT MEFs (more than one site per nucleus) and the higher level of transcription in the Suv39dn1,2, double knockout cells (more than seven sites per nucleus). When the percentage of cells meeting these criteria was scored, it revealed a clear reduction in the number of highly transcribing cells upon chromocenter replication (Fig. 4 B).
Figure 4.
Cell cycle regulation of γ satellite transcription in WT and Suv39h1,2 double knockout MEFs. Asynchronously growing WT and Suv39h1,2 double null cell line (D15) MEFs were subjected to PCNA and γ satellite RNA-FISH analysis as in Fig. 3. (A) The percentage of cells with RNA-FISH signals and the range of signals per cell were scored and plotted as in Fig. 3. (B) The percentage of cells with at least one (WT) or seven (D15) sites of γ satellite transcription revealed as RNA-FISH foci were scored as in Fig. 3. (C) RNA-FISH foci were scored in prophase/metaphase (P/M) and anaphase/telophase (A/T) cells as in Fig. 2 B.
We conclude that the elevated γ satellite transcript levels detected in D15 result from transcription taking place simultaneously at an increased number of sites on mouse chromocenters, rather than from elevated transcription from a similar number of sites or a disruption of cell cycle regulation. Moreover, the increased number of sites did not appear to result from a disruption of centromere clustering because the size and number of chromocenters was similar in WT versus D15 (unpublished data). Hence, cell cycle regulation of γ satellite transcription is independent of the Suv39h1,2-related features of heterochromatin.
Transcription of γ satellite requires activation of Cdk and passage through the restriction point
The very low levels of transcription during early G1 phase raised the intriguing possibility that transcription of pericentric heterochromatin might require passage through the restriction point and commitment to cell division. Hence, we examined cells that were arrested in G0 by contact inhibition. For all cell lines (C127, WT, and D15), very little transcription could be detected in arrested cells (Fig. 5 A). To distinguish whether long-term arrest in quiescence resulted in transcription down-regulation or if transcription was not induced because cells were prevented from passing through the restriction point, C127 cells were synchronized in mitosis as in Fig. 1 and released into G1 phase in the presence of various concentrations of serum in the medium or into a complete medium to which the Cdk inhibitor roscovatine was added 2 h after release into G1 phase. All cell populations were then allowed to proceed to 7 h after mitosis, when substantial up-regulation of γ satellite transcription was observed in control cells (Fig. 5 B). Both serum deprivation and roscovitine treatment severely inhibited γ satellite transcription. We conclude from this experiment that transcription of mouse pericentric heterochromatin is dependent on passage through the restriction point.
Figure 5.
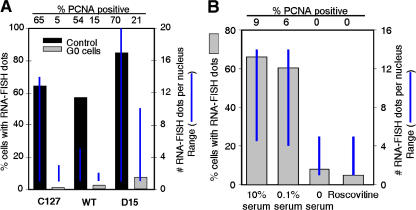
Transcription from pericentric heterochromatin is proliferation dependent. (A) C127 cells, MEFs, and Suv39h1,2 double null MEFs (D15) were rendered quiescent by contact inhibition and subjected to PCNA immunofluorescence and RNA-FISH with a γ satellite probe, along with asynchronously growing control cells. Because there was variability in the response of the different cells to contact inhibition, we focused on cells that were negative for PCNA in all populations (including asynchronous cells). The percentage of PCNA-negative cells with any number of RNA-FISH signals for contact-inhibited (gray) or control cells and the range of signals per cell were scored and plotted as in Fig. 3. (B) C127 cells were synchronized in mitosis by shakeoff as in Fig. 1 and released into G1 phase for 7 h with the indicated concentrations of serum or in 10% serum with roscovitine added at 2 h after release. RNA-FISH was quantified as in Fig. 2 C and shows the percentage positive and number of transcription sites per nucleus. For both A and B, the percentage of PCNA-positive cells are indicated above each graph. Two independent experiments gave the same results.
Discussion
We show that at least two different populations of RNA molecules are expressed from mouse pericentric heterochromatin at different times during the cell cycle. Transcription was Cdk dependent, indicating that cells do not synthesize these transcripts until after they commit to proliferation. Moreover, the transcripts were short-lived. Together, our results provide a satisfying explanation for why such transcripts were not detected in many studies that examined quiescent or slowly growing tissue but were found in tissues that contain proliferating cells. Moreover, they provide evidence for provocative links between heterochromatin and cellular proliferation that warrant further investigation.
Genesis and functions of pericentric transcripts
Although we cannot rule out the possibility that these heterochromatic transcripts result from cryptic transcription, possibly because of a cell cycle–specific change in chromatin structure, an alternative possibility is that there are specific functional promoters within the γ satellite repeats. In fact, specific 300-bp, GC-rich non–γ satellite DNA sequence motifs are peppered within the mouse γ satellite repeats (Kuznetsova et al., 2006). Moreover, transcription factors YY1 (Shestakova et al., 2004) and C/EBPα (Liu et al., 2007) appear to bind to DNA sequences within the mouse major satellite, and in the case of YY1, this interaction is proliferation dependent. Such promoters need not be abundant. Indeed, relatively few sites of transcription occur within pericentric heterochromatin at any moment in time, and given the large fraction of genomic DNA that corresponds to γ satellite DNA (∼5%; Waring and Britten, 1966; Prashad and Cutler, 1976), even the induced levels of transcription are not robust (Fig. 1 G).
Cell cycle regulation of both late G1/S phase and mitosis-specific transcripts was independent of Suv39h1,2. Hence, although our results do not address the role of these RNAs, they suggest that transcription is upstream of Suv39h1,2 and has the potential to drive heterochromatin formation during the cell cycle. It is tempting to speculate that transcription during S phase and mitosis might assist with the reassembly of some structural components of heterochromatin that are disrupted during these phases of the cell cycle. Mammalian heterochromatin replicates late during S phase of the cell cycle, and late replication seeds the assembly of hypoacetylated chromatin (Zhang et al., 2002). The events occurring at the replication fork likely contribute to the propagation of heterochromatin structure, which in turn may dictate late replication in the following cell cycle, thus forming a self-reinforcing loop (Wu et al., 2006a). It is possible that transcription after cells commit to DNA replication is somehow involved in preparing heterochromatin for reassembly at the replication fork. Although fission yeast pericentromeric heterochromatin is replicated early in the cell cycle (Kim et al., 2003), there is no a priori reason why a similar mechanism couldn't be operating at a different time during S phase.
The vast majority of transcription is shut down in mitosis because of the eviction of transcription factors (Prasanth et al., 2003), making the mitotic transcription of heterochromatin a particularly intriguing finding. What role if any such transcripts might play during mitosis is difficult to imagine; however, there may be a renewed requirement to reinforce heterochromatin structure during the late stages of mitosis when most cohesin has been removed (Dai et al., 2006). It is also possible that the eviction of one or more factors from heterochromatin allows for its transcription. In fact, HP1 is evicted from heterochromatin during mitosis (Fischle et al., 2005; Wu et al., 2006a), and an RNA component is required to tether HP1 to pericentric heterochromatin (Maison et al., 2002), so it is possible that mitotic heterochromatin transcription is induced by HP1 loss and/or assists in the reloading of HP1, which occurs during anaphase (Wu et al., 2006a). Mitotic transcription may also assist in the maintenance of centromere structure, as it has recently been shown that interactions with a single-stranded RNA are required for the integrity of kinetochore structure during mitosis in human cells (Wong et al., 2007). Finally, a more speculative possibility is that these RNAs may be components of the RNA helicase p68 and CENP-B–containing interchromosomal connections during mitosis (Kuznetsova et al., 2007).
In short, it is now of considerable importance to identify the promoter elements involved in regulating the transcription of both the S phase and mitotic transcripts and the functional consequences of perturbing this regulation.
Fission yeast and mammals: similarities and differences
Despite the conservation of most heterochromatin structural components from fission yeast to mammals, a requirement for transcription in the assembly of mammalian heterochromatin has been difficult to ascertain. In addition to inconsistent detection of γ satellite transcription, key components of the fission yeast posttranscriptional silencing machinery have not been detected (Huisinga et al., 2006). Although Dicer mutants in mice exhibit elevated levels of γ satellite transcripts (Fukagawa et al., 2004; Kanellopoulou et al., 2005), this has no consequence on histone or DNA methylation in heterochromatin (Murchison et al., 2005). Elevated levels of γ satellite transcripts detected in Suv39h1,2 knockouts could be interpreted as resulting from the disruption of repressive heterochromatin. However, we also see intermediate levels of these transcripts in C127 cells that have apparently normal pericentric heterochromatin (Wu et al., 2005). The elevated transcription in C127 cells may be a consequence of the more rapidly proliferating state of C127 cells, but this could also reflect transient changes in heterochromatin structure that might occur during the cell cycle.
Transcription of both species of RNA described here is mediated by RNA polymerase II, which is similar to heterochromatin transcription in fission yeast. However, we do not find evidence for siRNA-sized molecules at any time during the cell cycle, suggesting that if the RNA species we discuss here are involved in heterochromatin structure, important differences with the fission yeast system must exist. One notable difference is the apparent lack of an RNA-dependent RNA polymerase in mammals (Huisinga et al., 2006) that could amplify and maintain heterochromatic RNA after transcription, as in fission yeast. Because the mammalian transcripts have a short half-life and we find no evidence for their editing to undetectable forms, either there is some transcription throughout the cell cycle that has gone undetected in our experiments or, unlike fission yeast, these transcripts are only required transiently, perhaps to initiate rather than to maintain heterochromatin. It is now of considerable interest to know if fission yeast heterochromatin transcription mediated by RNA polymerase II is under cell cycle control, which could provide a novel direction with which to investigate parallels between fission yeast and mammalian heterochromatin.
Materials and methods
Cell synchronization
Mouse C127 cells were synchronized in mitosis by mechanical shakeoff after a brief and fully reversible nocodazole treatment (Sigma-Aldrich) as described previously (Wu et al., 1997). Similar results were obtained in experiments repeated without the use of nocodazole. For G1/S synchronization, 10 μg/ml aphidicolin (Calbiochem) was added 5 h after release from mitosis for an additional 10–12 h. Where roscovitine (Calbiochem) was used, 40 μM was added at 2 h after mitosis. For serum deprivation, mitotic cells were plated directly into a medium containing either 0.1% or no serum. For contact inhibition, cells were further cultured for 7 d after reaching confluence with fresh media every day. For BrdU pulse labeling, 15 μg/ml BrdU (Sigma-Aldrich) was added to medium for 30 min before fixation.
Northern hybridization and nuclear run-on
Total RNA was prepared using an mirVana microRNA isolation kit (Ambion) and treated with DNase (Promega). In parallel, <200 nt RNA (small) fractions were separated from total RNA using the same kit. To rule out any possibility of DNA contamination in our samples, we performed RT-PCR analysis using γ satellite–specific primers. Only reverse transcribed samples gave ladderlike PCR bands, and RNase A (Sigma-Aldrich) treatment completely eliminated the product. Total RNA was resolved via electrophoresis with a denaturing agarose gel, whereas <200 nt fractions were resolved with a denaturing 15% polyacrylamide gel. RNAs were then transferred to a nylon membrane. The γ satellite probe was plasmid pγSat (Lundgren et al., 2000) containing eight copies of the 234-bp repeat as a template (provided by N. Dillon, Imperial College London, London, UK), which was labeled with α-[32P]dATP using a random labeling kit (Invitrogen). Total and small RNA hybridization was done at 60 and 25°C, respectively. Nuclear run-on with equal numbers of cells (10 million) was performed as described previously (Sasaki et al., 2006), except that cells were permeabilized with digitonin (Sigma-Aldrich) as described previously (Wu et al., 1997) to maintain the integrity of mitotic chromosomes and allow detection of transcription during mitosis.
RNA-FISH and immuno–RNA-FISH
The RNA-FISH procedure was performed as described previously (Tam et al., 2002), using cells that were either grown on coverslips or centrifuged from suspension onto coverslips using a cytocentrifuge (Cytospin 2; Shandon). In brief, cells were washed with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 10 mM Pipes, pH 6.8), followed by CSK + 0.5% Triton X-100 (Sigma-Aldrich) permeablization for 5 min and 4% PFA fixation for 10 min on ice, and then stored in 70% ethanol at −20°C for no longer than 2 d. Slides were hybridized with a digoxigenin (Roche Applied Science) nick-translated γ satellite probe overnight at 37°C followed by fluorescent antibody detection as described previously (Li et al., 2001). In the case of immuno–RNA-FISH, PCNA staining, and RNA-FISH, PCNA immunostaining was performed using a monoclonal PCNA antibody (Oncogene Research Products) after RNA-FISH detection. For RNase A treatment, cells were treated with RNase A after permeabilization and before fixation.
Microscopy
Images were captured with an image restoration microscope system (DeltaVision; Applied Precision) attached to a fluorescence microscope (IX-71; Olympus) equipped with an oil objective lens (PlanApo 60×, 1.42 NA; Olympus) using a charge-coupled device camera (Coolsnap HQ; Photometrics) at RT. Approximately 40 optical sections (with 0.2-μm spacing) were taken and enhanced using the SoftWorx (Applied Precision) constrained iterative deconvolution process.
Immunostaining
BrdU staining was performed as described previously (Wu et al., 2006b). For vigilin staining, cells grown on coverslips were fixed with cold 70% ethanol. After blocking with 10% normal goat serum in phosphate buffer for 30 min, cells were then incubated with polyclonal antibodies against N and C termini of vigilin (gift of G. Neu-yilik, University of Heidelberg, Heidelberg, Germany) for 1 h at RT, followed by incubation with FITC-conjugated secondary antibodies.
RT-PCR
For conventional RT-PCR, RNA samples were reverse transcribed using either poly-dT, major satellite-specific, or random primer and subjected to PCR with γ satellite primers (5′-CATATTCCAGGTCCTTCAGTGTGC-3′ and 5′-GACGACTTGAAAAATGACGAAATC-3′). For the attempt to detect A-to-I–edited RNA, we used a degenerate primer (5′-CGGAATTCGAAAAY [A/C]GAGAAAC-3′) or primers from unlikely-to-be-edited regions (5′-GGAAAATGAGAAACATCCAC-3′) for reverse transcription and secondary primers (5′-CGGGATCCGTTTTCTCGCC-3′ or 5′-TTTTCAGTTTTCTCGCC-3′) for amplification.
Acknowledgments
We thank N. Dillon for providing the γ satellite DNA plasmid (pγSat), G. Neu-yilik for providing vigilin antibodies, P. Fraser for help with RNA-FISH, M. Davidson, G. Almouzni, and A. Probst for helpful discussions, and S. Jia for critical reading of the manuscript.
This work was supported by National Institutes of Health grant GM57233 to D.M. Gilbert.
Abbreviations used in this paper: DRB, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole; HP, heterochromatin protein; MEF, mouse embryonic fibroblast; PCNA, proliferating cell nuclear antigen; WT, wild type.
References
- Becker, M., C. Baumann, S. John, D.A. Walker, M. Vigneron, J.G. McNally, and G.L. Hager. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3:1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, B.S., T.B. Nesterova, E. Thompson, A. Hertweck, E. O'Connor, J. Godwin, C.B. Wilson, N. Brockdorff, A.G. Fisher, S.T. Smale, and M. Merkenschlager. 2005. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201:1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A.K., T.Y. Huh, and C.W. Helleiner. 1973. Transcription of satellite DNA in mouse L-cells. Can. J. Biochem. 51:529–532. [DOI] [PubMed] [Google Scholar]
- Dai, J., B.A. Sullivan, and J.M. Higgins. 2006. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 11:741–750. [DOI] [PubMed] [Google Scholar]
- Dimitrova, D.S., and D.M. Gilbert. 2000. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp. Cell Res. 254:321–327. [DOI] [PubMed] [Google Scholar]
- Fischle, W., B.S. Tseng, H.L. Dormann, B.M. Ueberheide, B.A. Garcia, J. Shabanowitz, D.F. Hunt, H. Funabiki, and C.D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 438:1116–1122. [DOI] [PubMed] [Google Scholar]
- Flamm, W.G., P.M. Walker, and M. McCallum. 1969. Some properties of the single strands isolated from the DNA of the nuclear satellite of the mouse (Mus musculus). J. Mol. Biol. 40:423–443. [DOI] [PubMed] [Google Scholar]
- Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784–791. [DOI] [PubMed] [Google Scholar]
- Gaubatz, J.W., and R.G. Cutler. 1990. Mouse satellite DNA is transcribed in senescent cardiac muscle. J. Biol. Chem. 265:17753–17758. [PubMed] [Google Scholar]
- Gilbert, D.M., and S.N. Cohen. 1987. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell. 50:59–68. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I., and S.C. Elgin. 2007. Transcription and RNA interference in the formation of heterochromatin. Nature. 447:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, S.I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 8:35–46. [DOI] [PubMed] [Google Scholar]
- Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel, J., N. Hanania, H. Tapiero, and L. Harel. 1968. RNA replication by nuclear satellite DNA in different mouse cells. Biochem. Biophys. Res. Commun. 33:696–701. [DOI] [PubMed] [Google Scholar]
- Huisinga, K.L., B. Brower-Toland, and S.C. Elgin. 2006. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 115:110–122. [DOI] [PubMed] [Google Scholar]
- Jia, S., T. Yamada, and S.I. Grewal. 2004. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell. 119:469–480. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou, C., S.A. Muljo, A.L. Kung, S. Ganesan, R. Drapkin, T. Jenuwein, D.M. Livingston, and K. Rajewsky. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, H., D.B. Goto, R.A. Martienssen, T. Urano, K. Furukawa, and Y. Murakami. 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 309:467–469. [DOI] [PubMed] [Google Scholar]
- Kim, S.M., D.D. Dubey, and J.A. Huberman. 2003. Early-replicating heterochromatin. Genes Dev. 17:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli, N., Y.M. Sun, S. van der Sar, P.B. Singh, and J.P. Brown. 2005. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochem. Biophys. Res. Commun. 337:901–907. [DOI] [PubMed] [Google Scholar]
- Kuznetsova, I., O.I. Podgornaya, and M.A. Ferguson-Smith. 2006. High-resolution organization of mouse centromeric and pericentromeric DNA. Cytogenet. Genome Res. 112:248–255. [DOI] [PubMed] [Google Scholar]
- Kuznetsova, I.S., N.I. Enukashvily, E.M. Noniashvili, A.N. Shatrova, N.D. Aksenov, V.V. Zenin, A.P. Dyban, and O.I. Podgornaya. 2007. Evidence for the existence of satellite DNA-containing connection between metaphase chromosomes. J. Cell. Biochem. 101:1046–1061. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Y. Ueda, A.A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A.H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192–1200. [DOI] [PubMed] [Google Scholar]
- Leonhardt, H., H.P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M.C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky, J.M., S.M. Shenoy, R.C. Pezo, and R.H. Singer. 2002. Single-cell gene expression profiling. Science. 297:836–840. [DOI] [PubMed] [Google Scholar]
- Li, F., J. Chen, M. Izumi, M.C. Butler, S.M. Keezer, and D.M. Gilbert. 2001. The replication timing program of the Chinese hamster β-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J. Cell Biol. 154:283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., B. Wu, J. Szary, E.M. Kofoed, and F. Schaufele. 2007. Functional sequestration of transcription factor activity by repetitive DNA. J. Biol. Chem. 282:20868–20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren, M., C.M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell. 103:733–743. [DOI] [PubMed] [Google Scholar]
- Maio, J.J., and D.M. Kurnit. 1974. Transcription of mammalian satellite DNAs by homologous DNA-dependent RNA polymerases. Biochim. Biophys. Acta. 349:305–319. [DOI] [PubMed] [Google Scholar]
- Maison, C., D. Bailly, A.H. Peters, J.P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329–334. [DOI] [PubMed] [Google Scholar]
- Martens, J.H., R.J. O'Sullivan, U. Braunschweig, S. Opravil, M. Radolf, P. Steinlein, and T. Jenuwein. 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt, C., M. Guilleme, J.S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison, E.P., J.F. Partridge, O.H. Tam, S. Cheloufi, and G.J. Hannon. 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 102:12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, C.S., L. Chakalova, K.E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J.A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 107:323–337. [DOI] [PubMed] [Google Scholar]
- Pidoux, A.L., and R.C. Allshire. 2004. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 12:521–534. [DOI] [PubMed] [Google Scholar]
- Prasanth, K.V., P.A. Sacco-Bubulya, S.G. Prasanth, and D.L. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 14:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashad, N., and R.G. Cutler. 1976. Percent satellite DNA as a function of tissue and age of mice. Biochim. Biophys. Acta. 418:1–23. [DOI] [PubMed] [Google Scholar]
- Rizzi, N., M. Denegri, I. Chiodi, M. Corioni, R. Valgardsdottir, F. Cobianchi, S. Riva, and G. Biamonti. 2004. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell. 15:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudert, F., S. Bronner, J.M. Garnier, and P. Dolle. 1995. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome. 6:76–83. [DOI] [PubMed] [Google Scholar]
- Samuel, C.E. 2003. RNA editing minireview series. J. Biol. Chem. 278:1389–1390. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., S. Ramanathan, Y. Okuno, C. Kumagai, S.S. Shaikh, and D.M. Gilbert. 2006. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol. Cell. Biol. 26:1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova, E.A., Z. Mansuroglu, H. Mokrani, N. Ghinea, and E. Bonnefoy. 2004. Transcription factor YY1 associates with pericentromeric {gamma}-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res. 32:4390–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K., and A.K. Panigrahi. 2002. RNA editing: complexity and complications. Mol. Microbiol. 45:591–596. [DOI] [PubMed] [Google Scholar]
- Tam, R., C. Johnson, L. Shopland, J. McNeil, and J.B. Lawrence. 2002. Applications of RNA FISH for visualizing gene expression and nuclear architecture. In FISH (Fluorescence In Situ Hybridization). B.G. Beatty, S. Mai, and J.A. Squire, editors. Oxford University Press, Oxford. 93–110.
- Valgardsdottir, R., I. Chiodi, M. Giordano, F. Cobianchi, S. Riva, and G. Biamonti. 2005. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell. 16:2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Z. Zhang, K. Blackwell, and G.G. Carmichael. 2005. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr. Biol. 15:384–391. [DOI] [PubMed] [Google Scholar]
- Waring, M., and R.J. Britten. 1966. Nucleotide sequence repetition: a rapidly reassociating fraction of mouse DNA. Science. 154:791–794. [DOI] [PubMed] [Google Scholar]
- Wei, X., J. Samarabandu, R.S. Devdhar, A.J. Siegel, R. Acharya, and R. Berezney. 1998. Segregation of transcription and replication sites into higher order domains. Science. 281:1502–1506. [DOI] [PubMed] [Google Scholar]
- Wong, L.H., K.H. Brettingham-Moore, L. Chan, J.M. Quach, M.A. Anderson, E.L. Northrop, R. Hannan, R. Saffery, M.L. Shaw, E. Williams, and K.H. Choo. 2007. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17:1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.R., G. Yu, and D.M. Gilbert. 1997. Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods. 13:313–324. [DOI] [PubMed] [Google Scholar]
- Wu, R., A.V. Terry, P.B. Singh, and D.M. Gilbert. 2005. Differential subnuclear localization and replication timing of histone h3 lysine 9 methylation States. Mol. Biol. Cell. 16:2872–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R., P.B. Singh, and D.M. Gilbert. 2006. a. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 174:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R., A.V. Terry, and D.M. Gilbert. 2006. b. Observing S-phase dynamics of histone modifications with fluorescently labeled antibodies. Methods Mol. Biol. 325:139–148. [DOI] [PubMed] [Google Scholar]
- Zaratiegui, M., D.V. Irvine, and R.A. Martienssen. 2007. Noncoding RNAs and gene silencing. Cell. 128:763–776. [DOI] [PubMed] [Google Scholar]
- Zhang, J., F. Xu, T. Hashimshony, I. Keshet, and H. Cedar. 2002. Establishment of transcriptional competence in early and late S phase. Nature. 420:198–202. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and G.G. Carmichael. 2004. Methods for the analysis of adenosine-to-inosine editing in RNA. Methods Mol. Biol. 257:75–84. [DOI] [PubMed] [Google Scholar]