Abstract
The spindle orientation checkpoint (SPOC) of budding yeast delays mitotic exit when cytoplasmic microtubules (MTs) are defective, causing the spindle to become misaligned. Delay is achieved by maintaining the activity of the Bfa1–Bub2 guanosine triphosphatase–activating protein complex, an inhibitor of mitotic exit. In this study, we show that the spindle pole body (SPB) component Spc72, a transforming acidic coiled coil–like molecule that interacts with the γ-tubulin complex, recruits Kin4 kinase to both SPBs when cytoplasmic MTs are defective. This allows Kin4 to phosphorylate the SPB-associated Bfa1, rendering it resistant to inactivation by Cdc5 polo kinase. Consistently, forced targeting of Kin4 to both SPBs delays mitotic exit even when the anaphase spindle is correctly aligned. Moreover, we present evidence that Spc72 has an additional function in SPOC regulation that is independent of the recruitment of Kin4. Thus, Spc72 provides a missing link between cytoplasmic MT function and components of the SPOC.
Introduction
The budding yeast spindle pole body (SPB) is the functional equivalent of the mammalian centrosome. The mitotic exit network (MEN) is an SPB-associated signaling cascade that controls mitotic exit, which is the transition from mitosis into G1 phase of the cell cycle (Gruneberg et al., 2000; Pereira and Schiebel, 2001; Stegmeier and Amon, 2004). The Ras-like GTPase Tem1 functions at the top of the MEN (Shirayama et al., 1994). The putative guanine nucleotide exchange factor Lte1 (an activator of the MEN) and the GTPase-activating protein (GAP) complex Bfa1–Bub2 (a MEN inhibitor) regulate Tem1 (Bardin et al., 2000; Pereira et al., 2000; Geymonat et al., 2002). Tem1 interacts with the Pak-like kinase Cdc15 (Asakawa et al., 2001), which, in turn, activates the Dbf2–Mob1 kinase complex (Mah et al., 2001). Ultimately, the MEN controls the activity of the conserved phosphatase Cdc14 (Stegmeier and Amon, 2004) and, thereby, mitotic exit (Visintin et al., 1998).
In yeast cells, the mother-bud junction determines the site of cytokinesis (Segal and Bloom, 2001). Therefore, cells with an anaphase spindle that is inappropriately positioned within the mother cell would cause cytokinesis to occur parallel to the plane of the spindle and, thus, result in aneuploidy. To prevent this from happening, the spindle orientation checkpoint (SPOC) senses (in an unknown manner) spindle orientation defects and actively inhibits the MEN of cells with a misaligned anaphase spindle. In cells with a correctly aligned anaphase spindle, phosphorylation of Bfa1 by Cdc5 polo kinase reduces Bfa1–Bub2 GAP activity to promote mitotic exit. However, when the spindle is misplaced, the SPOC prevents the Cdc5-dependent phosphorylation of Bfa1. Therefore, the Bfa1–Bub2 GAP complex remains active, and cells fail to exit mitosis and arrest in anaphase instead (Hu et al., 2001; Geymonat et al., 2003).
The protein kinase Kin4 is an additional component of the SPOC. On the basis of genetic data, it would appear that KIN4 functions upstream of BFA1 and BUB2 (D'Aquino et al., 2005; Pereira and Schiebel, 2005). A striking feature of Kin4 is its SPB distribution in relationship to the Bfa1–Bub2 complex. In cells with a correctly aligned spindle, the Bfa1–Bub2 GAP complex binds preferentially to the budward-directed SPB (Pereira et al., 2000, 2001), whereas Kin4 associates with the SPB that faces the mother cell body (Pereira and Schiebel, 2005). In contrast, Kin4 and the Bfa1–Bub2 GAP colocalize at both SPBs when the anaphase spindle becomes mispositioned. This recruitment of Kin4 and Bfa1–Bub2 to the same SPBs may be important for the cell cycle arrest response to spindle alignment defects (Pereira and Schiebel, 2005). The observation that the targeting of Bub2 to both SPBs causes defects in mitotic exit even when the anaphase spindle is correctly positioned is consistent with this notion (Fraschini et al., 2006).
How the SPOC senses spindle alignment defects and the molecular role of Kin4 in this process are currently unclear. In this study, we present evidence that the γ-tubulin complex receptor protein Spc72 provides a regulated binding site that recruits Kin4 to both SPBs whenever the anaphase spindle is mispositioned. This relocalization enables Kin4 to phosphorylate Bfa1, thereby protecting the Bfa1–Bub2 complex from inactivation by Cdc5 kinase. Thus, the SPB component Spc72 links cytoplasmic microtubules (MTs) with SPOC components and, therefore, could function as part of the sensors of spindle orientation defects.
Results
Local regulation of Cdc5 kinase at SPBs
The SPOC prevents the phosphorylation of Bfa1 by Cdc5 pololike kinase when the anaphase spindle becomes misaligned (Hu et al., 2001). This regulation may occur at SPBs because both Bfa1 and Cdc5 associate with this structure (Shirayama et al., 1998; Pereira et al., 2001). If this was the case, it could occur at two levels. It could arise from a reduction in the amount of Cdc5 that associates with the SPB or from a specific impairment of the capability of Cdc5 to phosphorylate Bfa1 at SPBs.
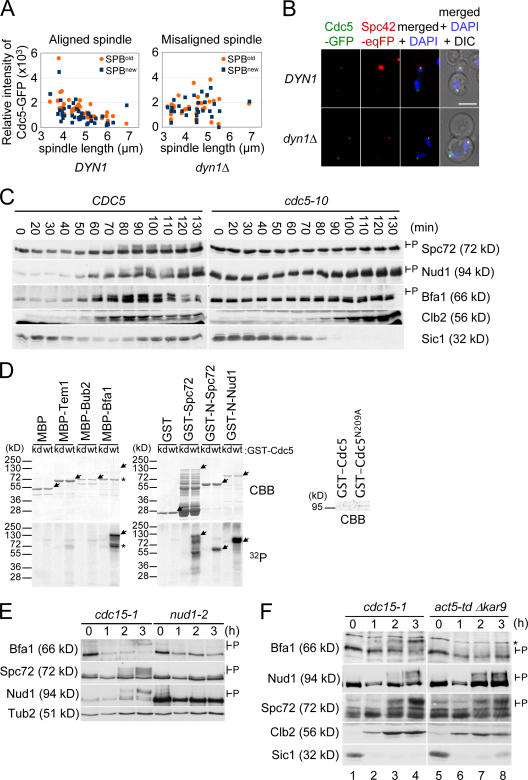
To address whether the amount of Cdc5 at SPBs is regulated by orientation of the anaphase spindle, we compared the relative fluorescence intensity of Cdc5-GFP signal at SPBs in wild-type cells with a correctly aligned anaphase spindle with that of dynein-deficient cells (dyn1Δ cells) in which the anaphase spindle is misaligned within the mother cell body (Fig. 1 A; Li et al., 1993). Because SPOC proteins show a polarized association with the SPBs (Pereira et al., 2001; Pereira and Schiebel, 2005), we measured the Cdc5-GFP signal at both SPBs. In dyn1Δ cells with a misaligned anaphase spindle, the Cdc5-GFP signal at both SPBs of a cell was of similar intensity (1,646 ± 836 [old SPB] and 1,535 ± 758 [new SPB]; P > 0.05 by t test; Fig. 1, A and B). In contrast, in wild-type cells, the Cdc5-GFP signal from the SPB in the bud was slightly stronger than that from the SPB in the mother cell (1,498 ± 880 vs. 1,086 ± 823; P < 0.05 by t test). Importantly, the mean intensity of the Cdc5-GFP signal at the SPBs of misaligned spindles was not significantly different (P > 0.05 by t test) from that at the SPBs of correctly aligned spindles (Fig. 1 A). Thus, spindle misalignment did not lower the amount of Cdc5 at SPBs.
Figure 1.
The SPOC regulates the phosphorylation of Bfa1 by Cdc5 at SPBs. (A and B) DYN1 CDC5-GFP and dyn1Δ CDC5-GFP cells in anaphase with a correctly aligned or misoriented anaphase spindle were analyzed by fluorescence microscopy. (A) Measurement of the relative fluorescence intensity of the Cdc5-GFP SPB signal of DYN1 CDC5-GFP cells with a correctly aligned anaphase spindle and dyn1Δ CDC5-GFP cells with the anaphase spindle misaligned in the mother cell body. The signals associated with the preexisting old (orange) and new (blue) SPBs were measured. (B) Cdc5-GFP signal at SPBs of DYN1 CDC5-GFP and dyn1Δ CDC5-GFP cells. Spc42-eqFP611 was used as an SPB marker. Note that Spc42-eqFP611 labeled the old SPB in the mother cell body more strongly because of a delay in the folding of the eqFP611 molecule (Pereira et al., 2001; Wiedenmann et al., 2002). DNA was stained with DAPI. (C) CDC5 and cdc5-10 cells were synchronized in G1 with α factor at 23°C and released at 37°C, the restrictive temperature of cdc5-10 cells. Samples were analyzed by immunoblotting for Spc72, Nud1, and Bfa1 phosphorylation (indicated as P). Budding index and FACS analysis are shown in Fig. S1 (A–C). (D) Purified MBP, MBP-Tem1, MBP-Bub2, MBP-Bfa1, GST, GST-Spc72, GST-N-Spc72, and GST-N-Nud1 were incubated with pololike kinase Cdc5 (wt) and the kinase-dead Cdc5N209A (kd; see CBB-stained gel on the right) in the presence of γ-[32P]ATP. Proteins were separated by SDS-PAGE, and phosphorylation was analyzed by autoradiography (32P) after CBB staining. Full-length proteins are marked by arrows, and the Bfa1 degradation product is marked by asterisks. (E) α factor–synchronized cdc15-1 and nud1-2 cells were incubated at 37°C for the indicated times. Cell extracts were analyzed by immunoblotting. The hyperphosphorylated protein bands are marked as P. (F) cdc15-1 and act5-td kar9Δ cells were synchronized with α factor in G1 at 23°C and released at 37°C. Cells were analyzed by immunoblotting. P marks hyperphosphorylated proteins. The asterisk marks a protein band cross reacting with the anti-Bfa1 antibodies. Cell morphology of cells arrested in anaphase are shown in Fig. S1 D (available at http://www.jcb.org/cgi/content/full/jcb.200705197). Bar, 5 μm.
Next, we determined whether the activity of Cdc5 at SPBs is regulated by the SPOC. For this analysis, we wanted to compare the ability of Cdc5 to phosphorylate SPB components, including Bfa1, in cells in which the spindle was correctly aligned with its ability to phosphorylate these substrates when anaphase spindles were misaligned. To perform this experiment, we sought SPB components in addition to Bfa1 that are Cdc5 kinase substrates. The SPB components Nud1 and Spc72 have both been shown to be phosphoproteins that bind to Cdc5 (Gruneberg et al., 2000; Ho et al., 2002). As such, they are excellent candidates for being targets of Cdc5. Consistently, phosphorylation of both Nud1 and Spc72 was dependent on CDC5 in vivo, as the slower migrating phosphobands of each protein seen during mitosis of wild-type cells were missing from blots of SDS-PAGE gels from cdc5-10 cells (Fig. 1 C; cell cycle profiles of cells of this experiment are shown in Fig. S1, A–C; available at http://www.jcb.org/cgi/content/full/jcb.200705197/DC1). In addition, Cdc5 was able to phosphorylate Bfa1, Nud1, and Spc72 directly in vitro (Fig. 1 D, 32P). In contrast, Cdc5 did not strongly modify maltose- binding protein (MBP), GST, Bub2, or Tem1 (Fig. 1 D). Together, these data suggest that Nud1 and Spc72 join Bfa1 as being substrates of Cdc5.
Although Spc72, Nud1, and Bfa1 are clearly targets of Cdc5, the aforementioned data do not determine whether their phosphorylation by Cdc5 takes place at SPBs or in the cytoplasm. To distinguish between these possibilities, we compared the phosphorylation pattern of Bfa1, Nud1, and Spc72 in cdc15-1 and nud1-2 cells. Both cell types arrest at the restrictive temperature in anaphase (Schweitzer and Philippsen, 1991; Gruneberg et al., 2000), but the two strains differ in one important respect: the cytoplasmic side of the SPB persists in cdc15-1 cells and plays host to Bfa1, Nud1, and Spc72 (Pereira et al., 2000), whereas this SPB substructure has disassembled and, thus, is absent from nud1-2 cells (Gruneberg et al., 2000). Bfa1, Nud1, and Spc72 were hyperphosphorylated in cdc15-1 cells (Fig. 1 E). In contrast, all three proteins were hypophosphorylated in nud1-2 cells. This correlation between the phosphorylation status and presence of a cytoplasmic SPB structure suggests that Bfa1, Nud1, and Spc72 become phosphorylated by Cdc5 at SPBs.
We determined whether the ability of Cdc5 to phosphorylate proteins at SPBs is affected by orientation of the anaphase spindle. For this experiment, we used synchronized cdc15-1 cells and act5-td kar9Δ cells, both of which arrest at the restrictive temperature in anaphase (>97% and ∼85%) because of an inactive MEN (Visintin et al., 1999; Hu et al., 2001). However, in cdc15-1 cells, the anaphase spindle was correctly aligned along the mother-bud axis, whereas it was misaligned in act5-td kar9Δ cells because of the failure of the redundant Kar9- and dynein-dependent spindle orientation pathways (for phenotypes, see Fig. S1 D; Segal and Bloom, 2001). In both cdc15-1 and act5-td kar9Δ cells, Nud1 and Spc72 became phosphorylated as soon as the B-type cyclin Clb2 accumulated in mitosis (Fig. 1 F, after 2 h). Thus, orientation of the mitotic spindle did not affect the activity of Cdc5 toward Nud1 and Spc72. This was different for Bfa1. In cdc15-1 cells, Bfa1 became hyperphosphorylated (Fig. 1 F, lane 4). In stark contrast, Bfa1 failed to acquire any modifications in act5-td kar9Δ cells and persisted in a hypophosphorylated isoform throughout the experiment (Fig. 1 F, lane 8; Hu et al., 2001). These data suggest that the overall kinase activity of Cdc5 at SPBs is not regulated by the SPOC. Rather, a spindle orientation–dependent mechanism located at SPBs specifically protects Bfa1 from Cdc5 kinase when the anaphase spindle is misaligned.
Kin4 kinase phosphorylates Bfa1
Kin4 kinase functions upstream of the Bfa1–Bub2 complex (D'Aquino et al., 2005; Pereira and Schiebel, 2005) and, therefore, is ideally positioned to act as a regulator of Bfa1 function. To address the molecular role of Kin4 in detail, we first tested whether Kin4 kinase activity was required for SPOC signaling. Cells in which Kin4 was mutated to abolish its kinase activity (Kin4T209A; D'Aquino et al., 2005) had no SPOC checkpoint (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200705197/DC1). However, this lack of checkpoint function was not caused by deficiencies in Kin4T209A expression or localization, as both were indistinguishable from those of wild-type active Kin4. Thus, Kin4 recruitment to the cell cortex and SPB did not require its kinase activity, but the SPOC did (Fig. S2).
To identify Kin4 targets in SPOC signaling, we adopted a candidate approach and focused on SPB-associated proteins because Kin4 binding to SPBs turned out to be essential for SPOC function (see Fig. 7). We tested whether Kin4 could phosphorylate the MEN scaffold protein Nud1 (Gruneberg et al., 2000), the Bfa1–Bub2 complex, pololike kinase Cdc5, a catalytically inactive version of Cdc5, Cdc5N209A (Cheng et al., 1998), the γ-tubulin complex receptor protein Spc72 (Knop and Schiebel, 1998), and the GTPase Tem1 (Shirayama et al., 1994). In all assays, a catalytically inactive kinase-dead Kin4T209A was used as a control to exclude the possibility that a contaminating kinase was responsible for the phosphorylation of a candidate protein (Fig. 2 A and Fig. S3, A and B, right; available at http://www.jcb.org/cgi/content/full/jcb.200705197/DC1).
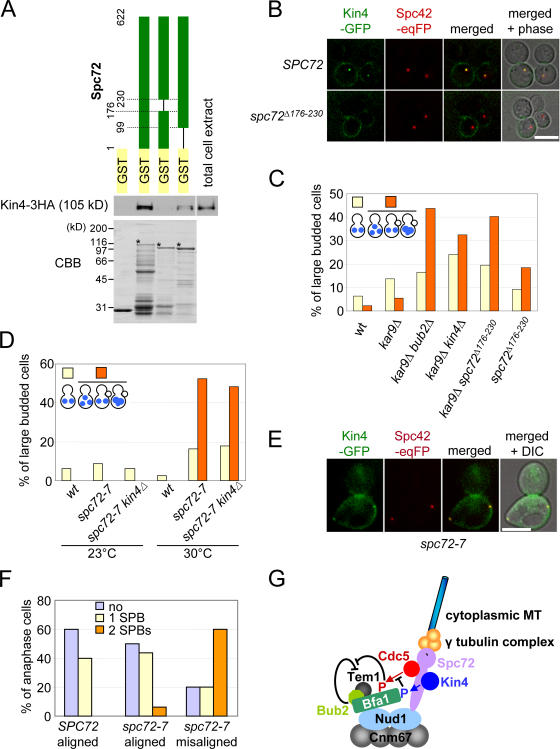
Figure 7.
A role of the γ-tubulin complex receptor protein Spc72 in regulation of the SPOC. (A) Kin4 binds to the SPB component Spc72. Kin4 pull-down assays with recombinant GST, GST-Spc72, GST-Spc72Δ176–230, and GST-Spc72Δ1–98. Total cell extract prepared from a yeast strain carrying KIN4-3HA was incubated with the purified recombinant proteins bound to glutathione beads (CBB-stained gel). After washing of the beads, Kin4-3HA was detected by immunoblotting. Asterisks mark the full-length Spc72 proteins. (B) In spc72 Δ176–230 cells, Kin4-GFP fails to bind to SPBs. SPC72 and spc72 Δ176–230 cells with KIN4-GFP SPC42-eqFP611 were treated with nocodazole. Kin4-GFP localization was determined by fluorescence microscopy. (C) spc72 Δ176–230 cells are SPOC deficient. Wild-type (wt), kar9Δ, kar9Δ bub2Δ, kar9Δ kin4Δ, kar9Δ spc72Δ 176–230, and spc72 Δ176–230 cells were grown to mid-log phase in YPAD at 30°C and were shifted to 37°C for 3 h. Cells were fixed, and the DNA was stained with DAPI. Large-budded cells of wild-type, kar9Δ, kar9Δ bub2Δ, kar9Δ kin4Δ, kar9Δ spc72 Δ176–230, and spc72 Δ176–230 cells were 27.9%, 33.1%, 42.0%, 29.5%, 35.8%, and 33.0%, respectively. n > 100 large-budded cells per strain. (D) spc72-7 cells are SPOC deficient. Wild-type (wt), spc72-7, and spc72-7 kin4Δ cells were grown at 23 and 30°C to mid-log phase. Cells were fixed, and DNA was stained with DAPI. >200 cells per strain. Large-budded cells of wild-type, spc72-7, and spc72-7 kin4Δ cells were 23.8%, 27.4%, and 31.8% at 23°C and 24.0%, 49.2%, and 47.3% at 30°C, respectively. (E) Kin4-GFP binds to SPBs in spc72-7 cells. SPC72 and spc72-7 cells with KIN4-GFP SPC42-eqFP611 were synchronized with α factor at 23°C and released at 30°C. Anaphase cells were analyzed. (F) Quantification of E. n > 30 anaphase cells per strain. (G) Model for the function of Kin4. See Discussion for details. Bars, 5 μm.
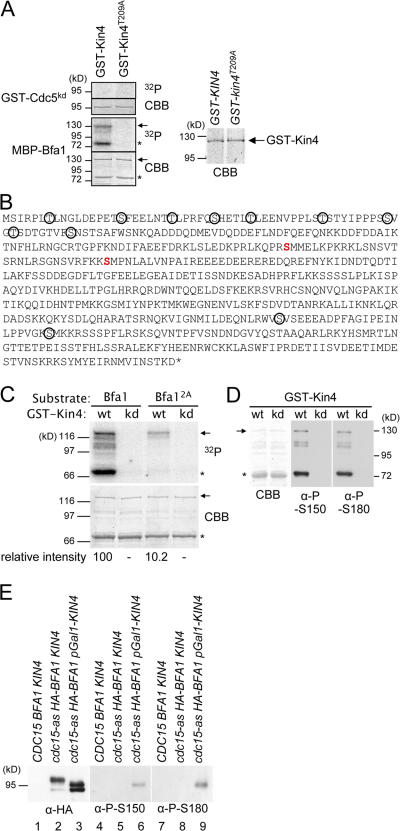
Figure 2.
Bfa1 is a substrate of Kin4. (A) Bfa1 but not Cdc5 is a substrate of Kin4. Equal amounts of purified GST-Kin4 and GST-Kin4T209A (CBB strain gel on the right) were incubated with the kinase-dead GST-Cdc5kd and MBP-Bfa1. Proteins were analyzed by autoradiography (32P). (B) Kin4 phosphorylation sites in Bfa1. The two Kin4 sites, which were determined by MALDI-TOF/TOF, are shown in red. Ser and Thr residues corresponding to the Cdc5 phosphorylation sites of Bfa1 are circled (Hu et al., 2001). (C) Phosphorylation of Bfa12A by Kin4 is strongly reduced. Purified MBP-Bfa1 and MBP-Bfa12A were incubated with Kin4 (wt) and Kin4T209A (kd) in the presence of γ-[32P]ATP. Phosphorylation of Bfa1 was determined by autoradiography. (D) The anti–P-S150 and –P-S180 phosphospecific antibodies only detect MBP-Bfa1 that was phosphorylated in vitro by Kin4 (wt) but not the kinase-dead Kin4T209A (kd). (A, C, and D) Full-length Bfa1 protein is indicated by arrows, and the Bfa1 degradation product is marked with asterisks. (E) Kin4 phosphorylates Bfa1 in vivo. Wild-type (lanes 1, 4, and 7), cdc15-as HA-BFA1 (lanes 2, 5, and 8), and cdc15-as HA-BFA1 pGal1-KIN4 cells (lanes 3, 6, and 9) were grown in YPR (yeast extract, peptone, and raffinose) medium to mid-log phase. Cells were arrested in G1 phase with α factor, and galactose was added for 1 h to induce pGal1-KIN4 expression. Cells were released into YPRG (YPR + galactose) medium containing 10 μM PP1 analogue 8, the ATP analogue that inhibits Cdc15-as, and were incubated for 3 h until cdc15-as cells were arrested uniformly with large buds. Precipitated proteins with anti-HA antibodies were probed by immunoblotting with anti-HA, –P-S150, and –P-S180 antibodies.
Kin4 but not the kinase-dead Kin4T209A phosphorylated both full-length recombinant Bfa1 (Fig. 2 A, arrows) and a C-terminal degradation product of Bfa1 (Fig. 2 A, asterisks). Cdc5 polo kinase was weakly phosphorylated when incubated with Kin4 or Kin4T209A (Fig. S3 B). However, the kinase-dead Cdc5kd was not phosphorylated by Kin4 (Fig. 2 A). This latter result indicates that at least some of the modifications of Cdc5 (Fig. S3) were caused by autophosphorylation. The other candidates that we tested were not strongly phosphorylated by Kin4 (Fig. S3). Together, this experiment identified Bfa1 as a clear in vitro substrate of Kin4.
We next mapped the sites on Bfa1 that were phosphorylated by Kin4 as a step toward understanding the functional significance of this modification. We used Kin4 that had been purified from yeast and phosphorylated recombinant MBP-Bfa1 in vitro. Serine 150 and 180 of Bfa1 were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF)/TOF as Kin4-directed phosphorylation sites (Fig. 2 B). We mutated both of these serine residues to alanine (BFA12A) to prevent phosphorylation. Phosphorylation of MBP-Bfa12A by purified Kin4 was reduced by ∼90% when compared with the levels seen with wild-type Bfa1 (Fig. 2 C), suggesting that both serine residues are indeed the major Kin4 kinase sites in our in vitro assay.
To determine whether Kin4 phosphorylates Bfa1 residues 150 and 180 in vivo, we raised phosphospecific antibodies against P-S150 and P-S180. The affinity-purified anti–P-S150 and –P-S180 antibodies only detected Bfa1 after it had been incubated with Kin4 but not after incubation with kinase-dead Kin4T209A (Fig. 2 D). This demonstrated that the two antibodies specifically recognized Kin4-phosphorylated Bfa1 and were unable to recognize unmodified Bfa1.
With these antibodies in hand, we could show that Kin4 phosphorylates Bfa1 in vivo. cdc15-as HA-BFA1 cells were arrested in anaphase by addition of the Cdc15-as inhibitor PP1. In these cells, Bfa1 accumulated in its hyperphosphorylated form. In contrast, in cdc15-as HA-BFA1 cells overexpressing KIN4, the overall phosphorylation of Bfa1 was reduced (Fig. 2 E, lane 3; D'Aquino et al., 2005). In anti-Bfa1 immunoprecipitates of cells with pGal1-KIN4 expression, the anti–P-S150 and –P-S180 antibodies detected Bfa1 (Fig. 2 E, lanes 6 and 9). This was not the case without pGal1-KIN4 expression (Fig. 2 E, lanes 5 and 8). Thus, Kin4 phosphorylates S150 and S180 of Bfa1 in vivo.
Bfa12A shows normal SPB localization and provides spindle assembly checkpoint function
We next compared the properties of cells expressing BFA1 with those expressing BFA12A. BFA1 and BFA12A were integrated into the BFA1 locus, where they were expressed as sole copies of BFA1 from the endogenous promoter. BFA1 and BFA12A accumulated to similar levels (Fig. S4 A, 0 h; available at http://www.jcb.org/cgi/content/full/jcb.200705197/DC1), both proteins localized in a polar manner preferentially with the budward-directed SPB (Fig. 3 A; Pereira et al., 2001), both responded to MT depolymerization as Bfa1 and Bfa12A were recruited to both SPBs in the presence of nocodazole (Fig. 3 B; Pereira et al., 2001), and both proteins recruited the partner protein Bub2 to the SPB (Fig. S4 B). Thus, in terms of SPB localization and the ability to recruit Bub2, the Bfa12A protein behaved as the wild-type Bfa1 molecule.
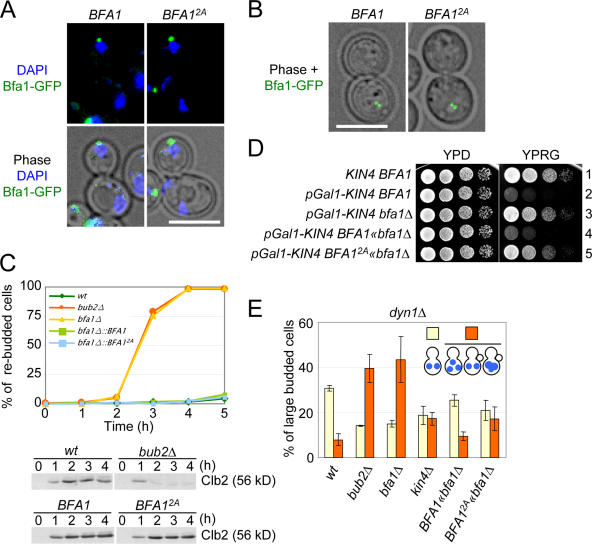
Figure 3.
Kin4 phosphorylation sites of Bfa1 are essential for SPOC function. SPB binding and relocalization to both SPBs after nocodazole treatment of BFA12A cells was indistinguishable from that of BFA1 cells. (A) Bfa12A associates in the same polar manner with the SPB in the bud as wild-type Bfa1. SPB localization of Bfa1-GFP and Bfa12A-GFP was determined by fluorescence microscopy. (B) Bfa12A binds to both SPBs in response to MT depolymerization in the same way as wild-type Bfa1. BFA1-GFP and BFA2A-GFP cells were treated with nocodazole. (C) BFA1 2A cells are spindle assembly checkpoint proficient. The function of the spindle assembly checkpoint was tested in wild-type (wt), bub2Δ, bfa1Δ, BFA1, and BFA1 2A cells. Cells were synchronized in G1 with α factor and released into medium containing nocodazole. The number of cells with multiple buds was determined over time. n > 100 cells per strain. Clb2 levels were determined by immunoblotting. (D) BFA12A suppresses the toxicity of KIN4 overexpression. BFA1 wild-type cells with (row 2) and without pGal1-KIN4 (row 1) and bfa1Δ (row 3), BFA1 (row 4), and BFA12A cells (row 5) with pGal1-KIN4 were incubated as serial dilutions on YPRG (induction of the pGal1 promoter) and YPD plates (repression of the pGal1 promoter) at 30°C for 2 d. (E) BFA12A cells are SPOC deficient. Wild-type (wt), bub2Δ, bfa1Δ, kin4Δ, BFA1, and BFA12A cells carrying the dyn1Δ allele were incubated in YPAD at 14°C. DNA was stained with DAPI. More than 100 large-budded cells were analyzed per strain and categorized as illustrated in the figure. Error bars indicate SD. Bars, 5 μm.
The spindle assembly checkpoint regulates the metaphase-anaphase transition because it inhibits the anaphase-promoting complex, when chromosome association with the spindle is defective (Hwang et al., 1998). For unknown reasons, it requires BFA1 but not KIN4 to prevent mitotic exit (D'Aquino et al., 2005; Pereira and Schiebel, 2005). Therefore, if Bfa12A is only defective in its ability to be regulated by Kin4, it should sustain the metaphase arrest of cells in response to spindle assembly checkpoint activation to a similar degree to that achieved by cells containing wild-type Bfa1. To test this notion, we analyzed the ability of synchronized BFA12A cells to maintain a metaphase arrest in the presence of the MT-depolymerizing drug nocodazole. BFA1, bfa1Δ, and bub2Δ cells were included as controls for cells with and without Bfa1–Bub2 function. BFA1 and BFA12A cells arrested in metaphase as large-budded cells with high Clb2 levels (Fig. 3 C). The cell cycle arrest was robust and lasted for at least 4 h. In contrast, the bfa1Δ and bub2Δ cells were unable to maintain a metaphase arrest, as indicated by the rebudding of cells and the degradation of Clb2 (Fig. 3 C and not depicted). These data indicate that the Bfa12A mutations do not compromise the transmission of the spindle assembly checkpoint signal by Bfa1 to inhibit mitotic exit in response to MT depolymerization.
Kin4 functions through the phosphorylation of Bfa1
Overexpression of KIN4 is toxic to cells. This growth defect is suppressed by the deletion of either BFA1 or BUB2 (D'Aquino et al., 2005). Thus, if Kin4 acts via the phosphorylation of serine residues 150 and 180 of Bfa1, BFA12A cells should be resistant to KIN4 overexpression. As reported previously (D'Aquino et al., 2005), pGal1-KIN4 BFA1 cells failed to grow on galactose plates (Fig. 3 D, rows 2 and 4). In contrast, pGal1-KIN4 bfa1Δ cells grew (Fig. 3 D, row 3). Importantly, BFA12A cells were resistant to KIN4 overexpression (Fig. 3 D, compare row 5 with rows 3 and 4). This implies that KIN4 acts by phosphorylating serines 150 and 180 of Bfa1.
If BFA12A cells are insensitive to Kin4 regulation, BFA12A cells should show a similar SPOC defect to that exhibited by kin4Δ cells. This prediction was tested by the introduction of a dyn1Δ background mutation. In 30% of large-budded dyn1Δ BFA1 cells, the anaphase spindle was misaligned in the mother cell body (Fig. 3 E). Only 6% of cells with a misaligned spindle progressed through the cell cycle, as indicated by the accumulation of abnormal cell types with three DAPI-staining regions (Fig. 3 E). The number of large-budded cells with an SPOC-deficient phenotype increased in dyn1Δ bub2Δ and dyn1Δ bfa1Δ cells to ∼40% and in dyn1Δ kin4Δ cells to ∼20% (Pereira et al., 2000). In our hands, dyn1Δ kin4Δ cells consistently showed a less pronounced SPOC defect than either dyn1Δ bfa1Δ or dyn1Δ bub2Δ cells. Importantly, dyn1Δ BFA12A cells behaved in an identical manner to dyn1Δ kin4Δ cells (Fig. 3 E). Together, Kin4 functions in SPOC regulation through modifying serines 150 and 180 of Bfa1.
Kin4 fails to protect Bfa12A from phosphorylation by Cdc5
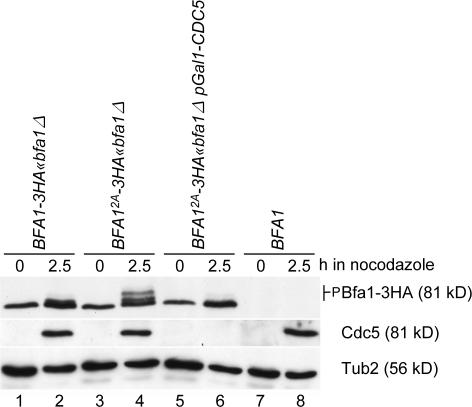
Phosphorylation of Bfa1 by Kin4 may inhibit the ability of Cdc5 to modify Bfa1 and may stop Cdc5 from inducing mitotic exit (Pereira and Schiebel, 2005). To test this model, we incubated BFA1 and BFA12A cells with nocodazole. The ensuing MT depolymerization brought Bfa1, Kin4, and Cdc5 together at both SPBs. Kin4 could then be able to phosphorylate Bfa1, which could then prevent subsequent modifications of Bfa1 by Cdc5. In contrast, Bfa12A should behave differently in this model. Specifically, even in the presence of Kin4, Bfa12A should still become phosphorylated by Cdc5 because the Bfa12A mutant protein would no longer be able to act as a substrate for Kin4 kinase and would not be able to confer the protection from Cdc5 that phosphorylation on these sites normally imparts. Consistent with this model, Bfa1 of wild-type cells was hypophosphorylated after nocodazole treatment (Fig. 4, lane 2), whereas Bfa12A (Fig. 4, lane 4) accumulated as hyperphosphorylated protein in a similar manner to the accumulation of Bfa1 in kin4Δ cells (Pereira and Schiebel, 2005). In the absence of Cdc5, Bfa12A remained hypophosphorylated (Fig. 4, compare lane 4 with lane 6), demonstrating that Cdc5 was the kinase that was responsible for the reduced mobility of Bfa12A. We conclude that phosphorylation by Kin4 kinase protects Bfa1 from inhibitory phosphorylation by Cdc5.
Figure 4.
Kin4 fails to protect Bfa12A from phosphorylation by Cdc5. BFA1-3HA (lanes 1 and 2), BFA12A-3HA (lanes 3 and 4), BFA12A-3HA pGal1-CDC5 (lanes 5 and 6), and BFA1 cells (lanes 7 and 8) grown in YPRG medium were synchronized in G1 with α factor and released in YPD medium containing nocodazole. Cells were analyzed by immunoblotting with the indicated antibodies. Note that Cdc5 becomes degraded with mitotic exit (Shirayama et al., 1998), explaining why Cdc5 was not detected in G1 cells (lanes 1, 3, 5, and 7). Cdc5 was efficiently depleted after the addition of glucose (lane 6).
Plasma membrane–bound Kin4 is SPOC deficient
Kin4 associates with the cortex of the mother cell in mid- anaphase and with the SPB in the mother cell body but not the SPB in the bud (D'Aquino et al., 2005; Pereira and Schiebel, 2005). However, after SPOC activation, Kin4 associates with both SPBs (Pereira and Schiebel, 2005). Thus, the control of Bfa1 by Kin4 that we outline here could arise from a modification of Bfa1 at either the SPB, the cell cortex, or at both locations. Therefore, we asked whether Kin4 at the plasma membrane is able to promote SPOC function. For this analysis, KIN4 was fused to the C terminus of RAS2 (amino acids 301–322; KIN4-pr). This domain of Ras2 can target proteins to the plasma membrane through prenylation of cysteine 318 (Pryciak and Huntress, 1998). To exclude the possibility that the fusion protein itself affects the function of Kin4, cysteines 318 and 319 of the membrane-targeting element of Ras2 were mutated to serine (KIN4-pr-SS).
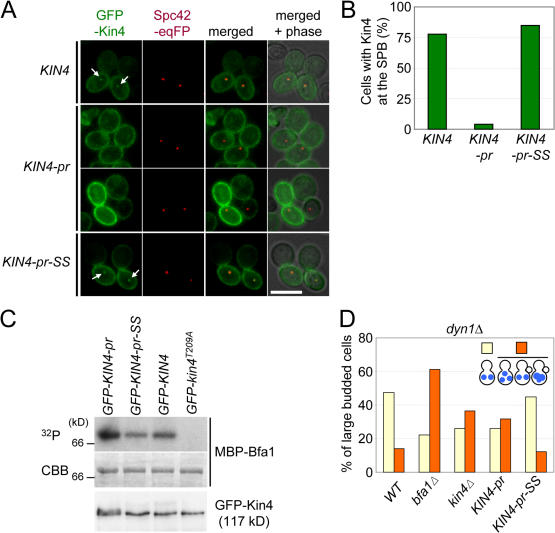
First, we analyzed the localization of Kin4-pr and Kin4- pr-SS. To simplify the analysis, we arrested cells with nocodazole in metaphase and briefly induced expression of the pGal1-GFP-KIN4 derivatives by the addition of galactose. GFP-Kin4 associated with the cortex of the mother cell and, as expected for nocodazole-treated cells, associated with both SPBs (Fig. 5 A, arrows; Pereira and Schiebel, 2005). GFP–Kin4-pr-SS behaved as GFP-Kin4. In contrast, the entire cell cortex, including that of the bud, was decorated by GFP–Kin4-pr. This is probably a reflection of the fusion of the membrane-targeting sequence of Ras2. It is noteworthy that the GFP–Kin4-pr signal at the mother cortex was comparable with, or even stronger than, that of GFP-Kin4 or GFP–Kin4-pr-SS (Fig. 5 A). In addition, anchorage to the cortex effectively blocked the SPB association of GFP–Kin4-pr (Fig. 5, A and B). A weak GFP–Kin4-pr SPB signal could be detected in only ∼4% of cells (Fig. 5 B), and, when this signal was seen, it had an intensity that was <20% of the SPB signal observed in GFP-KIN4 cells. Thus, in contrast to Kin4, Kin4-pr is restricted to the cell cortex and does not accumulate on SPBs.
Figure 5.
Plasma membrane–bound Kin4 fails to promote SPOC function. (A) Localization of GFP-Kin4, GFP–Kin4-pr, and GFP–Kin4-pr-SS. The indicated cells carrying SPC42-eqFP611 were grown in YPR medium. After 2 h of incubation with nocodazole, galactose was added for 1 h to induce the expression of the pGal1-GFP-KIN4 constructs. Cells were analyzed by fluorescence microscopy. The arrows point toward SPB-associated Kin4-GFP signals. (B) Quantification of A. n > 50 large-budded cells per strain. (C) GFP–Kin4-pr, GFP–Kin4-pr-SS, GFP-Kin4, and GFP-Kin4T209A were immunoprecipitated with anti-GFP antibodies. Kin4 kinase activity was determined using purified MBP-Bfa1 as a substrate. A C-terminal degradation product of Bfa1 is shown. The bottom panel shows the GFP-Kin4 levels determined by immunoblotting with anti-GFP antibodies. (D) KIN4 and KIN4-pr-SS cells are checkpoint proficient, whereas kin4Δ and KIN4-pr cells are SPOC deficient. The indicated cell types carrying dyn1Δ were incubated at 14°C. DNA was stained with DAPI. The indicated cell types were determined for >100 large-budded cells per strain. Bar, 5 μm.
In vitro measurements using Bfa1 as a substrate (Fig. 5 C, Coomassie brilliant blue [CBB]) demonstrated that the kinase activity of Kin4, Kin4-pr, and Kin4–pr-SS were similar (Fig. 5 C, 32P). Therefore, modification of the C terminus of Kin4 did not affect kinase activity.
Measurements using dyn1Δ cells showed that KIN4-pr-SS was SPOC proficient. In contrast, KIN4-pr behaved in the same checkpoint-deficient manner as kin4Δ cells (Fig. 5 D). This indicates that membrane-bound Kin4 is unable to sustain the SPOC. Thus, either continuous turnover at the cell cortex or association with the SPB is essential for the SPOC function of Kin4.
Kin4 anchored to the SPB delays mitotic exit
The aforementioned data indicate that membrane-tethered Kin4 fails to elicit SPOC function. Implicit in this observation is an essential role of the SPB-associated Kin4 in SPOC regulation. To obtain further evidence that Kin4's function in cell cycle regulation is at the SPB, we examined whether Kin4 tethered to both SPBs interferes with mitotic exit. This would be expected because it would enable Kin4 to phosphorylate Bfa1 and, thereby, prevent inactivation of the Bfa1–Bub2 complex by Cdc5 polo kinase that would otherwise promote mitotic exit.
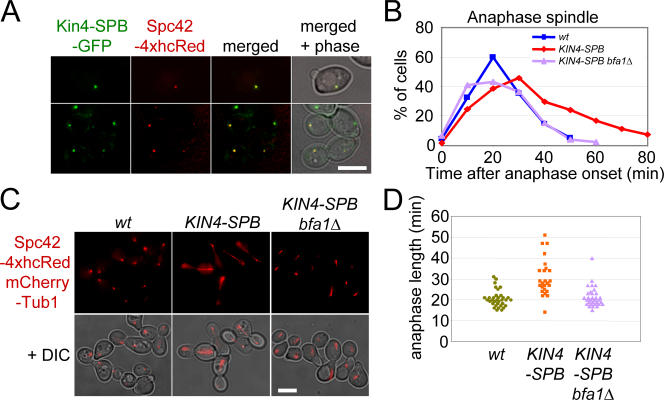
To target Kin4 to both SPBs, the region coding for amino acids 177–622 of SPC72 was fused with KIN4 (hereafter, we refer to this gene as KIN4-SPB). SPC72 was used as a fusion module because the C-terminal domain of Spc72 is able to target proteins efficiently to the cytoplasmic side of the SPB (Knop and Schiebel, 1998; Usui et al., 2003). KIN4-SPB was expressed from the endogenous KIN4 promoter as a sole copy of KIN4. Kin4-SPB protein localized to SPBs throughout the cell cycle (Fig. 6 A). It is important to note that Kin4-SPB associated with both the budward- and motherward-directed SPBs even when these KIN4-SPB cells had correctly aligned anaphase spindles (Fig. 6, A and C). This symmetric SPB localization was never observed for the wild-type Kin4 protein in cells with a correctly aligned spindle (Pereira and Schiebel, 2005).
Figure 6.
SPB-anchored Kin4 inhibits mitotic exit in cells with a correctly aligned spindle. (A) SPB localization of Kin4-SPB–GFP. KIN4-SPB–GFP SPC42-4xhcRed cells were grown to mid-log phase in YPAD medium at 30°C. (B) Duration of anaphase in wild-type (wt), KIN4-SPB, and KIN4-SPB bfa1Δ cells. Wild-type, KIN4-SPB, and KIN4-SPB bfa1Δ cells all carrying SPC42-4xhcRed mCherry-TUB1 were synchronized with α factor in G1 and released at 30°C. Samples were fixed with 4% PFA. Anaphase onset of wild-type, KIN4-SPB, and KIN4-SPB bfa1Δ cells was set to 0 min. (C) KIN4-SPB and KIN4-SPB bfa1Δ cells have normal spindle morphology and orientation. Images 40 min after anaphase onset in B are shown. (D) Anaphase length of wild-type (wt), KIN4-SPB, and KIN4-SPB bfa1Δ cells. Duration of anaphase in wild-type, KIN4-SPB, and KIN4-SPB bfa1Δ cells carrying SPC42-4xhcRed mCherry-TUB1 was determined by live cell imaging. The start of anaphase was defined by spindle elongation, and the end of anaphase was defined by disassembly of the spindle. For wild-type, KIN4-SPB, and KIN4-SPB bfa1Δ cells, 34, 25, and 32 cells were analyzed, respectively. Note that the duration of the experiment was 60 min. Therefore, nine KIN4-SPB cells that initiated anaphase and persisted for >30 min but did not disassemble the spindle within the experiment were excluded from the analysis. Thus, the mean duration of anaphase of KIN4-SPB cells is probably an underestimation. Bars, 5 μm.
Anaphase progression was analyzed in α factor–synchronized KIN4-SPB cells carrying SPC42-4xhcRed mCherry-TUB1. In KIN4-SPB cells, the correctly aligned anaphase spindles persisted for a longer time than in wild-type cells (Fig. 6 B). This phenotype was suppressed by the deletion of BFA1, suggesting that Kin4-SPB delays mitotic exit via modification of Bfa1 activity (Fig. 6 B). To further confirm this observation, we measured the duration of anaphase in KIN4-SPB cells by live cell imaging. The duration of anaphase was extended by a mean of 10 min when the timing in KIN4-SPB cells was compared with wild-type cells (wild type, 20.5 ± 4.0 min, n = 34; KIN4-SPB, 30.8 ± 8.8 min, n = 25). In contrast, the duration of anaphase in KIN4-SPB bfa1Δ cells (21.4 ± 4.8 min, n = 32) was indistinguishable from wild type (Fig. 6 D). Consistently, degradation of the mitotic cyclin Clb2 that accompanies mitotic exit was delayed in KIN4-SPB cells (unpublished data). These results suggest that if Kin4 is permanently recruited to both SPBs, it is capable of inhibiting mitotic exit regardless of the orientation of the spindle. This highlights the significance of the restricted association of Kin4 to the motherward SPB in undisturbed wild-type cells and further supports the notion that Kin4 as Cdc5 regulates Bfa1 at SPBs.
Roles of the γ-tubulin receptor protein Spc72 in regulation of the SPOC
To better understand the role of the SPB in mediating the regulation of Bfa1 by Kin4, we sought SPB proteins that could physically interact with Kin4. Kin4 showed a strong two-hybrid interaction with Spc72 (unpublished data). In addition, Kin4 bound to Spc72 and Spc72Δ1–98 in pull-down experiments but was unable to associate with truncated Spc72Δ176–230 (Fig. 7 A). This suggested that Spc72 targets Kin4 to SPBs. Consistently, Kin4 failed to localize to SPBs in spc72 Δ176–230 cells, whereas the cell cortex association was indistinguishable from that of wild-type cells (Fig. 7 B). Thus, Kin4 binding to the SPB requires amino acids 176–230 of Spc72.
We are now able to use spc72 Δ176–230 cells, which are devoid of Kin4 at SPBs (Fig. 7 B), to ask whether the association of Kin4 with the SPB is important for SPOC function. To increase the percentage of cells with misaligned spindles in our assays, spc72 Δ176–230 was combined with the kar9Δ mutation. The cold-sensitive dyn1Δ was not used in this experiment because spc72 Δ176–230 cells displayed a cold-sensitive growth defect (unpublished data). Only a small proportion of the population of kar9Δ cells with misaligned anaphase spindles continued to progress through the cell cycle, as indicated by the accumulation of cell types (5.5%) characteristic for SPOC failure (Fig. 7 C, kar9Δ). spc72 Δ176–230 cells also had misaligned anaphase spindles (∼10%) as a result of defective cytoplasmic MTs (Usui et al., 2003). Interestingly, a substantial number of spc72 Δ176–230 cells failed to arrest cell cycle progression in anaphase. Instead, 18.4% of large-budded cells accumulated with a SPOC-deficient phenotype (Fig. 7 C). Furthermore, kar9Δ spc72 Δ176–230 as well as kar9Δ kin4Δ and kar9Δ bub2Δ cells exhibited similar degrees of SPOC defects. The inability of kar9Δ spc72 Δ176–230 cells to arrest in anaphase clearly demonstrates a functionalinput from Spc72 in SPOC regulation, probably through the recruitment of Kin4 to SPBs.
Finally, we asked whether Spc72 influences SPOC function solely via its interaction with Kin4 or whether it exerts an additional influence on SPOC function. To this end, we analyzed spc72-7 cells at the semipermissive temperature of 30°C. When grown at 30°C, a considerable fraction of spc72-7 cells enter anaphase with misaligned spindles and accumulate large-budded cells with multiple DAPI-staining regions, which are a hallmark of SPOC-deficient cells (Fig. 7 D). This phenotype was not enhanced by the deletion of KIN4, suggesting that SPC72 functions downstream of KIN4 in the SPOC pathway. Importantly, spc72-7 cells with misaligned anaphase spindles carried Kin4-GFP at both SPBs (Fig. 7, E and F), indicating that the absence of Kin4 at SPBs was not responsible for the SPOC defect of spc72-7 cells. We conclude that Spc72 influences SPOC function by both the targeting of Kin4 to SPBs, which is revealed by the spc72 Δ176–230 allele, and by a separate mechanism that is defective in spc72-7 cells grown at 30°C.
Discussion
In this paper, we provide the first molecular understanding of how the kinase Kin4 functions in the SPOC. We show that the γ-tubulin complex adaptor protein Spc72 provides a SPOC- regulated binding site for the kinase Kin4. Kin4 bound to Spc72 then phosphorylates Bfa1. This Kin4-modified Bfa1 is protected from further phosphorylation by Cdc5 pololike kinase. Finally, we provide the first experimental evidence that recruitment of Kin4 to the SPB that carries the Bfa1–Bub2 GAP complex is sufficient to delay mitotic exit, illustrating the importance of the spatial SPB localization of SPOC components (Figs. 6 and 7).
Phosphorylation by Kin4 protects Bfa1 from Cdc5
In trying to understand how orientation of the anaphase spindle could regulate the ability of Cdc5 pololike kinase to phosphorylate Bfa1 in mid-anaphase, we first considered the possibilities that the amount or activity of Cdc5 at SPBs is controlled by orientation of the anaphase spindle. However, neither Cdc5 binding to SPBs nor Cdc5-dependent phosphorylation of the SPB components Nud1 and Spc72 were considerably affected by misalignment of the anaphase spindle (Fig. 1). Thus, a mechanism that is dependent on the orientation of the anaphase spindle must specifically regulate the activity of Cdc5 toward Bfa1.
This mechanism likely involves the kinase Kin4 because it functions upstream of the Bfa1–Bub2 GAP complex (D'Aquino et al., 2005; Pereira and Schiebel, 2005). Because the kinase activity of Kin4 is essential for its function in SPOC regulation (Fig. S2), we screened SPB-associated MEN and SPOC components for their ability to be phosphorylated by Kin4. This approach identified serine residues 150 and 180 of Bfa1 as phosphorylation sites of Kin4. Although there may be other Kin4 substrates, subsequent analysis of the phosphorylation mutant Bfa12A strongly suggested that Bfa1 is the main, if not the only, target of Kin4 in SPOC regulation.
Elimination of the Kin4-dependent phosphorylation of Bfa1, as achieved with the BFA12A or kin4Δ mutations, enables Cdc5 to modify Bfa1 in conditions in which this is normally prevented in wild-type cells (Fig. 4). This suggests that by phosphorylating Bfa1, Kin4 protects Bfa1 from the inactivating modifications that would otherwise be imposed by Cdc5. In this way, Kin4 counteracts Cdc5 and keeps Bfa1 active when the anaphase spindle becomes misaligned (Fig. 7 G). How the Kin4-modified Bfa1 is protected from Cdc5 is not fully understood. Bfa1 phosphorylated by Kin4 may no longer be a substrate of Cdc5. Alternatively, Kin4 modification of Bfa1 may create a binding site for an unknown factor that protects Bfa1 from Cdc5. Finally, in the context of the SPB, Kin4 modification may introduce a conformational change that renders the Bfa1 protein inaccessible to a further modification by the SPB-bound Cdc5. We favor one of the two latter possibilities because Bfa1 phosphorylated by Kin4 is still modified by Cdc5 in vitro (unpublished data). Measurements of conformational changes of SPOC components within the SPB will be required to further elucidate the mechanisms of how Bfa1–Bub2, Cdc5, Kin4, and Spc72 are regulated in response to spindle alignment defects.
The γ-tubulin complex receptor Spc72 provides a link between MT ends and the SPOC
The SPOC becomes active as soon as cytoplasmic MTs fail to interact in a differential manner with the distinct cortices of the mother and the bud (Pereira et al., 2001). Thus, it is probably not the misaligned spindle itself but rather the defective cytoplasmic MT–cortex interactions that are sensed by the SPOC. Given the localization of Bfa1–Bub2, Kin4, and Cdc5 to the SPB, it is likely that the sensory mechanism that detects these MT–cell cortex interaction defects is also associated with this organelle (Pereira et al., 2000; Pereira and Schiebel, 2005).
In this study, we identified the γ-tubulin complex receptor Spc72 (Knop and Schiebel, 1998) as a core component of the SPB that exerts a functional role in SPOC control. Support for this conclusion comes from two observations. First, Spc72 binds Kin4 to SPBs. This interaction is regulated because Kin4 only associates with both SPBs when cytoplasmic MTs are defective. Second, spc72-7 and spc72 Δ176–230 cells are SPOC deficient (Fig. 7). Importantly, spc72-7 cells are checkpoint deficient even though Kin4 is associated with both SPBs in these cells (Fig. 7). These latter data support the view that the SPB component Spc72 does not function solely as a binding scaffold for Kin4 but rather is also more actively involved in SPOC regulation.
Spc72 directly interacts with the yeast γ-tubulin complex, the kinases Kin4 and Cdc5, and the scaffold protein Nud1 (Fig. 7 G; Knop and Schiebel, 1998; Gruneberg et al., 2000). These multiple interactions place Spc72 in an ideal position to translate the positional information of the spindle that is relayed to the SPB by the interactions between the cytoplasmic MT and the cortex into a cell cycle regulating SPOC signal.
Spatial regulation of SPOC components
Whenever the spindle is correctly positioned, the Bfa1–Bub2 complex binds predominately to the budward-directed SPB, and Kin4 is restricted to the SPB in the mother cell (Pereira et al., 2001; Pereira and Schiebel, 2005). Cdc5 pololike kinase associates with both SPBs (Fig. 1 A) and phosphorylates Bfa1 in mid-anaphase, thereby reducing the Bfa1–Bub2 GAP activity to permit mitotic exit (Hu et al., 2001; Geymonat et al., 2003). However, when the anaphase spindle becomes misaligned, the γ-tubulin complex receptor protein Spc72 at the previously budward-directed SPB promotes the recruitment of Kin4. The signal that recruits Kin4 to the second SPB is presently unclear, but it may include conformational changes in Spc72 itself. These might be expected to arise as a result of alterations in the forces acting on the cytoplasmic MTs that are anchored to the SPB by this Spc72 scaffold protein. Bfa1–Bub2 also binds to the second SPB of cells with mispositioned anaphase spindles (Pereira and Schiebel, 2001). This colocalization allows Kin4 to modify Bfa1 (Fig. 2). As a consequence, Cdc5 is unable to phosphorylate Bfa1, and the Bfa1–Bub2 GAP complex remains active (Fig. 4; Geymonat et al., 2003).
Support for this spatial control model is provided by the cell cycle phenotype of KIN4-SPB cells, in which Kin4 is constitutively tethered to both SPBs via the SPB-targeting domain of Spc72. In these cells, the duration of anaphase becomes extended from 20 to ∼30 min in a manner that is dependent on the inhibitory modifications of Bfa1, which are, in turn, dependent on Kin4 activity. Thus, targeting of Kin4 to the SPB carrying the Bfa1–Bub2 GAP complex is sufficient to delay mitotic exit. Although Kin4 kinase activity may still be regulated by spindle orientation (D'Aquino et al., 2005), our results clearly demonstrate the necessity for a tight, special regulation of SPOC components.
It is important to note that Spc72 is related to the transforming acidic coiled-coil (TACC) protein of higher eukaryotes. Alterations in TACC function have been associated with transformation in the development of cancer (Raff, 2002). Thus, it is possible that TACC proteins also connect MT function to cell cycle regulators.
Materials and methods
Plasmids and yeast strains
Strains and plasmids are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200705197/DC1). Yeast strains were derivatives of YPH499 and were constructed by PCR-based methods (Janke et al., 2004). The red fluorescent eqFP611 from the sea anemone Entacmaea quadricolor (Wiedenmann et al., 2002) was used to mark SPBs through a fusion with SPC42 (Donaldson and Kilmartin, 1996). Alternatively, four copies of the far-red fluorescent protein HcRed, which was discovered through site-directed and random mutagenesis efforts on a nonfluorescent chromoprotein (hcCP) isolated from the Indo-Pacific Anthozoa species, was used in some experiments (Gurskaya et al., 2001). The Cherry-tubulin construct was described previously (Khmelinskii et al., 2007).
Cell cycle analysis and growth conditions
For synchronization, yeast cells were grown in YPAD (yeast extract, peptone, adenine, and dextrose) medium and arrested in G1 by treatment with 10 μg/ml α factor for 2.5 h or 2 h at 23°C or 30°C, respectively, until >95% of cells showed a mating projection. Cells were then washed with prewarmed growth medium to remove α factor and were resuspended in YPAD medium at the indicated temperatures. Cells with CDC5 under the control of the pGal1 promoter were grown in YPA + 3% raffinose and 2% galactose medium. For the depletion of Cdc5, pGal1-CDC5 cells were synchronized with α factor and released into YPAD medium to repress the pGal1 promoter. Synthetic complete medium was used for live cell imaging experiments. To depolymerize MTs, cells were incubated in YPAD medium containing 15 μg/ml nocodazole at 30°C for 2.5 h. Cells carrying dyn1Δ were incubated at 14°C for 20 h before examination.
Preparation of recombinant proteins
GST and GST-fused proteins were expressed in Escherichia coli Rosetta (DE3) by adding 0.1 mM IPTG for 2 h at 37°C. MBP and all MBP-fused proteins were expressed in E. coli Rosetta (DE3) and purified according to the manufacturer's protocol (New England Biolabs, Inc.). After intensive washing steps, GST and GST fusion proteins bound to glutathione–Sepharose beads were used for in vitro pull-down assays. For in vitro kinase assays, all recombinant proteins were eluted either with glutathione in the case of GST fusion proteins or with maltose for MBP fusion proteins. Proteins were then dialyzed against a buffer containing 20 mM Hepes, pH 7.4, 0.5 mM EDTA, and 10% glycerol. All recombinant proteins were kept at −20°C except for MBP-Bfa1, which was stored at −80°C.
In vitro pull-down assay
Yeast total cell extracts were prepared from logarithmically growing cells in immunoprecipitation buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol, 0.2 mM NaVO3 100 mM β-glycerophosphate, 50 mM NaF, 1 mM PMSF, 1 mM DTT, 0.5% Triton X-100, and Complete EDTA-free protease inhibitor cocktail [Roche]). 300 μg of total cell extract was incubated with GST or GST-Spc72–bound beads for 2 h at 4°C. The beads were washed three times with immunoprecipitation buffer. Kin4-3HA was detected with the mouse monoclonal antibody 12CA5.
In vitro kinase assay
GST-Cdc5 and GST-Cdc5kd used in Figs. 1 D and 2 A were expressed in yeast strains provided by S. Sedgwick (National Institute for Medical Research, London, UK) and were purified as described previously (Geymonat et al., 2002). GST-Kin4 and GST-Kin4T209A used in Fig. 2 (A, C, and D) were purified from the yeast strains listed in Table S1 using the same protocol. Kin4-3HA, Kin4T209A-3HA, GFP-Kin4, and GFP-Kin4 derivatives in Figs. S3 and 5 C were enriched as follows. Total cell extracts were prepared in immunoprecipitation buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5% glycerol, 0.2 mM NaVO3, 100 mM β-glycerophosphate, 50 mM NaF, 1 mM PMSF, 1 mM DTT, 1% NP40, and Complete EDTA-free protease inhibitor cocktail [Roche]). Kin4 kinase was immunoprecipitated with mouse monoclonal antibody 12CA5 or affinity-purified rabbit anti-GFP antibody bound to protein A–coupled beads. Kin4 and Cdc5 kinase assays were performed as previously described (Geymonat et al., 2002; D'Aquino et al., 2005). Radioactivity was either detected with BioMax MS films (Kodak) or determined by a phosphoimager (FLA-300; Fujifilm) and quantified with Image Gauge version 3.45 (Fujifilm). To determine Kin4-dependent phosphorylation sites by MALDI-TOF/TOF, MBP-Bfa1 was incubated with GST-Kin4 purified from the strain provided by S. Sedwick in the same buffer as described for the other Kin4 kinase assays but in the presence of 6.7 mM ATP and was incubated at room temperature for 1 h.
Antibodies
Affinity-purified sheep anti-Cdc5, -Bfa1, -Clb2, and -Tub2 antibodies were described previously (Pereira et al., 2002). Anti-Nud1 and -Spc72 antibodies have been described previously (Gruneberg et al., 2000; Ho et al., 2002). Anti-Sic1 antibodies were raised in guinea pigs. Mouse monoclonal antibody 12CA5 was used to detect Bfa1-3HA and Kin4-3HA. Anti–P-S150 and anti–P-S180 antibodies of Bfa1 were raised in rabbits against peptides RLKQPRS(p)MMELK and VRFKKS(p)MPNL, respectively. The antibodies were purified using the nonphosphorylated peptide as preadsorption matrix followed by affinity purification using the immobilized phosphopeptides.
Fluorescence microscopy
Yeast cells with Kin4-GFP grown in YPAD were immediately analyzed by fluorescence microscopy without washing or fixation. Other cells were fixed with 70% ethanol, washed with PBS, and incubated in PBS containing DAPI to visualize DNA. CDC5-GFP, BFA1-GFP, and BUB2-GFP cells were analyzed by fluorescence microscopy after fixation with 4% PFA in 150 mM phosphate buffer, pH 6.5, for 10 min at room temperature. PFA-fixed cells were incubated with 1 μg/ml DAPI for 15 min at room temperature. Z-series images of 0.35-μm steps were captured with a microscope (Axiophot; Carl Zeiss MicroImaging, Inc.) equipped with a 100× NA 1.45 Plan-Fluar oil immersion objective (Carl Zeiss MicroImaging, Inc.) and a camera (Cascade:1K; Photometrics) and were processed with MetaMorph software (Universal Imaging Corp.; Figs. 3, A and B; 5, A and B; 6, A and C; 7 B; S2 A; and S4 B), or images were captured with DeltaVision (Applied Precision) equipped with GFP and TRITC filters (Chroma Technology Corp.), a 100× NA 1.4 plan Apo oil immersion objective (IX70; Olympus), and a camera (CoolSNAP HQ; Roper Scientific) and were quantified/processed with SoftWoRx 3.5.0 (Applied Precision; Figs. 1, A and B; 6 D; and 7 E). Projected images are shown. The fluorescence intensity of Cdc5-GFP was measured on a plane that has an SPB in focus with SoftWoRx 3.5.0. Time-lapse experiments were carried out in synthetic complete medium on a concanavalin A–coated glass-bottom dish (MatTek) with DeltaVision at 30°C. Z series at 0.4-μm steps (2 × 2 binning) were acquired every 1 min. Photoshop (Adobe) was used to mount the images and to produce merged color images. No manipulations other than contrast and brightness adjustments were used.
Online supplemental material
Fig. S1 shows the cell cycle progression of cells in Fig. 1. Fig. S2 presents an analysis of kin4T209A cells. Fig. S3 shows an in vitro kinase assay of Kin4. Fig. S4 presents an analysis of BFA12A cells. Table S1 presents the strains and plasmids used in this study. Video 1 shows anaphase of a wild-type cell, Video 2 shows anaphase of a KIN4-SPB cell, and Video 3 shows anaphase of a KIN4-SPB bfa1Δ cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705197/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Amon, Elledge, and Sedwick for antibodies, plasmids, and yeast strains and Drs. Gruss and Tavares for discussions. U. Jäkle and J. Reichert are acknowledged for excellent technical support, and we thank Dr. I. Hagan for critical comments on the manuscript.
The work of G. Pereira is supported by the Helmholtz Association Young Investigator grant HZ-NG-111, and E. Schiebel is supported by Deutsche Forschungsgemeinschaft Schi grant 295/3-1.
Abbreviations used in this paper: CBB, Coomassie brilliant blue; GAP, GTPase-activating protein; MALDI-TOF, matrix-assisted laser desorption ionization–time of flight; MBP, maltose-binding protein; MEN, mitotic exit network; MT, microtubule; SPB, spindle pole body; SPOC, spindle orientation checkpoint; TACC, transforming acidic coiled coil.
References
- Asakawa, K., S. Yoshida, F. Otake, and A. Toh-e. 2001. A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics. 157:1437–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin, A.J., R. Visintin, and A. Amon. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 102:21–31. [DOI] [PubMed] [Google Scholar]
- Cheng, L., L. Hunke, and C.F. Hardy. 1998. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase Cdc5p. Mol. Cell. Biol. 18:7360–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aquino, K.E., F. Monje-Casas, J. Paulson, V. Reiser, G.M. Charles, L. Lai, K.M. Shokat, and A. Amon. 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell. 19:223–234. [DOI] [PubMed] [Google Scholar]
- Donaldson, A.D., and J.V. Kilmartin. 1996. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol. 132:887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini, R., C. D'Ambrosio, M. Venturetti, G. Lucchini, and S. Piatti. 2006. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat, M., A. Spanos, S.J. Smith, E. Wheatley, K. Rittinger, L.H. Johnston, and S.G. Sedgwick. 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277:28439–28445. [DOI] [PubMed] [Google Scholar]
- Geymonat, M., A. Spanos, P.A. Walker, L.H. Johnston, and S.G. Sedgwick. 2003. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278:14591–14594. [DOI] [PubMed] [Google Scholar]
- Gruneberg, U., K. Campbell, C. Simpson, J. Grindlay, and E. Schiebel. 2000. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 19:6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya, N.G., A.F. Fradkov, A. Terskikh, M.V. Matz, Y.A. Labas, V.I. Martynov, Y.G. Yanushevich, K.A. Lukyanov, and S.A. Lukyanov. 2001. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 507:16–20. [DOI] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G.D. Bader, L. Moore, S.L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183. [DOI] [PubMed] [Google Scholar]
- Hu, F., Y. Wang, D. Liu, Y. Li, J. Qin, and S.J. Elledge. 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 107:655–665. [DOI] [PubMed] [Google Scholar]
- Hwang, L.H., L.F. Lau, D.L. Smith, C.A. Mistrot, K.G. Hardwick, E.S. Hwang, A. Amon, and A.W. Murray. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 279:1041–1044. [DOI] [PubMed] [Google Scholar]
- Janke, C., M.M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. [DOI] [PubMed] [Google Scholar]
- Khmelinskii, A., C. Lawrence, J. Roostalu, and E. Schiebel. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and E. Schiebel. 1998. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 17:3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.-Y., E. Yeh, T. Hays, and K. Bloom. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 90:10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, A.S., J.J. Jang, and R.J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 98:7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel. 2001. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr. Opin. Cell Biol. 13:762–769. [DOI] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel. 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell. 19:209–221. [DOI] [PubMed] [Google Scholar]
- Pereira, G., T. Höfken, J. Grindlay, C. Manson, and E. Schiebel. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6:1–10. [PubMed] [Google Scholar]
- Pereira, G., T.U. Tanaka, K. Nasmyth, and E. Schiebel. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p/Bub2p checkpoint protein complex. EMBO J. 20:6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., C. Manson, J. Grindlay, and E. Schiebel. 2002. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak, P.M., and F.A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12:2684–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, J.W. 2002. Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 12:222–225. [DOI] [PubMed] [Google Scholar]
- Schweitzer, B., and P. Philippsen. 1991. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 7:265–273. [DOI] [PubMed] [Google Scholar]
- Segal, M., and K. Bloom. 2001. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 11:160–166. [DOI] [PubMed] [Google Scholar]
- Shirayama, M., Y. Matsui, and A. Toh-e. 1994. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M-phase. Mol. Cell. Biol. 14:7476–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., W. Zachariae, R. Ciosk, and K. Nasmyth. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17:1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon. 2004. Closing mitosis: the function of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232. [DOI] [PubMed] [Google Scholar]
- Usui, T., H. Maekawa, G. Pereira, and E. Schiebel. 2003. The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 22:4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., K. Craig, E.S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718. [DOI] [PubMed] [Google Scholar]
- Visintin, R., E.S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 398:818–823. [DOI] [PubMed] [Google Scholar]
- Wiedenmann, J., A. Schenk, C. Röcker, A. Girod, K.-D. Spindler, and G.U. Nienhaus. 2002. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria). Proc. Natl. Acad. Sci. USA. 99:11646–11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.