Abstract
The survival motor neuron (SMN) complex functions in maturation of uridine-rich small nuclear ribonucleoprotein (RNP) particles. SMN mediates the cytoplasmic assembly of Sm proteins onto uridine-rich small RNAs, and then participates in targeting RNPs to nuclear Cajal bodies (CBs). Recent studies have suggested that phosphorylation might control localization and function of the SMN complex. Here, we show that the nuclear phosphatase PPM1G/PP2Cγ interacts with and dephosphorylates the SMN complex. Small interfering RNA knockdown of PPM1G leads to an altered phosphorylation pattern of SMN and Gemin3, loss of SMN from CBs, and reduced stability of SMN. Accumulation in CBs is restored upon overexpression of catalytically active, but not that of inactive, PPM1G. This demonstrates that PPM1G's phosphatase activity is necessary to maintain SMN subcellular distribution. Concomitant knockdown of unr interacting protein (unrip), a component implicated in cytoplasmic retention of the SMN complex, also rescues the localization defects. Our data suggest that an interplay between PPM1G and unrip determine compartment-specific phosphorylation patterns, localization, and function of the SMN complex.
Introduction
Removal of introns from primary RNA transcripts (splicing) takes place in specialized complexes called spliceosomes, in which factors needed for splicing of pre-mRNAs are enriched. Currently, >150 different proteins and several small RNAs have been identified as part of spliceosomes, which are organized in distinct subcomplexes. The most prominent spliceosome subunits are the uridine-rich small nuclear RNPs (U snRNPs) of the Sm class. They consist of an RNA component (uridine-rich small nuclear RNA [U snRNA]) and numerous proteins that are either common for all or specific for one particle (for review see Nilsen, 2003).
Even though splicing occurs in the nucleus, major parts of the biogenesis of U snRNPs take place in the cytoplasm. The nuclear-encoded m7G-capped U snRNA is transiently exported to the cytoplasm to allow binding of the common (Sm) proteins. This leads to the formation of the Sm core domain, the structural framework of all spliceosomal U snRNPs of the Sm class (Raker et al., 1996). Formation of the Sm core is required for cap hypermethylation and the subsequent nuclear import of U snRNPs (Hamm et al., 1990). Within the nucleus, U snRNPs are first targeted to subnuclear domains termed Cajal bodies (CBs), where additional modifications on the RNA occur and at least some specific proteins are added. Eventually, the mature U snRNPs migrate to perichromatin fibrils, the sites of transcription and splicing (for reviews see Meister et al., 2002; Matera et al., 2007).
Interestingly, recent studies indicated that several aspects of the biogenesis cycle of U snRNPs are factor mediated and regulated in vivo. The most prominent factor in this process is the survival motor neuron (SMN) complex, a macromolecular entity that actively mediates the binding of the common Sm proteins onto U snRNAs. This complex consists of nine major proteins, including the SMN gene product, Gemin2–8, and the unr-interacting protein (unrip; for reviews see Meister et al., 2002; Gubitz et al., 2004; Pellizzoni, 2007) (Carissimi et al., 2005; Grimmler et al., 2005b). The SMN complex is controlled by another complex, whose name-giving component is the type-II protein arginine methyltransferase 5. This unit, possibly in conjunction with other factors, converts arginine residues in some Sm proteins into symmetrical dimethylarginines, thereby enhancing their affinity for the SMN complex and stimulating U snRNP assembly (Brahms et al., 2001; Friesen et al., 2001; Meister et al., 2001b; Meister and Fischer, 2002). Furthermore, it has been shown that the SMN complex (or parts thereof) also participate in the subsequent nuclear import of U snRNPs (Narayanan et al., 2004; Shpargel and Matera, 2005). Once in the nucleus, both units migrate to CBs, where the SMN complex accumulates and U snRNPs are released to sites of splicing after additional maturation steps (Stanek and Neugebauer, 2006). These observations suggest that U snRNPs dissociate from SMN complexes in CBs and that the SMN complex returns as a separate unit to the cytoplasm at later stages.
Although the cytoplasmic role of the SMN complex is understood in some detail, its functions in the nucleus are only poorly characterized. Thus, it is still unclear how U snRNPs are separated from the SMN complex after nuclear import and how the return of the SMN complex to the cytoplasm is facilitated. An important player in this process might be unrip, which interacts with the SMN complex primarily in the cytoplasm. Knockdown of this factor leads to enhanced accumulation of SMN in nuclear bodies (Grimmler et al., 2005b), suggesting a role of unrip in the intracellular distribution of the SMN complex. The biogenesis of U snRNPs appears also to be influenced by phosphorylation of different components of the assembly machinery. Thus, it has been shown that SMN is highly phosphorylated when it is in the cytoplasm, whereas the nuclear pool is hypophosphorylated (Grimmler et al., 2005a). Compartment-specific determinants and the phosphorylation status of SMN (and potentially other SMN-complex components) could hence influence the biogenesis pathway of U snRNPs in the cytoplasm and in the nucleus.
Here, we show that the SMN complex specifically interacts with the nuclear phosphatase PPM1G/PP2Cγ (here referred to as PPM1G). This C-type, Mg2+/Mn2+-dependent phosphatase has previously been shown to be specifically involved in nuclear functions, including splicing (Murray et al., 1999; Allemand et al., 2007) and nucleosome assembly (Kimura et al., 2006). Knockdown of PPM1G led to reduced dephosphorylation of SMN-complex proteins and abolished accumulation of SMN proteins in CBs. Rescue experiments demonstrate that the catalytic activity of the phosphatase is necessary to maintain the specific localization of the SMN proteins in the nucleus. Our data support the idea that PPM1G is a major regulator of nuclear functions of the SMN complex: it determines the SMN complex's compartment-specific phosphorylation pattern and is required for its correct localization and stability.
Results
PPM1G is a nuclear phosphatase in vertebrates
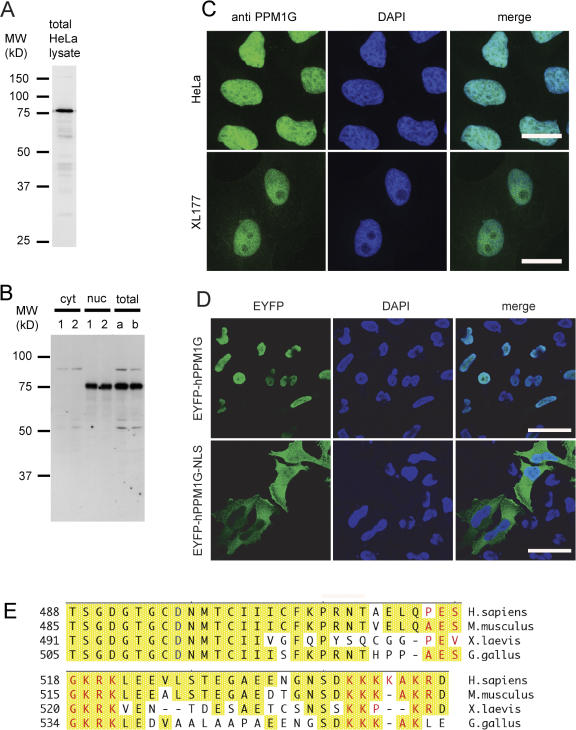
As published recently, the compartment-specific phosphorylation pattern of the SMN protein pointed to the activity of a nuclear phosphatase, which dephosphorylates proteins of the SMN complex (Grimmler et al., 2005a). One potential candidate that may govern SMN modifications in the nucleus is PPM1G, which has been suggested to localize to the nucleus and to function in splicing (Murray et al., 1999). Consistent with this, antibodies generated against the human (Fig. 1 A) or the Xenopus laevis orthologue of PPM1G (xPPM1G; Fig. 1 B) detected this protein in the nucleoplasm in both human HeLa and X. laevis XL177 cells when used for indirect immunofluorescence (Fig. 1 C). Sequence analysis revealed two stretches of basic amino acids in the C terminus of human PPM1G (hPPM1G), which could serve as an NLS. The region of the potential C-terminal NLS is conserved throughout most vertebrates (Fig. 1 E). Indeed, when we expressed the full-length or a truncated version of hPPM1G lacking the C-terminal NLS as EYFP fusions in human HeLa cells, only the full-length protein showed nuclear accumulation, whereas the truncated version did not (Fig. 1 D). Likewise, accumulation of fluorescently labeled xPPM1G in in vitro reconstituted nuclei was not seen for analogous C-terminal truncations of xPPM1G, although the truncation did not affect the catalytic activity of xPPM1G (unpublished data). Moreover, when we manually dissected X. laevis oocytes into cytosolic and nuclear fractions, we exclusively detected xPPM1G in the nuclear fraction using our xPPM1G antibody (Fig. 1 B). Using quantitative immunoblotting, we estimated the abundance of PPM1G in X. laevis oocyte nuclei to be in the range of 5 μM and at least 1 μM in HeLa cell nuclei, which is consistent with recently published data (Kimura et al., 2006). This shows that PPM1G is an abundant nuclear phosphatase in vertebrates.
Figure 1.
Nuclear localization of protein phosphatase PPM1G is conserved among vertebrates. (A) Immunoblot testing affinity-purified antibody raised against hPPM1G on total HeLa lysate. (B) Immunoblot testing antibodies against xPPM1G and determining the subcellular localization of PPM1G in X. laevis oocytes. 1 and 2 represent two different pools of nuclei (nuc) and corresponding cytosols (cyt) from manually dissected stage VI oocytes. a and b represent two different pools of complete lysates from stage VI oocytes. (C) Indirect immunofluorescence on HeLa cells using the antibodies against hPPM1G (top) or on XL177 cells using the antibody against xPPM1G (bottom). Bars, 20 μm. (D) Immunofluorescence on HeLa cells being transfected with monomeric pEYFP-C1 vectors encoding either full-length (EYFP-hPPM1G) or hPPM1G without the NLS (EYFP-hPPM1G-NLS). Cells were fixed 40 h after transfection and stained with an antibody against GFP. Bars, 40 μm. (E) C termini of PPM1G from four different vertebrate species were aligned using MegAlign software (DNA Star); conserved residues are highlighted in yellow; the glutamate residue critical for phosphatase activity (D496 in hPPM1G) is indicated in blue; and predicted NLS sequences (P-sort software; www.psort.org) are indicated in red. Sequences used are the following: Homo sapiens, O15355; Mus musculus, Q61074; X. laevis, Q7ZYR7; and Gallus gallus (nucleotides 173–1859 from GenBank/EMBL/DDBJ under accession no. XR_026917) after an additional nucleotide (G) had been inserted in position 1675 (see ChEST73b8, ChEST376n8, and others from BBSRC ChickEST Database).
Interaction of PPM1G with the SMN complex
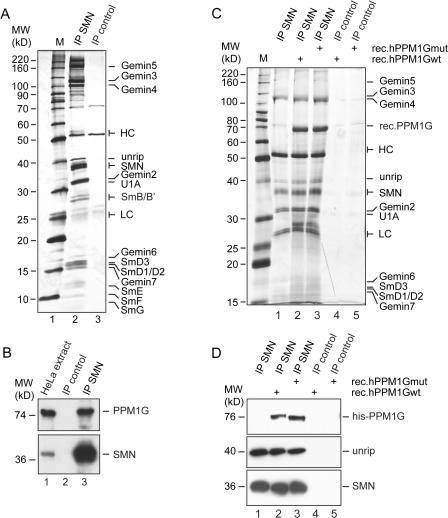
To address whether PPM1G might regulate nuclear functions of the SMN complex, we tested if PPM1G was directly associated with the SMN complex. We affinity-purified SMN-complex proteins from total cellular extracts using a monoclonal antibody directed against the N terminus of human SMN. Bound proteins were eluted and compared with an eluate of a control column by SDS-PAGE and silver staining. Known SMN-complex components (i.e., SMN, Gemin3–5, and unrip) were eluted from this column as determined by mass spectrometry and/or Western blotting (Fig. 2 A, compare lanes 2 and 3). Moreover, among the proteins coeluting with the SMN complex in the 70-kD range was the protein phosphatase PPM1G, which we identified on the basis of 21 peptides covering 32.9% of its sequence (not depicted). Immunoblot analysis with antibodies against hPPM1G confirmed that PPM1G specifically bound to the SMN complex but not to the control column (Fig. 2 B, compare lanes 2 and 3).
Figure 2.
Interaction of PPM1G and the SMN complex. (A) SMN complex was precipitated from HeLa cell extract with a monoclonal anti-SMN antibody (7B10), covalently linked to protein G–Sepharose (IP-SMN). An unrelated mouse serum, covalently linked to protein G–Sepharose, was used as control (IP control). Interacting proteins were eluted by boiling in 2× SDS sample buffer, separated by SDS-PAGE, and analyzed by silver staining. (B) Immunoprecipitations of SMN complex (lane 3) or controls (lane 2) were done as described in A and analyzed by immunoblotting using specific SMN or PPM1G antibodies, respectively. Lane 1 shows HeLa cell total extract. (C) Interaction of recombinant PPM1G with the SMN complex. Immunoprecipitated SMN complex (lanes 2 and 3) or controls (lanes 4 and 5) were incubated with 5 μg of 6× histidine-tagged, purified, recombinant PPM1Gwt (rec.PPM1Gwt; lanes 2 and 4) or catalytically inactive PPM1G (D496A, rec.PPM1Gmut; lanes 3 and 5). Bound proteins were eluted by boiling in 2× SDS sample buffer, resolved by SDS-PAGE, and analyzed by silver staining. (D) Samples generated as described in C were analyzed by immunoblotting using anti-histidine (top), anti-unrip (middle), or anti-SMN antibodies (bottom). Note that because of different gels and markers used in B–D, the apparent molecular mass of PPM1G is slightly different in these panels.
To further analyze binding of PPM1G to the SMN complex, we affinity purified the SMN complex, as described in the previous paragraph, and incubated the complex with recombinant His-tagged human wild-type (wt) PPM1G (rec.hPPM1Gwt). In parallel, we generated a mutant PPM1G (rec.PPM1Gmut) in which aa 496 was changed from an aspartate into an alanine (Murray et al., 1999). The recombinant mutant protein showed dramatically reduced catalytic activity (unpublished data). After incubation and removal of unbound proteins by extensive washing, the SMN containing immunocomplexes were analyzed by SDS-PAGE and silver staining (Fig. 2 C) and immunoblotting (Fig. 2 D). Detection of the SMN protein verified the specificity of the purification (Fig. 2, C and D [bottom]; note that SMN could only be detected upon immunopurification with the specific antibodies [IP SMN] but not in controls [IP control]). PPM1Gwt and catalytically inactive PPM1G were equally detectable in the bound fraction of purified SMN complexes but not in controls (Fig. 2, C and D [top]). Although the wt protein actively dephosphorylated the SMN protein, as indicated by a slightly changed mobility of SMN (Fig. 2, C [SMN] and D [bottom]), interaction of either wt or catalytically inactive PPM1G with the SMN complex was equally efficient but did not change the overall composition of the complex as judged by silver staining of immunocomplexes and immunoblot detection of SMN and unrip (Fig. 2, C and D). Collectively, these data identify the nuclear phosphatase PPM1G as a novel interaction partner of the SMN complex.
PPM1G is required for proper localization of the SMN complex in CBs
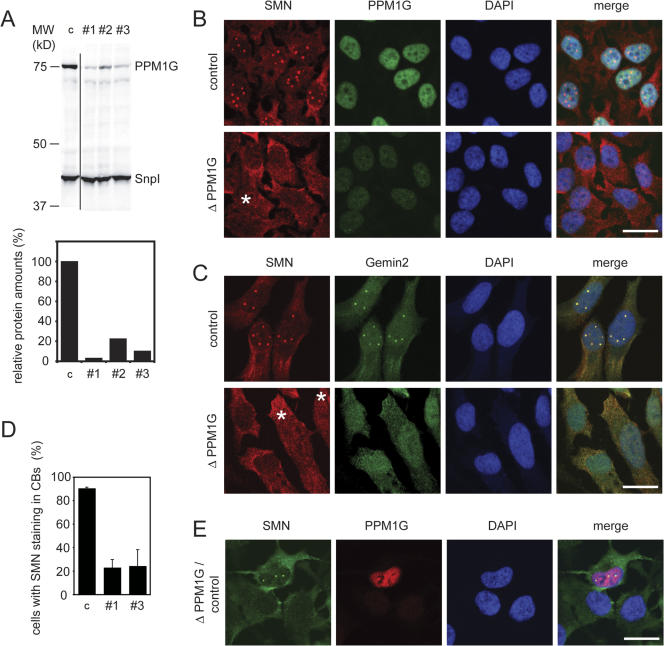
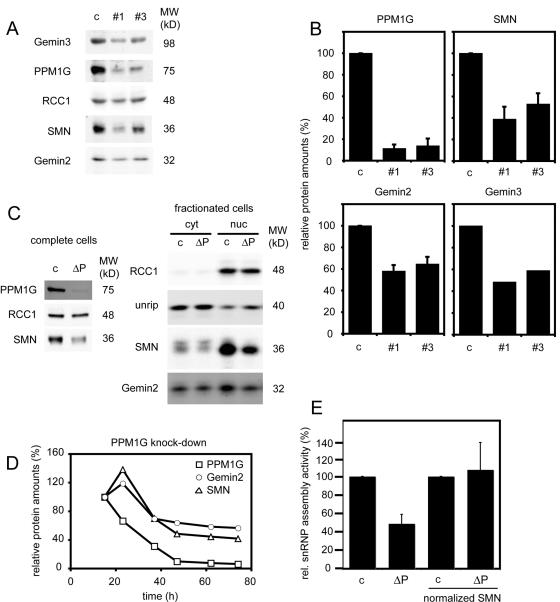
We next asked whether PPM1G was required for proper localization of the SMN complex. Based on immunofluorescence experiments, it had been reported that SMN is diffusely distributed in the cytoplasm but localized in the nucleus, where it is found to be highly enriched in specific bodies termed gems (Liu and Dreyfuss, 1996). However, later studies showed that these bodies are in most, but not all, cell lines equivalent to the well-known nuclear CBs (Carvalho et al., 1999; Dundr et al., 2004). In agreement with previous data, we observed strong accumulation of SMN in CBs of HeLa CCL2 cells (unpublished data). To analyze the role of PPM1G in the localization of SMN proteins, we knocked down PPM1G expression in HeLa CCL-2 cells by RNA interference using three different synthetic double-stranded siRNA oligonucleotides. As judged by immunoblotting, all three siRNAs efficiently reduced the protein amount of PPM1G (Fig. 3 A; the knockdown efficiency was typically 75–95%). Oligo pairs #1 and #3, however, were reproducibly the most efficient and therefore were used for further phenotypic analysis (Fig. 3 A). When SMN-complex localization was tested in siRNA-treated cells, we observed substantial reduction of the nuclear PPM1G in the majority of treated cells (Fig. 3 B). This was correlated with SMN disappearing from CBs (Fig. 3, B and D). Likewise Gemin2, a direct interaction partner of SMN, disappeared from CBs upon PPM1G knockdown (Fig. 3 C). Concomitantly, in ∼50% of all cells the diffuse nucleoplasmic signal of SMN or Gemin2 was increased (Fig. 3, B and C), whereas low nucleoplasmic SMN signals were observed in other cells (Fig. 3, B–D; and Fig. 4). Similarly, upon overexpression of PPM1G lacking the C-terminal NLS (Fig. 1) we frequently observed reduced cytoplasmic and increased diffuse nuclear staining of SMN. However, accumulation of SMN in CBs was not affected (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200704163/DC1). Collectively, these data suggest that nuclear PPM1G is required for correct nuclear-cytoplasmic distribution and subcellular localization of the SMN complex.
Figure 3.
siRNA-mediated knockdown of PPM1G from HeLa cells induces the loss of SMN complex from CBs. (A) Representative experiment showing the depletion efficiency of the three siRNA oligonucleotides (#1, #2, and #3) against PPM1G mRNA. An oligonucleotide against vimentin mRNA was used as a control (c). (top) Immunoblot detection of PPM1G levels 60 h after transfection of cells. Total protein was analyzed by Immunoblotting with antibodies as indicated (snurportin1 [SnpI], loading control). (bottom) Quantification of relative PPM1G amounts from the Immunoblot shown on top. (B) Immunofluorescence staining of HeLa cells after control treatment or knockdown of PPM1G (ΔPPM1G, shown here and in C after treatment with oligo #1; the same phenotype was observed with oligo #3). Cells were fixed 60 h after transfection with siRNA oligonucleotides and stained for SMN and PPM1G. (C) Knockdown of PPM1G as in B. Cells were stained for SMN and Gemin2. Asterisks show cells with increased nucleoplasmic SMN signal. (D) Quantification of cells showing loss of SMN staining in CBs (phenotypes after treatment with oligo #1 or #3). Bars represent the relative number of cells showing localized SMN staining in the nucleus after PPM1G knockdown. Relative numbers are given as means from three independent experiments and error bars represent SD. Absolute numbers of counted cells were as follows (1/2/3 experiment): control, 134/163/172; oligo #1, 99/204/149; and oligo #3, 104/236/158. (E) PPM1G knockdown and SMN delocalization are correlated on the single cell level. HeLa cells were transfected with control or PPM1G #1 siRNA oligonucleotides separately, mixed 15 h after transfection, and plated out together on coverslips. 80 h after transfection, cells were fixed and immunostained for SMN and PPM1G. Bars, 20 μm.
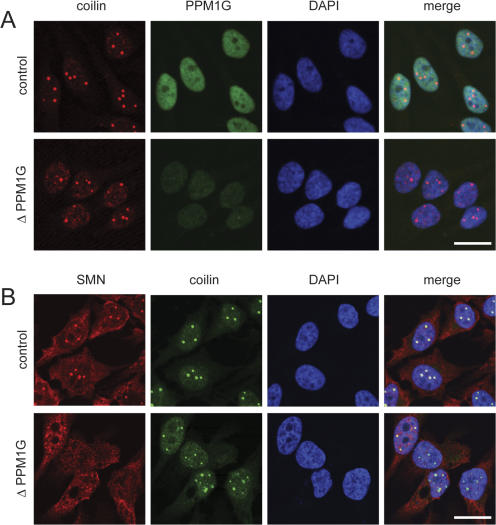
Figure 4.
Knockdown of PPM1G from HeLa cells does not compromise coilin accumulation in CBs. Immunofluorescence staining of HeLa cells after control treatment or knockdown of PPM1G (ΔPPM1G, oligo #1; the same phenotype was seen with oligo #3). Cells were fixed 60 h after transfection with siRNA oligonucleotides and stained for coilin and PPM1G (A) or coilin and SMN (B). Bars, 20 μm.
To confirm correlation of the efficiency of PPM1G knockdown and the loss of the localized nuclear SMN signal, we transfected cells with control siRNA oligos or siRNA oligos to knockdown PPM1G and mixed them after transfection. Indeed, when residual staining of PPM1G was hardly detectable, no accumulation of SMN in nuclear bodies was observed. In contrast, PPM1G was still clearly detectable in cells, which regularly showed normal SMN staining in the nucleus (Fig. 3 E).
Accumulation of SMN in CBs depends on the interaction of SMN with coilin (Hebert et al., 2001, 2002), one of the central molecular components of CBs (Stanek and Neugebauer, 2006). We therefore determined whether down-regulation of PPM1G affected coilin localization. Knockdown of PPM1G frequently resulted in the accumulation of additional small coilin-positive structures (Fig. 4, A and B). However, coilin still strongly accumulated in CBs upon PPM1G knockdown (Fig. 4, A and B). Importantly, this was observed in cells in which loss of SMN from CBs was verified by direct SMN detection (Fig. 4 B). Collectively, these data suggest that PPM1G is required for the accumulation of SMN and Gemin2 in CBs but is not a major determinant of overall CB integrity as judged by coilin staining.
PPM1G influences the stability and thus the function of the SMN complex but does not change the pattern of spliceosome speckles
In the course of our experiments, we noted that the expression levels of SMN and Gemin2 were reduced upon knockdown of PPM1G, which is consistent with a destabilization of SMN-complex proteins. We therefore determined the amounts of different SMN proteins by quantitative immunoblotting after PPM1G knockdown with the two most effective oligo pairs. When PPM1G was depleted by ∼90%, the levels of SMN, Gemin2, and Gemin3 were also considerably reduced. This effect was specific, as the level of the nonrelated protein regulator of chromosome condensation 1 (RCC1) was not affected (Fig. 5, A and B). To determine where SMN was primarily destabilized, we knocked down PPM1G (Fig. 5 C, left), fractionated the cells, and determined amounts of SMN-complex proteins in the nuclear and cytosolic fractions by immunoblotting. Although levels of both Gemin2 and SMN were reduced in both fractions, protein loss was more pronounced in nuclear fractions. Interestingly, levels of the cytoplasmic interaction partner of the SMN-complex unrip were not substantially changed (Fig. 5 C, right). When we followed the knockdown of PPM1G and the destabilization of SMN proteins over time, we observed that PPM1G preceded the depletion of SMN and Gemin2, suggesting that its knockdown is causative for the reduced amounts of SMN proteins (Fig. 5 D). To determine whether decreased SMN levels led to impaired SMN function, we tested assembly of U1 snRNPs by band-shift experiments (Meister et al., 2001a). As expected, reduced snRNP assembly was observed upon PPM1G knockdown (Fig. 5 E; 48.9 ± 10% compared with control extracts), but after normalizing SMN protein levels similar activities were again observed (Fig. 5 E, normalized SMN; 107 ± 29.4%), indicating that residual SMN complex after PPM1G knockdown was equally functional to that in control lysates.
Figure 5.
The SMN complex is destabilized upon PPM1G knockdown in HeLa cells. (A) Immunoblot detection of PPM1G, Gemin2, and Gemin3 60 h after transfection of HeLa cells with siRNA oligonucleotides #1 or #3 against PPM1G. Total protein from HeLa cells was analyzed by immunoblotting with antibodies as indicated. RCC1 detection was used as a loading control. (B) Quantification of relative protein amounts after PPM1G knockdown in HeLa cells as shown in A. Relative amounts of PPM1G, SMN, and Gemin2 were averaged from three independent experiments. Amounts of Gemin3 were quantified from the experiment shown in A. Error bars represent SD. (C) Reduction of SMN and Gemin2 in the cytoplasm and in the nucleus after siRNA-mediated knockdown of PPM1G. Control-treated (c) and PPM1G siRNAi-treated (ΔP) HeLa cells were fractionized into cytosol, and nuclei and fractions were immunoblotted for components of the SMN complex (unrip, Gemin2, and SMN) and for RCC1 (only nuclear) as a control for proper separation. (D) Destabilization of SMN and Gemin2 follow the knockdown of PPM1G from HeLa cells. Total cell lysates were analyzed by immunoblotting for relative amounts of PPM1G, SMN, and Gemin2 at indicated times after transfection with siRNA oligonucleotide #3 against PPM1G. □, relative amounts of PPM1G; ▵, amounts of SMN; ◯, amounts of Gemin2. Signals of proteins as analyzed 15 h after transfection were set to 100%. Amounts of the small GTPase Ran were analyzed and used to normalize quantifications of SMN and Gemin2 proteins. (E) U snRNP assembly activity of HeLa cytosolic extracts derived from control cells (c) or cells after knockdown of PPM1G (ΔP). Extracts with equal amounts of total protein (left) or extracts normalized by immunoblotting to identical amounts of SMN protein (right) were assessed for U snRNP assembly by using 32P-labeled U1 snRNA as previously described (Meister et al., 2001a) and quantified. The activity of control extracts was set to 100%. Error bars represent SD from three independent experiments.
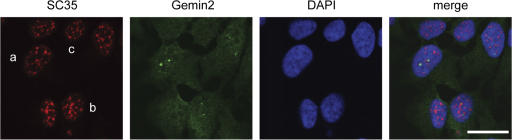
Previously published data describing a function of PPM1G in cell-free splicing assays (Murray et al., 1999) suggested an important role of PPM1G for the assembly of the complete spliceosome machinery. Defects in the integrity of the splicing machinery led to dramatic changes in the characteristic speckle-like localization of the arginine/serine-rich splicing factor 2 (SC35) protein (O'Keefe et al., 1994), an integral component of spliceosomes. Although PPM1G knockdown caused extensive or complete loss of the localized nuclear accumulation of Gemin2 as a marker for the SMN complex (Fig. 6, b and c), no obvious change in the overall pattern of SC35 staining was observed. This suggests that the overall structure of the splicing machinery was not readily affected after knockdown of PPM1G, whereas Gemin2 was lost from CBs.
Figure 6.
SMN delocalization from CBs induced by PPM1G depletion is not correlated with a change in SC35 localization. 72 h after knockdown of PPM1G, HeLa cells were fixed and stained for Gemin2 and SC35. Cells showing pronounced localized nuclear Gemin2 staining (a), a strongly reduced localized nuclear Gemin2 staining (b), or no localized nuclear Gemin2 staining (c) did not show a considerably different SC35 staining pattern. Bar, 20 μm.
The catalytic activity of PPM1G promotes accumulation of SMN in CBs
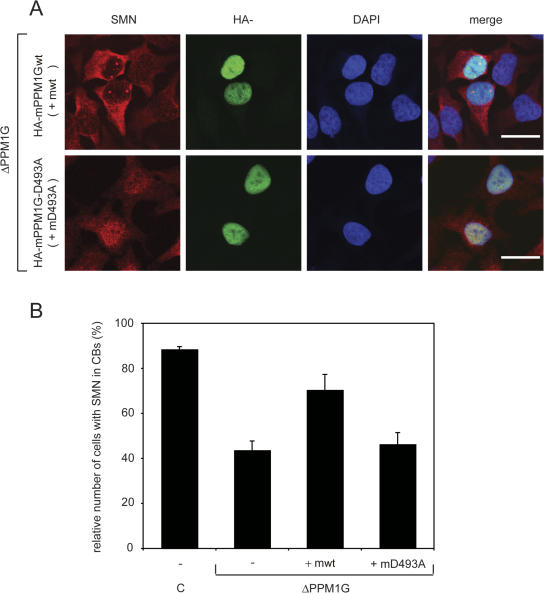
A possible explanation for the loss of SMN proteins from CBs is that PPM1G directly dephosphorylates a protein of the SMN complex and that this dephosphorylation is instrumental for efficient targeting or accumulation of the SMN complex in CBs. This hypothesis assumes that the loss of the catalytic activity of PPM1G is responsible for the observed phenotype. To directly test whether the catalytic activity of PPM1G is required for the stability of the SMN complex in nuclear CBs, we used the mouse orthologue of the human cDNA to complement the siRNA-induced phenotype. The mouse mRNA mismatches in two positions with the corresponding human mRNA, making it much less sensitive to the human siRNAs. Overexpression of the mouse HA-tagged wt protein upon transfection with siRNA oligos considerably increased the number of cells showing the characteristic localization of the SMN complex in CBs compared with cells treated with the siRNA only (Fig. 7, A and B; and not depicted). To analyze whether the catalytic activity of PPM1G was essential for rescue activity, we generated a catalytically inactive mutant of mouse PPM1G (mPPM1G) in which aspartate in position 493 was replaced by alanine (D493A) corresponding to the D496A mutant in hPPM1G (Murray et al., 1999; Fig. 1 E). Upon expression of the mutant protein, the number of cells showing SMN accumulation in CBs was not substantially different from the number in nonrescued controls (Fig. 7, A and B). This indicates that the catalytic activity of PPM1G promotes accumulation of the SMN complex in CBs and suggests that PPM1G is a major determinant of the SMN-complex localization in the nucleus.
Figure 7.
Expression of wt, but not that of catalytically inactive mPPM1G, can rescue correct nuclear localization of the SMN complex in CBs. HeLa cells were first transfected with siRNA oligonucleotide #1 against PPM1G mRNA or a control siRNA oligo (c). Next, cells were transfected with pCMV-HA vectors encoding either wt (HA-mPPM1Gwt or mwt) or phosphatase-dead (HA-mPPM1G-D493A or mD493A) mPPM1G, 12 h after the first transfection. 48 h after the first transfection, cells were fixed and immunostained with antibodies against SMN and the HA tag of mPPM1G. (A) Representative images of cells showing SMN, HA, and DAPI after knockdown of endogenous PPM1G and overexpression of mPPM1G versions as indicated. Bars, 20 μm. (B) Quantification of phenotypes. The relative number of cells showing localization of SMN protein in CBs is plotted. Numbers are means from three independent experiments and error bars represent SD. Absolute numbers of counted cells (1/2/3 experiment): control, 104/171/197; ΔhPPM1G, 139/180/187; ΔhPPM1G + mwt, 67/164/160; and ΔhPPM1G + mD496A, 75/160/171.
Reduction of PPM1G levels leads to impaired dephosphorylation of SMN and Gemin3
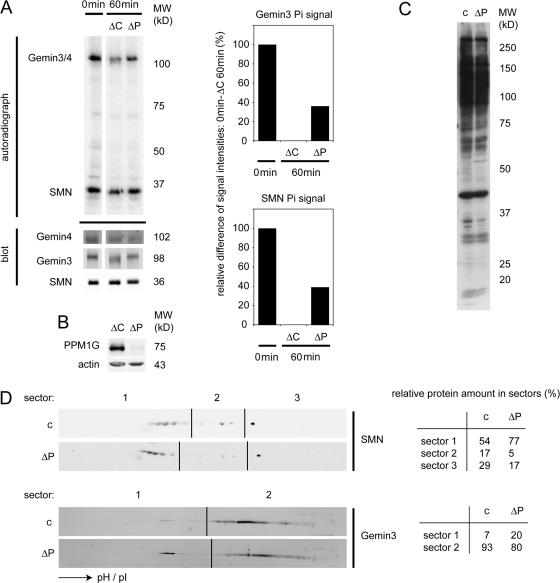
Given that phosphatase activity of PPM1G was required for SMN accumulation in CBs, we next analyzed the dephosphorylation of the major phosphoproteins of the SMN complex, SMN and Gemin3 (Grimmler et al., 2005a). We prepared control extracts, which had been incubated with immobilized unspecific IgG, and extracts from which PPM1G was efficiently immunodepleted using immobilized anti-PPM1G antibodies (Fig. 8, A and B). To measure the efficiency of dephosphorylation of SMN-complex proteins, we first immunopurified the SMN complex and labeled SMN-complex proteins by the addition of γ-[32P]ATP and a complete HeLa extract. Next, we compared the dephosphorylation of purified SMN-complex components in a chase reaction in extracts without ATP. Indeed, we observed considerably less efficient dephosphorylation upon depletion of PPM1G from extracts used for the dephosphorylation reaction (Fig. 8 A, compare ΔC and ΔP after 60 min). Moreover, the mobility of Gemin3 on SDS gels was reproducibly increased upon dephosphorylation in control extracts but not in PPM1G-depleted extracts, whereas the mobility of Gemin4 remained unchanged (Fig. 8 A). This suggested that PPM1G dephosphorylated both SMN and Gemin3 in cell-free extracts. To confirm this result in living cells, we determined the phosphorylation pattern of SMN- complex proteins upon knockdown of PPM1G. PPM1G knockdown did not change the overall phosphorylation pattern of cellular proteins as judged by immunoblotting using total lysates and an antibody against phosphoserine/threonine epitopes (Fig. 8 C). To determine the phosphorylation states of Gemin3 and SMN, we separated total proteins from lysates of control cells or cells after PPM1G knockdown on conventional 2D gels. Immunoblot analysis using antibodies against SMN and Gemin3 revealed that both proteins were phosphorylated as indicated by populations of more acidic forms of the proteins (Fig. 8 D). Strikingly, for both SMN and Gemin3, more acidic forms accumulated upon knockdown of PPM1G. Despite the overall reduction of SMN and Gemin3, relative levels of the less acidic forms of SMN (sector 2) were ∼3.5-fold decreased, whereas the relative levels of the more acidic forms (sector 1) were increased (Fig. 8 D). Similarly, more acidic forms of Gemin3 in sector 1 were increased approximately threefold (Fig. 8 D). Collectively, this strongly suggested that PPM1G directly dephosphorylates SMN-complex proteins in the nucleus.
Figure 8.
Substrates of PPM1G within the SMN complex. (A) Immunoprecipitated SMN complex from complete HeLa cell extracts was labeled with radioactive γ-[32P]ATP, and then dephosphorylated in ATP-free HeLa cell extracts immunodepleted with control rabbit IgG (ΔC) or with antibodies against hPPM1G (ΔP). An input sample (only radioactive ATP incubation) and samples after 60 min incubations in ATP-free extracts were loaded for analysis by autoradiography and quantitative immunoblotting. Immunoblotting signals were used to correct autoradiography for the quantification shown in the graphs. For quantification, the signal differences between input and ΔC 60 min were set as 100% difference. (B) Immunodepletion control for the extracts ΔC and ΔP used in A. (C) Control (c) and PPM1G (ΔP) siRNA knockdown HeLa cells as used for the immunoblot analysis of 2D gel patterns in D were analyzed using a phosphoserine/threonine antibody to detect changes in the overall pattern of bulk phosphorylated proteins. (D) Immunoblots of 2D gels (first dimension, isoelectric focusing; second dimension, SDS-PAGE) from control and PPM1G siRNA-treated HeLa cells as in C. LiCOR scans of decorations against SMN and Gemin3 are shown. Note that differences caused by destabilization of SMN complex components after siRNA knockdown of PPM1G were compensated by showing different exposures. Tables showing the relative intensities of signals in the corresponding image sectors are shown on the right. Note that the absolute reduction of SMN in sector 2 was approximately sevenfold, whereas the overall reduction of SMN protein mass was only twofold, indicating a 3.5-fold specific decrease in the amount of less phosphorylated forms of SMN.
Unrip, a cytoplasmic component of the SMN complex, works antagonistically to PPM1G
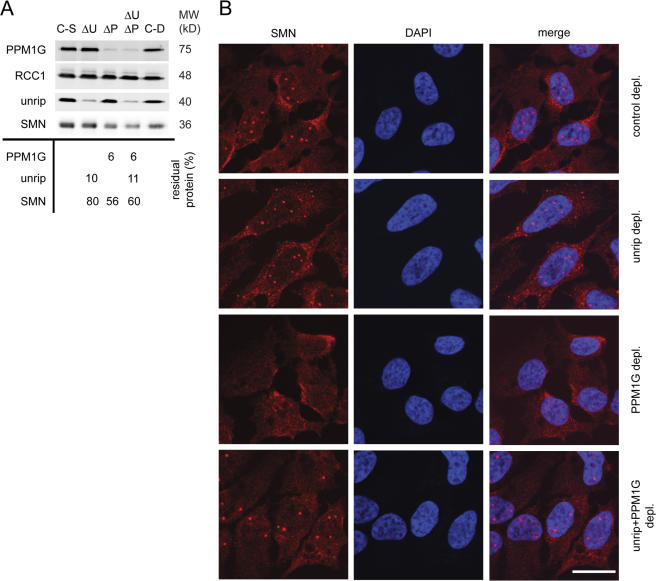
Unrip associates with the SMN complex primarily in the cytoplasm, and knockdown of unrip leads to the enhanced accumulation of the SMN complex in CBs in the nucleus (Carissimi et al., 2005; Grimmler et al., 2005b). This raised the possibility that PPM1G and unrip act antagonistically on the SMN complex. To test this, we knocked down PPM1G, unrip alone, or both gene products together by siRNA treatment, and then we analyzed SMN localization in CBs (Fig. 9). The efficiencies of the single or double knockdown were compared with controls and quantified by immunoblotting (Fig. 9 A). As published previously (Grimmler et al., 2005b), knockdown of unrip alone led to an increase in the number of nuclear bodies in which SMN accumulated (unpublished data). In addition, we observed the formation of cytoplasmic SMN aggregates in 10–20% of all cells (Fig. 9 B). Knockdown of PPM1G alone (oligo pair #1; Fig. 9, A and B, ΔP) led to the loss of the nuclear, CB-localized SMN signal in the majority of cells (Fig. 9 B, SMN signal in PPM1G-depleted cells). In contrast, knockdown of PPM1G and unrip together restored apparently normal SMN localization in CBs (Fig. 9 B). A similar number of cells displayed normal, punctate SMN staining in the nucleus (1–5 bright spots, little diffuse nucleoplasmic signal; Fig. 9 B) compared with untreated controls. Interestingly, cells showing cytoplasmic SMN aggregates upon knockdown of unrip alone (see above) were not observed anymore after double knockdown. However, the amount of SMN was reduced upon knockdown of PPM1G alone and of PPM1G together with unrip (Fig. 9 A, ΔU, ΔP, and ΔUΔP). Collectively, this suggested that unrip and PPM1G have compartment-specific and antagonistic activities toward the SMN complex.
Figure 9.
SiRNA-mediated codepletion of unrip and PPM1G compensates the SMN localization phenotypes of the individual depletions. (A) Immunoblot on total cell lysates after knockdown of PPM1G, unrip, or both (65 h after transfection), with antibodies as indicated. The table gives relative amounts of proteins shown in the blot after selective knockdown of gene products as indicated, compared with RCC1 as loading control (ΔP, PPM1G knockdown; ΔU, unrip knockdown; C-S, control for single gene knockdown; and C-D, control for double gene knockdown). No number was indicated when the amounts of protein were <15% different from controls. (B) Immunofluorescence images of HeLa cells after control treatment (top), knockdown (depl.) of unrip (second row), PPM1G (third row), or both (bottom) using antibodies against SMN and unrip, DAPI, and merge as indicated. Bar, 20 μm.
Discussion
The biogenesis of U snRNPs is a complicated process and requires activities both in the nucleus and in the cytoplasm. Newly transcribed and exported U snRNAs recruit Sm proteins to assemble the Sm core and are subsequently hypermethylated in the cytoplasm. The partially assembled and modified RNPs are then imported into the nucleus, where additional maturation steps lead to the completion of mature U snRNPs (Stanek and Neugebauer, 2006). It is clear from several studies that the SMN complex acts as a key assembler of U snRNPs in the cytoplasm (Meister et al., 2002; Gubitz et al., 2004; Pellizzoni, 2007). Thus, this entity not only facilitates Sm core formation but also serves as a binding platform for the cap-methyltransferase TGS1/PIMT (Mouaikel et al., 2003) and nuclear import factors (Rollenhagen and Pante, 2006). In fact, it has been shown that the SMN complex travels into the nucleus together with the U snRNP (Narayanan et al., 2004; Shpargel and Matera, 2005). In the initial phase in the nucleus, the U snRNPs appear to stay associated with SMN complexes until they reach CBs. At that point, U snRNPs and SMN complexes are believed to dissociate. Whereas U snRNPs disperse in nuclear interchromatin granules and perichromatin fibrils (the sites of storage and splicing, respectively), the SMN complex is likely to return to the cytoplasm, although this has not been shown directly. Hence, the question arises of how the different functions of the SMN complex, its interactions with different partners, and the dynamic equilibrium of its subcellular localizations are regulated. Several components of the SMN complex were shown to be phosphorylated, particularly SMN, which displayed a compartment-specific phosphorylation pattern. This suggested that posttranslational modifications could be involved in the regulation of SMN-complex functions (Grimmler et al., 2005a).
Here, we have shown that the nuclear phosphatase PPM1G specifically copurifies with the SMN complex. We observed that reduction of PPM1G levels in cell-free extracts, as well as in living cells, led to reduced dephosphorylation of SMN and Gemin3. Collectively, this strongly suggests that PPM1G directly dephosphorylates SMN and Gemin3 in the nucleus. Moreover, decreased levels of PPM1G cause dramatic changes in the subcellular localization of SMN as shown by RNA-interference studies. These studies revealed that the loss of PPM1G was strictly correlated with the loss of SMN accumulation in CBs. PPM1G knockdown caused a similar phenotype for the SMN-interacting protein Gemin2, suggesting that accumulation of the whole SMN complex in CBs was affected. A partial rescue of this phenotype was observed when we subsequently overexpressed mPPM1Gwt, but not upon overexpression of a catalytically inactive variant. Interestingly, the loss of SMN accumulation in CBs was not accompanied by the accumulation of SMN in other nuclear bodies (i.e., gems). This might suggest that reversible phosphorylation controls the ability of the SMN complex to accumulate in nuclear bodies in general. We therefore propose that PPM1G phosphatase activity is needed to maintain compartment-specific phosphorylation patterns of SMN-complex proteins in the nucleus, which are required for accumulation of the SMN complex in nuclear bodies.
It has been shown previously that accumulation of the SMN complex in CBs depends on the direct interaction of SMN and coilin (Hebert et al., 2001, 2002). Consistent with that, knockdown of the SMN protein inhibits targeting of other proteins of the SMN complex to CBs. At the same time, however, it induces some fragmentation of CBs into numerous smaller foci likely because of compromised ongoing U snRNP biogenesis (Shpargel and Matera, 2005; Girard et al., 2006; Lemm et al., 2006). Although the complete loss of localized SMN signal from CBs in our experiments was not correlated with the loss of coilin accumulation in CBs, we still observed an increased number of small coilin-positive structures. Moreover, PPM1G knockdown in our experiments also resulted in decreased levels of SMN, Gemin2, and Gemin3, both in the cytoplasm and in the nucleus. PPM1G knockdown might therefore generate a mild version of the phenotype seen upon direct SMN knockdown. When we determined the pattern of U snRNPs using the Y12 antibody after PPM1G knockdown, no obvious differences to control cells were observed (unpublished data). Also, colocalization of the U2 snRNP–specific factor U2B with coilin in CBs could readily be detected in the vast majority of PPM1G knockdown cells in which SMN did not accumulate in CBs (unpublished data). As PPM1G knockdown does not cause changes in the overall pattern of U snRNP localization and has only mild effects on coilin accumulation, this raises the possibility that accumulation of SMN in nuclear bodies is not required for U snRNP biogenesis.
In many PPM1G knockdown cells, the loss of SMN from CBs was accompanied by an increase in the levels of diffuse nucleoplasmic SMN. This suggests that deregulated SMN localization in the nucleus is not a consequence of reduced levels of SMN, but that it might actually cause reduced SMN levels. Reduced cytoplasmic SMN levels explain the reduced capacity of extracts from PPM1G knockdown cells in U1 snRNP assembly. Yet, the specific activity of the SMN complex in cytosolic extracts from PPM1G knockdown cells was not affected and localization of U snRNPs was not changed. If PPM1G knockdown did not affect U snRNP biogenesis, it would likely also not influence the capacity of cells in splicing of pre-mRNAs. However, it had been demonstrated previously that PPM1G plays a role in splicing if tested in cell-free extracts from HeLa nuclei. Still, PPM1G seemed not to be directly essential for splicing in the complete system, suggesting the presence of a redundant activity in complete splicing extracts (Murray et al., 1999). Consistent with that, specific knockdown of PPM1G from a cellular system in our hands did not induce differences in the morphology of splicing speckles as judged by immunofluorescence with antibodies against SC35. However, we did not directly test the efficiency of splicing in living cells with reduced levels of PPM1G. It is conceivable that after PPM1G knockdown, with time, general splicing defects become apparent or that splicing of a certain subset of substrates is immediately impaired, as has now been shown for alternative splicing of the CD44 pre-mRNA (Allemand et al., 2007).
Our results on changed phosphorylation patterns of SMN components implicate the existence of a kinase, which phosphorylates proteins of the SMN complex. Like unrip, this kinase is supposedly a cytoplasmic interaction partner of the SMN complex, which is spatially separated from the activity of PPM1G. To date, such an activity has not been identified. However, we showed that delocalization of SMN from CBs upon knockdown of PPM1G was compensated upon concomitant knockdown of unrip. Conversely, generations of additional SMN-positive aggregates in the nucleus and in the cytoplasm seen upon unrip knockdown were not observed when PPM1G was knocked down in parallel. Given an antagonistic subcellular localization of unrip and PPM1G, this not only indicates that proteins of the SMN complex can exchange between the nucleus and the cytoplasm, but it might also suggest that unrip is involved in cytoplasmic SMN phosphorylation.
Previously, PPM1G has been reported to regulate diverse nuclear functions in splicing (Murray et al., 1999; Allemand et al., 2007), histone maturation (Kimura et al., 2006), and cell cycle progression (Guthridge et al., 1997; Suh et al., 2006). Our data now demonstrate that PPM1G dephosphorylates proteins of the SMN complex in the nucleus and is an important regulator of the subcellular localization of SMN-complex proteins. Many steps in SMN localization and dynamics demand regulation, such as timing of import and export of components, interactions with binding partners in the specific subcellular domains, and time of residence in a particular compartment.
We consider dephosphorylation in the nucleus likely to follow import of the SMN complex into the nucleus to precede nuclear accumulation and targeting to CBs, dissociation from U snRNPs, and potentially retransport to the cytoplasm. It will be interesting in the future to determine if the dephosphorylation of SMN-complex proteins mediates efficiency, order, and timing of these events.
Materials and methods
PPM1G expression constructs
His-hPPM1G and His-xPPM1G were cloned into pQE32 (SphI–KpnI; QIAGEN). A PCR template for hPPM1G constructs, clone FLEXO833D0920D (RZPD), was used. His-xPPM1G was amplified by PCR from complete cDNA of X. laevis eggs. His-hPPM1G D496A was generated by site-directed mutagenesis. EYFP-hPPM1G was generated by PCR and cloned into a monomeric pEYFP-C1 vector (BglII–KpnI; gift from J. Ellenberg, European Molecular Biology Laboratory, Heidelberg, Germany). EYFP-hPPM1G-NLS was generated by cutting the EYFP-hPPM1G full-length construct with BglII and SacI and cloning into the corresponding sites of the same vector. For the HA-mPPM1G construct, mPPM1G was amplified from IRAKp961L079Q (RZPD) and cloned into the pCMV-HA vector (SalI–BglII; CLONTECH Laboratories, Inc.). HA-mPPM1G D493A was generated using site-directed mutagenesis.
Protein expression and purification
PPM1G fusion proteins were expressed in Escherichia coli BL21 pRep4 (QIAGEN) at 37°C and purified via Ni2+-NTA (QIAGEN) in 20 mM Hepes, pH 7.5, 500 mM KCl, 8 mM imidazole, 5% glycerol, 2 mM MgCl2, and protease inhibitor mix (1 μg/ml of pepstatin A, aprotinin, and leupeptin [PPM1G buffer]). Bound proteins were eluted in PPM1G buffer containing an additional 250 mM imidazole and further purified on a poly-Lysine column (bound in PPM1G buffer with 650 mM KCl and eluted with increasing KCl concentrations). Purified PPM1G was dialyzed to 100 mM KCl in PPM1G buffer (without imidazol and protease inhibitors).
Generation of antibodies against xPPM1G and hPPM1G and Gemin2
Rabbits were immunized using purified 6× histidine-tagged xPPM1G or hPPM1G. Antibodies were affinity purified via matrix-coupled (Affi-Gel; Bio-Rad Laboratories) antigens. Antibodies against Gemin2 were raised in rabbit against full-length 6× histidine-tagged human Gemin2 protein, affinity purified using the respective GST-tagged antigen, and covalently linked to a glutathione-Sepharose matrix (GE Healthcare).
siRNA treatments
Final oligonucleotide concentration was 75 nM in all experiments except those shown in Fig. 9. Here, final oligonucleotide concentration was 50 nM for oligonucleotides against each target, unrip and PPM1G mRNA. siRNA oligonucleotides were transfected with a mixture of RNA oligonucleotides and Lipofectamine 2000 (Invitrogen) in opti-MEM (1 μl Lipofectamine to 0.8 μg RNA).
The following double-stranded RNA oligonucleotides against human mRNA sequences were used: control, agacgcauugucaacauccugucug (vimentin; Invitrogen); PPM1G #1, ucacauuccagaugccaucacaggc (Invitrogen); #2, uuaaagagcuggcacagaudtdt (Dharmacon); and #3, aggcuaccaugacuauugadtdt (Dharmacon). Against unrip, a 1:1 mixture of two oligonucleotides was used (Grimmler et al., 2005b).
Oligonucleotide #1, which mismatches with the mRNA of mPPM1G in the positions indicated in bold in the sequence above, was used for RNAi rescue experiments. Cells were transfected with siRNAs as described above. 12 h later, the medium was exchanged and cells were transfected a second time with pCMV-HA plasmids as described above.
Immunofluorescence and image acquisition
HeLa CCL2 (American Type Culture Collection) and X. laevis XL177 cells were used for immunofluorescence experiments. Cells on glass coverslips were washed and fixed with 3% paraformaldehyde. The following primary antibodies were used: monoclonal SMN antibody (clone 7B10; Meister et al., 2001a); affinity-purified polyclonal Gemin2 antibody (this study); affinity-purified polyclonal coilin antibody (a gift from M. Platani, European Molecular Biology Laboratory; Bohmann et al., 1995); polyclonal affinity-purified anti-HA tag antibody (ab9110; Abcam); polyclonal affinity-purified anti-GFP antibody (a gift from D. Görlich, Zentrum für Molekulare Biologie, Heidelberg, Germany); polyclonal affinity-purified antibodies against xPPM1G and hPPM1G (this study); polyclonal antibody against unrip (Grimmler et al., 2005b); monoclonal antibody against SC35 (Novus Biologicals, Inc.); monoclonal antibody against coilin (5P10; gift from K. Neugebauer, Max Planck Institute of Cell Biology and Genetics, Dresden, Germany; Almeida et al., 1998); and a monoclonal antibody against Sm proteins (Y12; gift from I. Mattaj, European Molecular Biology Laboratory).
Alexa Fluor 488 goat anti–rabbit antibody (Invitrogen), Cy2 or Cy3 goat anti–rabbit antibody (Jackson ImmunoResearch Laboratories), and Cy2 or Cy3 goat anti–mouse antibody (Jackson ImmunoResearch Laboratories) were used as secondary antibodies together with DAPI. Coverslips were mounted using Fluoromount-G (Southern Biotechnology Associates, Inc.).
Images were acquired on fixed samples at room temperature with a laser scanning confocal microscope (TCS SP2 or SP5; Leica) using 63× objectives (1.2, water immersion; or 1.4, oil immersion). Images were recorded using confocal software (Leica) and were exported as TIFF. Figures were then generated using Photoshop 7.0 and Illustrator CS2 (Adobe).
Immunoblotting
Samples were separated by SDS gel electrophoresis (Laemmli, 1970) and transferred to nitrocellulose membranes. The following primary antibodies were used: against PPM1G, SMN, and Gemin2 as described for immunofluorescence, monoclonal Gemin3 antibody from rat (a gift from F.A. Grässer, Uniklinik, Homburg/Saar, Germany), anti-RCC1 sera from rabbit (Hetzer et al., 2000), affinity-purified anti-snurportin antibody from rabbit (Paraskeva et al., 1999), and monoclonal anti-Histidine antibody (clone HIS-1; Sigma-Aldrich).
Alexa Fluor 680 goat anti–rabbit IgG (Invitrogen) and IRDye 800 anti–mouse IgG (Rockland Immunochemicals, Inc.) were used as secondary antibodies. Fluorescence signals from immunoblots were detected and quantified using an infrared scanner (LI-COR; Odyssey). Anti-xPPM1G (Fig. 1 A) was detected with horseradish peroxidase–coupled anti–rabbit antibody (Sigma-Aldrich). Gemin3 antibody from rat was detected with horseradish peroxidase–coupled anti–rat antibody (Sigma-Aldrich), and secondary antibody signals were detected using ECL reagent (Pierce Chemical Co.). For quantification of Gemin3 signals, Advanced Image Data Analyzer software (Raytest) was used.
In vitro dephosphorylation of γ-[32P]ATP–labeled SMN complex
HeLa cells were lysed in 20 mM Hepes, pH 7.5, 500 mM KCl, 8.7% glycerol, 2 mM MgCl2, 1% NP-40, and protease inhibitor mix (1 μg/ml of pepstatin A, aprotinin, and leupeptin) by dounce homogenization.
The lysate was precleared by centrifugation in a rotor (SS34; Sorvall) at 16,000 rpm for 30 min and dialyzed against the same buffer as was used for cell lysis but without NP-40 and with 100 mM KCl. After dialysis, the extract was cleared by centrifugation in a rotor (TLA 100.3; Beckman Coulter) at 40,000 rpm for 30 min. Part of the extract was immunodepleted using either rabbit control IgGs (Sigma-Aldrich) or affinity-purified anti-PPM1G antibody (this study).
For immunoprecipitation of the SMN complex, complete extract was incubated with anti-SMN (7B10) antibody prebound to protein G Dynabeads (Invitrogen). Beads were washed with buffer, as for dialysis of extract, but containing 0.01% Triton X-100 and 200 mM KCl. Beads were incubated at 32°C for 30 min in extract supplied with a mixture of cold and γ-[32P]ATP, washed again, divided, and incubated in control-depleted extract or PPM1G-depleted extract without ATP at 32°C for 60 min. Beads were washed with buffer as for dialysis of extract, but containing 5 mM EDTA instead of MgCl2 and incubated in SDS-PAGE sample buffer. Samples were analyzed by immunoblotting and autoradiography.
2D gel immunoblotting analysis of whole cell lysates from HeLa cells
After siRNA treatment, cell pellets were resuspended in isoelectric focusing buffer containing 6 M urea, 2 M thiourea, 2% CHAPS, 0.15% DTT, and 0.5% ampholytes (Pharmalytes; GE Healthcare). In this buffer, cells were sonified and centrifuged in a rotor (TLA 100.3) for 30 min at 40,000 rpm. Supernatant was used for isoelectric focusing with Immobiline Dry Strips, pH 3–10 (nonlinear; GE Healthcare), followed by SDS-PAGE and immunoblot.
Calculation of PPM1G concentration in nuclei of X. laevis oocytes and HeLa cells
Immunoblotting of dilution series of recombinant-purified xPPM1G and hPPM1G compared with complete X. laevis oocyte extract or complete HeLa lysates was used to estimate PPM1G concentrations to be 700 nM in X. laevis oocytes, 5 μM in X. laevis oocyte nuclei, 300 nM in HeLa, and at least 1 μM in HeLa nuclei.
Transfection of cells with plasmids
Approximately 2 × 105 HeLa CCL2 cells were transfected with a mixture of 0.7 μg DNA and 1.4 μl Lipofectamine 2000 (Invitrogen) in OPTI-MEM medium (Invitrogen) according to the manufacturer's protocol.
HeLa cell extract, fractionation, immunoprecipitation, and protein–protein interaction assay
HeLa S3 cells were resuspended in 2.5 vol PBS containing 0.01% IgePal (Rhodia Inc.), lyzed by sonication, and cleared by centrifugation (SW 32 Ti; Beckman Coulter) for 10 min at 10,000 rpm. The supernatant (cell extract) was passed through a 0.2-μm low-protein–binding filter.
Immunoprecipitations were performed with 7B10 monoclonal anti-SMN antibody (Meister et al., 2001a) or normal mouse serum covalently linked to protein G–Sepharose. After washing, immunoprecipitated proteins were eluted by boiling in 2× SDS sample buffer.
The protein–protein interaction assay consisted of the following: immunoprecipitated SMN complex from HeLa cell extract was incubated with 5 μg of recombinant hPPM1G in 50 μl of buffer containing 20 mM Hepes, 100 mM KCl, 5 mM MgCl2, 2 mM MnCl2, and 2% glycerol, pH 7.5. After washing, interacting proteins were eluted from the Sepharose matrix by boiling in 2× SDS sample buffer.
The Qproteome nuclear protein kit (QIAGEN) was used to separate nuclei and cytosols according to the manufacturer's protocol. Cytoplasmic supernatants were precipitated in 80% −20°C acetone, and isolated nuclei were directly lyzed in 1.5× SDS gel sample buffer.
Online supplemental material
Fig. S1 shows that expression of EYFP-PPM1G without NLS sequence (EYFP-PPM1G-NLS), but not expression of PPM1Gwt (EYFP-PPM1G), changes localization of the SMN complex in HeLa cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704163/DC1.
Acknowledgments
We thank Melpi Platani, Karla Neugebauer, Dirk Görlich, and Iain Mattaj for antibodies, Jan Ellenberg for the EYFP fusion vector, Markus T. Bohnsack for help with oocyte dissections, and Kerstin Hupfeld for excellent technical assistance. We are grateful to Hartmut Krischke and Thomas Ruppert for support with 2D gel electrophoresis.
This work was supported by the Zentrum für Molekulare Biologie and the Deutsche Forschungsgemeinschaft (GR1737/4-1/2 to O.J. Gruss and FI573/4-1 and SFB581 to U. Fischer).
Abbreviations used in this paper: CB, Cajal body; RCC1, regulator of chromosome condensation 1; SMN, survival motor neuron; U snRNA, uridine-rich small nuclear RNA; U snRNP, uridine-rich small nuclear RNP; unrip, unr-interacting protein; wt, wild type.
References
- Allemand, E., M.L. Hastings, M.V. Murray, M.P. Myers, and A.R. Krainer. 2007. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Struct. Mol. Biol. 14:630–638. [DOI] [PubMed] [Google Scholar]
- Almeida, F., R. Saffrich, W. Ansorge, and M. Carmo-Fonseca. 1998. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142:899–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann, K., J. Ferreira, N. Santama, K. Weis, and A.I. Lamond. 1995. Molecular analysis of the coiled body. J. Cell Sci. Suppl. 19:107–113. [DOI] [PubMed] [Google Scholar]
- Brahms, H., L. Meheus, V. de Brabandere, U. Fischer, and R. Luhrmann. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 7:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carissimi, C., J. Baccon, M. Straccia, P. Chiarella, A. Maiolica, A. Sawyer, J. Rappsilber, and L. Pellizzoni. 2005. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 579:2348–2354. [DOI] [PubMed] [Google Scholar]
- Carvalho, T., F. Almeida, A. Calapez, M. Lafarga, M.T. Berciano, and M. Carmo-Fonseca. 1999. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 147:715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., M.D. Hebert, T.S. Karpova, D. Stanek, H. Xu, K.B. Shpargel, U.T. Meier, K.M. Neugebauer, A.G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, W.J., S. Massenet, S. Paushkin, A. Wyce, and G. Dreyfuss. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 7:1111–1117. [DOI] [PubMed] [Google Scholar]
- Girard, C., H. Neel, E. Bertrand, and R. Bordonne. 2006. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res. 34:2925–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler, M., L. Bauer, M. Nousiainen, R. Korner, G. Meister, and U. Fischer. 2005. a. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 6:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler, M., S. Otter, C. Peter, F. Muller, A. Chari, and U. Fischer. 2005. b. Unrip, a factor implicated in cap-independent translation, associates with the cytosolic SMN complex and influences its intracellular localization. Hum. Mol. Genet. 14:3099–3111. [DOI] [PubMed] [Google Scholar]
- Gubitz, A.K., W. Feng, and G. Dreyfuss. 2004. The SMN complex. Exp. Cell Res. 296:51–56. [DOI] [PubMed] [Google Scholar]
- Guthridge, M.A., P. Bellosta, N. Tavoloni, and C. Basilico. 1997. FIN13, a novel growth factor-inducible serine-threonine phosphatase which can inhibit cell cycle progression. Mol. Cell. Biol. 17:5485–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm, J., E. Darzynkiewicz, S.M. Tahara, and I.W. Mattaj. 1990. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 62:569–577. [DOI] [PubMed] [Google Scholar]
- Hebert, M.D., P.W. Szymczyk, K.B. Shpargel, and A.G. Matera. 2001. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 15:2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, M.D., K.B. Shpargel, J.K. Ospina, K.E. Tucker, and A.G. Matera. 2002. Coilin methylation regulates nuclear body formation. Dev. Cell. 3:329–337. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., D. Bilbao-Cortes, T.C. Walther, O.J. Gruss, and I.W. Mattaj. 2000. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell. 5:1013–1024. [DOI] [PubMed] [Google Scholar]
- Kimura, H., N. Takizawa, E. Allemand, T. Hori, F.J. Iborra, N. Nozaki, M. Muraki, M. Hagiwara, A.R. Krainer, T. Fukagawa, and K. Okawa. 2006. A novel histone exchange factor, protein phosphatase 2Cγ, mediates the exchange and dephosphorylation of H2A–H2B. J. Cell Biol. 175:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lemm, I., C. Girard, A.N. Kuhn, N.J. Watkins, M. Schneider, R. Bordonne, and R. Luhrmann. 2006. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 17:3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., and G. Dreyfuss. 1996. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Matera, A.G., R.M. Terns, and M.P. Terns. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220. [DOI] [PubMed] [Google Scholar]
- Meister, G., and U. Fischer. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21:5853–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G., D. Buhler, R. Pillai, F. Lottspeich, and U. Fischer. 2001. a. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3:945–949. [DOI] [PubMed] [Google Scholar]
- Meister, G., C. Eggert, D. Buhler, H. Brahms, C. Kambach, and U. Fischer. 2001. b. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11:1990–1994. [DOI] [PubMed] [Google Scholar]
- Meister, G., C. Eggert, and U. Fischer. 2002. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12:472–478. [DOI] [PubMed] [Google Scholar]
- Mouaikel, J., U. Narayanan, C. Verheggen, A.G. Matera, E. Bertrand, J. Tazi, and R. Bordonne. 2003. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 4:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.V., R. Kobayashi, and A.R. Krainer. 1999. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 13:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, U., T. Achsel, R. Luhrmann, and A.G. Matera. 2004. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell. 16:223–234. [DOI] [PubMed] [Google Scholar]
- Nilsen, T.W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 25:1147–1149. [DOI] [PubMed] [Google Scholar]
- O'Keefe, R.T., A. Mayeda, C.L. Sadowski, A.R. Krainer, and D.L. Spector. 1994. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva, E., E. Izaurralde, F.R. Bischoff, J. Huber, U. Kutay, E. Hartmann, R. Luhrmann, and D. Gorlich. 1999. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni, L. 2007. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 8:340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker, V.A., G. Plessel, and R. Luhrmann. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Rollenhagen, C., and N. Pante. 2006. Nuclear import of spliceosomal snRNPs. Can. J. Physiol. Pharmacol. 84:367–376. [DOI] [PubMed] [Google Scholar]
- Shpargel, K.B., and A.G. Matera. 2005. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 102:17372–17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek, D., and K.M. Neugebauer. 2006. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 115:343–354. [DOI] [PubMed] [Google Scholar]
- Suh, E.J., T.Y. Kim, and S.H. Kim. 2006. PP2Cgamma-mediated S-phase accumulation induced by the proteasome-dependent degradation of p21(WAF1/CIP1). FEBS Lett. 580:6100–6104. [DOI] [PubMed] [Google Scholar]