Abstract
The regulation of cytoskeletal components in the dendritic shaft core is critical for dendrite elongation and branching. Here, we report that a brain-specific Ras guanine nucleotide exchange factor (RasGEF) carrying two kinase non-catalytic C-lobe domains (KINDs), very-KIND (v-KIND), regulates microtubule-associated protein 2 (MAP2). v-KIND is expressed in developing mouse brain, predominantly in the cerebellar granule cells. v-KIND not only activates Ras small GTPases via the C-terminal RasGEF domain, but also specifically binds to MAP2 via the second KIND domain (KIND2), leading to threonine phosphorylation of MAP2. v-KIND overexpression suppresses dendritic extension and branching of hippocampal neurons and cerebellar granule cells, whereas knockdown of endogenous v-KIND expression promotes dendrite growth. These findings suggest that v-KIND mediates a signaling pathway that links Ras and MAP2 to control dendrite growth.
Introduction
Dendrite morphogenesis requires an intrinsic differentiation program that is guided by extracellular cues and electrical activity. These different forces converge to control the gene expression and cytoskeletal dynamics that specify dendrite growth and branching, as well as dendritic spine formation (Scott and Luo, 2001; Whitford et al., 2002; Jan and Jan, 2003; Miller and Kaplan, 2003; Van Aelst and Cline, 2004). Various small GTPases act as key regulators in dendrite development (Scott and Luo, 2001; Gao and Bogert, 2003). The Rho family GTPases, such as RhoA, Rac1, and Cdc42, regulate actin dynamics and profoundly influence dendrite morphogenesis (Cline, 2001; Redmond and Ghosh, 2001; Luo, 2002). Another small GTPase Ras family is also important for activity-dependent filopodia formation and dendritic complexity via the activation of a MAPK signaling pathway (Wu et al., 2001; Koh et al., 2002; Redmond et al., 2002; Goldin and Segal, 2003; Alonso et al., 2004). Recent studies indicate that the Ras–phosphatidylinositol 3-kinase–Akt–mammalian target of the rapamycin signaling pathway has a pivotal role in regulating dendrite formation (Jaworski et al., 2005; Kumar et al., 2005). These small GTPases undergo structural changes in response to alternative binding of GDP and GTP: activation by GTP requires guanine nucleotide exchange factor (GEF), and inactivation requires GTPase-activating protein (GAP) (Cherfils and Chardin, 1999). The GTP-bound active form interacts with downstream effectors to activate cell signaling pathways, including that in dendrite morphogenesis (Goi et al., 1999; Yang and Mattingly, 2006).
Various signaling pathways underlying dendrite development eventually affect the dynamics of actin filaments or microtubules (MTs) or both during dendrite growth. The high molecular weight MAP2 (referred to hereafter as MAP2) family proteins are an abundant group of MT-associated proteins that are predominantly expressed in neurons (Friedrich and Aszodi, 1991) and are selectively enriched in the soma and dendrites (Bernhardt and Matus, 1984). In dendrites, MAP2 is a major component of the cross-bridges between MTs, or between MTs and other cytoskeletal components (Shiomura and Hirokawa, 1987; Hirokawa et al., 1988). In MAP2-deficient mice, the dendrites have a reduced MT density, and are shorter in hippocampal neurons (Harada et al., 2002). Moreover, MAP2 can be highly phosphorylated and is thought to act as a phosphorylation- dependent modulator of MTs during dendrite development (Sanchez et al., 2000). The details of the molecular mechanisms underlying these cytoskeletal regulations, however, are not fully understood.
Very-KIND protein (v-KIND) was first characterized as a nervous system–specific protein carrying two kinase noncatalytic C-lobe domains (KINDs), a RasGEF N-terminal domain (RasN), and a putative RasGEF domain (Mees et al., 2005). The functional role of this new class of RasGEF v-KIND is largely unknown. In this study, we identified v-KIND as a transcript with up-regulated, brain-specific, and cerebellum- dominant expression profiles by transcriptomic analysis of postnatal mouse cerebellum (Cerebellar Development Transcriptome Database [CDT-DB] project, http://www.cdtdb.brain.riken.jp). v-KIND was expressed during mouse brain development and interacted with Ras and MAP2. Overexpression of v-KIND suppressed dendrite growth, leading to simple branching patterns. In contrast, knockdown of v-KIND expression by RNA interference (RNAi) led to more complicated dendrite branching patterns. The results of the present study suggest that v-KIND is important for the control of dendrite growth. v-KIND is the first RasGEF known to bind to MAP2, thereby providing a new mechanism to link the Ras GTPase signaling pathway and MAP2-MT cytoskeletal organization during dendrite morphogenesis.
Results
v-KIND is a novel RasGEF protein predominantly expressed in postnatal mouse brain
We first identified a transcript with an up-regulated and brain-specific expression profile (CDT-DB clone number CD01268; GenBank/EMBL/DDBJ accession no. BP427523) in the transcriptomic analysis of cerebellar development, and cloned it as a mouse v-KIND gene (GenBank accession no.: AJ580324). v-KIND encoded a 1,742-amino acid protein that included two KIND domains (KIND1 and KIND2), a coiled-coil region, a Ras GEF N-terminal (RasN) domain, and a RasGEF domain (Fig. 1 A).
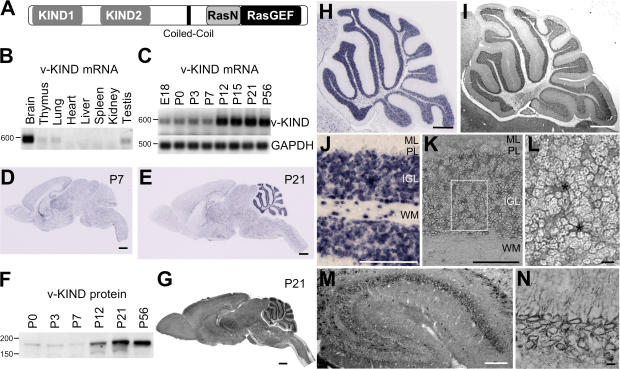
Figure 1.
v-KIND was predominantly expressed in developing mouse cerebellum. (A) Domain structure of v-KIND protein. (B) RT-PCR analysis of v-KIND mRNA expression in P21 mouse tissue. (C) RT-PCR analysis of v-KIND mRNA expression in postnatal cerebellum at six different stages. RT-PCR for GAPDH was used as an internal control. (D and E) In situ hybridization analysis of v-KIND distribution in the P7 (D) and P21 (E) mouse brain. Bar, 1 mm. (F) Immunoblotting of v-KIND in the postnatal cerebellum. Equal amounts of protein lysates from mouse cerebellum at six postnatal stages (P0, P3, P7, P12, P21, and P56) were subjected to immunoblotting analysis by a specific antibody generated against v-KIND protein. (G) Immunohistochemical staining of v-KIND in P21 mouse brain sections. Bar, 1 mm. (H) In situ hybridization analysis of v-KIND distribution in whole cerebellum; Bar, 1 mm. (I) Immunolabeling of v-KIND protein in mouse cerebellum. (J) In situ hybridization analysis of v-KIND distribution in the cerebellar cortex; Bar, 100 μm. ML, molecular layer; PL, Purkinje cell layer; IGL, internal granule cell layer; WM, white matter. (K) Immunolabeling of v-KIND protein in cerebellar cortex. (L) Magnified image of granule cells in the IGL as indicated in the rectangle in (K); Bar, 10 μm. Asterisks indicate synaptic glomeruli. (M and N) Immunolabeling of v-KIND protein in mouse hippocampus. M, whole hippocampus; Bar, 100 μm. N, v-KIND immunolabeling in pyramidal neurons in the CA1 region; Bar, 10 μm.
Analysis of eight postnatal day (P) 21 mouse tissues showed that v-KIND mRNA was predominantly expressed in the brain (Fig. 1 B). Expression of v-KIND mRNA in P7 mouse brain was very low and barely detected in the other tissues (unpublished data). Temporal expression analysis in developing mouse cerebellum revealed that the expression of v-KIND mRNA was sharply up-regulated between P7 and P12 (Fig. 1 C).
Cellular localization of v-KIND mRNA in P7 and P21 mouse brain was analyzed by in situ hybridization (Fig. 1, D and E, H, and J). At P7, v-KIND mRNA expression levels were generally low throughout the brain, but high in hippocampus, thalamus, and the cerebellar white matter (Fig. 1 D). At P21, v-KIND mRNA was detected in nearly all fields at varying intensities (Fig. 1 E): it was predominantly localized in the cerebellum, and was observed at intermediate levels in the hippocampus and thalamus (Fig. 1 E). In the cerebellum, v-KIND mRNA was concentrated in the internal granular layer (IGL) and expression was higher in the anterior lobe than in the posterior lobe (Fig. 1 H). v-KIND mRNA was predominantly observed in granule cells, and was expressed in cells within the white matter (Fig. 1 J). These developmentally regulated and region-specific expression patterns suggest that v-KIND is responsible for specific developmental events in postnatal mouse brain.
Distribution of v-KIND protein in P21 mouse brain
We investigated v-KIND protein expression in developing mouse brains using an antibody that specifically recognizes v-KIND protein. The specificity of the antibody is shown in Fig. S1 (available at http://www.jcb.org/cgi/content/full/jcb.200702036/DC1). The v-KIND protein level was greatly increased by P21 (Fig. 1 F), which almost paralleled the mRNA expression pattern (Fig. 1 C). Subcellular fractionation analysis of P7 and P21 mouse cerebella shows that v-KIND protein is a soluble protein but appears to associate with precipitable components, such as cell membranes or cytoskeletal elements (Fig. S1 B).
The cellular distribution of v-KIND protein in mouse brain was analyzed using immunohistochemistry (Fig. 1, G, I, and K–N). Widespread immunolabeling patterns of v-KIND were observed in the P21 mouse brain (Fig. 1 G). Intense immunolabeling signals were detected in the IGL of the cerebellum (Fig. 1 I) and moderate immunolabeling signals were observed in the hippocampus (Fig. 1 M), which was consistent with the Western blotting data, showing that v-KIND is more abundant in cerebella than hippocampus (Fig. S1 C). In the cerebellum, there was more v-KIND protein in the anterior lobe than in the posterior lobe (Fig. 1 I), which was similar to the mRNA distribution pattern (Fig. 1 H). In the IGL, v-KIND protein was predominantly observed around the periphery of the granule cells and in the glomerular rosette areas, indicating that v-KIND protein expressed in granule cells is largely localized in the somatodendritic region (Fig. 1 L). In the hippocampus, v-KIND protein was distributed in the CA1-CA2-CA3 region (Fig. 1 M) and was localized in the soma and dendrites of CA1 pyramidal neurons (Fig. 1 N).
v-KIND has GEF activity for Ras and binds to activated Ras
To investigate the function of the RasGEF domain of v-KIND, we analyzed the interaction between v-KIND and Ras. v-KIND coimmunoprecipitated with Ras from cerebellar lysates (Fig. 2 A). This interaction was verified by an exogenous expression system in COS7 cells (Fig. 2 B). The expressed FLAG-tagged v-KIND was coimmunoprecipitated with endogenous Ras by the anti-Ras antibody, whereas the mutant lacking the RasGEF domain (ΔGEF), failed to be precipitated. These results suggest that v-KIND is associated with Ras through the RasGEF domain.
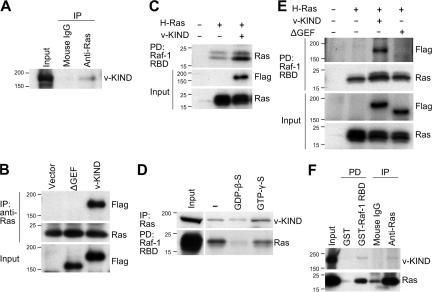
Figure 2.
v-KIND protein acts as a Ras GEF in mouse cerebellum. (A) Coimmunoprecipitation of Ras with v-KIND from mouse cerebellar lysates. Equal amounts of cerebellar lysates from P21 mice were immunoprecipitated with the anti-Ras antibody and immunoblotted with the anti-v-KIND antibody. (B) RasGEF domain was responsible for v-KIND-Ras association. Equal amounts of protein lysates from COS7 cells expressing FLAG-tagged v-KIND or a v-KIND mutant lacking the RasGEF domain (ΔGEF) were immunoprecipitated with the anti-Ras antibody and immunoblotted with the anti-FLAG antibody (top panel). Middle panel, Ras in the immunoprecipitates. Bottom panel, FLAG-tagged v-KIND and ΔGEF in whole lysates. (C) v-KIND activated H-Ras. Activated H-Ras (GTP-Ras) in cells with or without FLAG-tagged v-KIND coexpression was analyzed by pull-down assay using GST-Raf-1- RBD. The protein complex was then subjected to immunoblotting with anti-Ras and anti-FLAG antibody. (D) Binding of v-KIND to Ras-GDP and Ras-GTP. Equal amounts of mouse hippocampal protein lysate were treated by GDP-β-S or GTP-γ-S, and then investigatedby Raf-1 RBD pull-down assay. Top panel, v-KIND binding with Ras in the presence of GDP-β-S or GTP-γ-S; Bottom panel, the quantity of GTP-Ras. (E) GST-Raf-1-RBD pulled down activated GTP-Ras (second panel) together with FLAG-tagged v-KIND, but not ΔGEF (top panel). (F) v-KIND protein was pulled down with GTP-Ras by GST-Raf-1-RBD from mouse cerebellum. Top panel, v-KIND in the pull-down lysate; bottom panel, activated Ras in the pull-down lysate.
The RasGEF activity of v-KIND was then investigated by the pull-down assay using the GST-fused Raf-1 Ras binding domain (RBD), which specifically binds to the activated GTP-bound form of Ras (Ras-GTP). Coexpression of H-Ras with v-KIND in COS7 cells showed increased immunoreactivity for H-Ras-GTP pulled-down by the Raf-1 RBD in comparison with expression of H-Ras alone (Fig. 2 C), indicating that coexpressed v-KIND has potential RasGEF activity.
The RasGEF domain has binding affinity for both Ras-GDP and Ras-GTP (Cherfils and Chardin, 1999). We investigated the binding of v-KIND to Ras in hippocampal lysates in the presence of either GDP-β-S or GTP-γ-S (Fig. 2 D). In general, GDP-β-S inhibits the production of Ras-GTP, whereas GTP-γ-S activates it (Taylor et al., 2001). The quantity of Ras-GTP was measured by Raf-1 RBD pull-down assay. As expected, the immunoreactivity for Ras-GTP pulled-down by the Raf-1 RBD was higher in the presence of GTP-γ-S than in the presence of GDP-β-S (Fig. 2 D, bottom). In addition, the presence of GTP-γ-S increased the amount of v-KIND coimmunoprecipitated with anti-Ras antibody in comparison with that of GDP-β-S (Fig. 2 D, top), indicating that v-KIND binds to both Ras-GDP and Ras-GTP, but binds more preferentially to Ras-GTP.
We compared the binding specificity of v-KIND and ΔGEF to H-Ras by the Raf-1 RBD pull-down assay. v-KIND together with H-Ras was pulled down by Raf-1 RBD from lysates of COS7 cells coexpressing H-Ras and v-KIND, whereas ΔGEF was not pulled down by Raf-1 RBD from lysates of cells coexpressing H-Ras and ΔGEF (Fig. 2 E). These results suggest that the RasGEF domain of v-KIND has RasGEF activity and a major region responsible for the interaction with the activated Ras.
We verified the in vivo association of endogenous v-KIND and activated Ras in mouse hippocampal lysates (Fig. 2 F). In contrast to GST alone (as a negative control), GST-Raf-1 RBD pulled down v-KIND as well as Ras. Similarly, in contrast to control mouse IgG, the anti-Ras antibody coimmunoprecipitated v-KIND and Ras. Collectively, these results suggest that v-KIND acts as a RasGEF in mouse brain.
Localization of v-KIND in dendrites and soma of primary cultured neurons
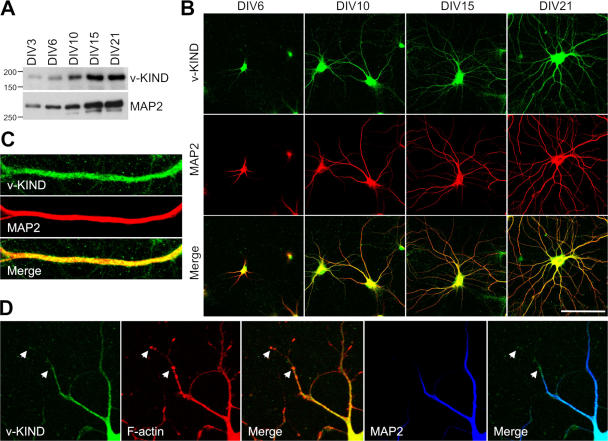
Subcellular localization of v-KIND was analyzed by immunocytochemistry in primary cultured neurons. In cerebellar granule cells at 14 days in vitro (DIV), v-KIND was distributed prominently in the soma and punctately in dendrites as marked by anti-MAP2 antibodies (Fig. S1 D). Because the dendrites of hippocampal neurons are easier to visualize than the dendrites of cerebellar granule cells, we compared the expression patterns of v-KIND with those of MAP2 at various differentiation stages in hippocampal primary cultures (DIV3, 6, 10, 15, and 21). Immunoblotting analysis revealed that the expression level of v-KIND gradually increased and peaked at DIV15, which was similar to that of MAP2 (Fig. 3 A). Immunocytochemical analysis showed v-KIND immunoreactivity almost overlapped with MAP2 immunoreactivity in soma and dendrites and was not localized to axons throughout differentiation (Fig. 3 B). Immunostaining patterns around the dendrite shafts differed between v-KIND and MAP2: v-KIND immunoreactivity appeared as many small puncta around the dendrites, although MAP2 immunoreactivity appeared very heavily and smoothly on the dendrites (Fig. 3 C). In DIV10 cultures, v-KIND immunoreactivity was also observed in the more distal tips of the dendrites, which were MAP2-negative, but F-actin positive (Fig. 3 D).
Figure 3.
v-KIND protein is enriched in the dendritic shafts in primary cultured neurons. (A) Immunoblotting of v-KIND protein in primary cultured hippocampal neurons at DIV3, DIV6, DIV10, DIV15, and DIV21. MAP2 was used as a control. (B) v-KIND protein (green) distribution in primary cultured hippocampal neurons at DIV6, DIV10, DIV15, and DIV21. Dendrites were marked by anti-MAP2 antibody (red). Images were captured by confocal microscopy (Carl Zeiss MicroImaging, Inc.). Bar, 100 μm. (C) Costaining of v-KIND (green) with MAP2 (red) in the dendritic shaft of hippocampal neurons at DIV21. (D) Costaining of v-KIND (green) with MAP2 (blue) and F-actin (phalloidin, red) in dendrites of hippocampal neurons at DIV10. Arrow, colocalized v-KIND and F-actin.
Overexpression of v-KIND impairs dendritic growth of hippocampal neurons
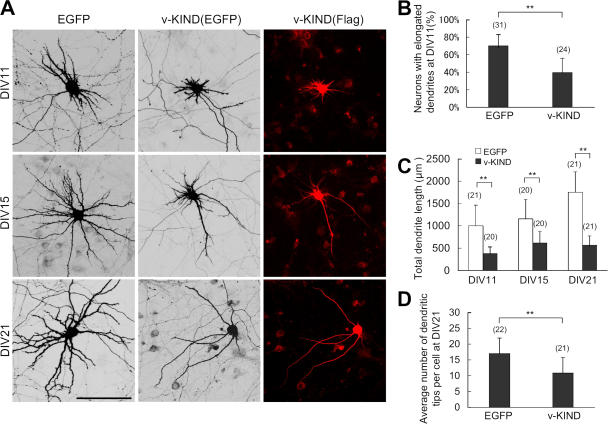
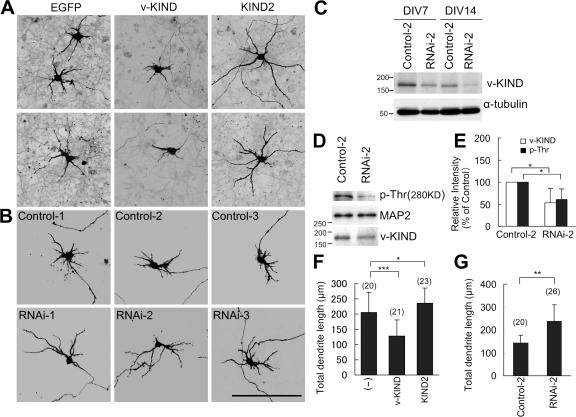
Because Ras family proteins regulate dendrite morphogenesis, we investigated the function of v-KIND in dendrite growth by overexpressing v-KIND in hippocampal primary cultures. Based on the fact that peak dendritic outgrowth occurs at DIV6-7 (Dotti et al., 1988), and taking into account transfection efficiency and post-transfection expression levels, hippocampal cultures were cotransfected with pCAG-v-KIND plus pCAG-EGFP (as a conventional reporter for cell morphology) at DIV3, and their dendrite morphology was observed at DIV11, DIV15, and DIV21. At DIV11, cells expressing EGFP alone exhibited normal dendrite growth with long branches, whereas v-KIND plus EGFP-overexpressing cells had a severely retarded dendritic pattern with very short protrusions (Fig. 4 A). At DIV11, v-KIND overexpression significantly reduced the number of cells with dendrites longer than 50 μm long (∼39% of cells with v-KIND overexpression vs ∼70% with EGFP alone; Fig. 4 B; P < 0.001) and significantly decreased total dendrite length per cell (378 ± 153 μm with v-KIND vs 999 ± 463 μm with EGFP alone; Fig. 4 C; P < 0.001). At DIV15, v-KIND–overexpressing cells had a few long, but less-branched dendrites and many short dendrites, whereas cells expressing EGFP alone had longer, branched dendrites (Fig. 4 A). Total dendrite length of individual v-KIND-overexpressing cell (599 ± 258 μm) was approximately two times shorter than that of cells expressing EGFP alone (1,156 ± 433 μm; Fig. 4 C). At DIV21, cells expressing EGFP alone had almost mature arborization patterns with complex and long branched dendrites, whereas cells overexpressing v-KIND had very simple dendritic arborization patterns with a few long and less-branched dendrites (Fig. 4 A). Total dendrite length of v-KIND-overexpressing cells (566 ± 216 μm) was less than that of cells expressing EGFP alone (1753 ± 421 μm; Fig. 4 C; P < 0.001). In addition, the number of dendritic branch tips per cell (branches with a distance from soma to tips longer than 20 μm were counted) was reduced by v-KIND overexpression (11 ± 5) compared with expression of EGFP alone (17 ± 5; Fig. 4 D; P < 0.001). These results demonstrate that overexpression of v-KIND induces significant dendrite growth defects, including reduced elongation and branching.
Figure 4.
v-KIND overexpression in hippocampal neurons suppressed dendrite growth. Primary cultured hippocampal neurons were cotransfected by pCAG-v-KIND plus pCAG-EGFP at DIV3. Neurons at DIV11, DIV15, and DIV21 were fixed and observed. Neurons overexpressing pCAG-EGFP alone were used as controls. (A) Images of neurons overexpressing v-KIND plus EGFP. Bar, 100 μm. (B) Statistical analysis of percentage of neurons with elongated dendrites. (C and D) Statistical analysis of total dendrite length (C) and dendrite arbor numbers (D) of individual neurons expressing v-KIND plus EGFP at DIV11, DIV15, and DIV21. In statistical analysis, data from at least three independent experiments are presented as mean ± SD. **, P < 0.001(t test). The number of neurons counted is shown in the parentheses above the columns.
v-KIND suppresses dendrite growth during development
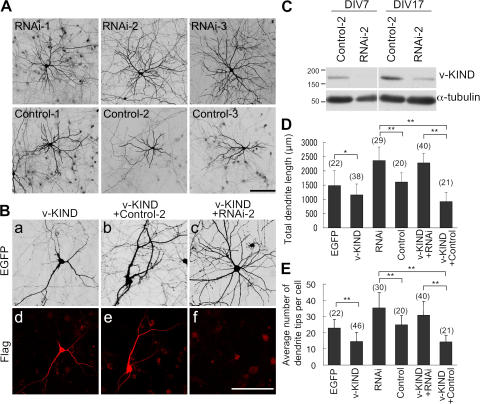
To further analyze the function of v-KIND, we used RNAi to knockdown v-KIND expression. The knockdown efficiency of three types of stealth v-KIND RNAi constructs (RNAi-1, 2, 3) was confirmed in COS7 cells by cotransfecting cells with pEGFP-v-KIND (Fig. S2, A–D, available at http://www.jcb.org/cgi/content/full/jcb.200702036/DC1). Hippocampal cultures were transfected with each v-KIND RNAi construct together with EGFP at DIV3 and the dendritic patterns of EGFP-positive neurons were analyzed at DIV21 (Fig. 5 A). In contrast to three control RNAi constructs (Control-1, -2, -3), all v-KIND RNAi constructs induced the formation of complex dendritic patterns with significantly increased dendrite branching and length (Fig. 5 A). The efficiency of v-KIND RNAi knockdown was verified by transfecting with the RNAi construct at DIV3 followed by analyzing endogenous v-KIND levels at DIV7 and DIV17. The representative data of RNAi-2 effects are shown in Fig. 5 (C and D). In contrast to the Control-2, the RNAi-2 significantly reduced v-KIND immunoreactivity at both DIV7 and DIV17, indicating that the RNAi was effective for v-KIND knockdown for at least 2 wk after transfection. For statistical analyses, we combined all data obtained from the experiments transfecting with each RNAi construct (RNAi-1, RNAi-2, or RNAi-3) and those from the control experiments transfecting with each control construct (Control-1, Control-2, or Control-3). The average total dendrite length per cell transfected with the v-KIND RNAi construct and control RNAi was 2,359 ± 475 and 1,610 ± 316 μm, respectively (Fig. 5 E). The average number of dendritic branch tips per cell was 35 ± 9.7 for v-KIND RNAi-transfected cells and 24 ± 6 for control RNA-transfected cells (Fig. 5 F). These data indicated that RNAi-induced knockdown of cellular v-KIND expression enhanced dendrite growth (Fig. 5 E, P < 0.001) and complexity in hippocampal neurons (Fig. 5 F, P < 0.001).
Figure 5.
Knockdown of v-KIND in hippocampal neurons promoted dendrite growth. Neurons were transfected at DIV3, fixed, immunolabeled, and observed at DIV21. Bar, 100 μm. (A), EGFP images of hippocampal neurons transfected with single v-KIND RNAi construct (RNAi-1, -2, and -3) or control construct (Control-1, -2, and -3) plus pCAG-EGFP. (B) Hippocampal neurons overexpressing v-KIND, v-KIND plus RNAi-2 (v-KIND+RNAi-2), or v-KIND plus Control-2 (v-KIND+Control-2). (a–c) EGFP images of neurons; (d–f) neurons immunolabeled by anti-FLAG antibody. (C) v-KIND RNAi-2 effectively knocked down v-KIND expression in hippocampal neurons. Hippocampal neurons were transfected soon after dissociation (see Materials and methods). Cells were lysated at DIV7 and DIV17 as indicated. v-KIND expression levels were analyzed by immunoblotting with the anti-v-KIND antibody (top panel) and the anti-α-tubulin antibody was used as a quantity control (bottom panel). (D and E) Statistical analysis of total dendrite length (D) and dendrite arbor numbers (E) of individual neurons. Data obtained from single RNAi experiments (RNAi-1, -2, and -3) were combined for statistical analysis of RNAi effects, and those from single control experiments (Control-1, -2 and -3) were combined for statistical analysis of control effects. Data from at least three independent experiments are presented as mean ± SD. *, P < 0.01, and **, P < 0.001 (t test). The number of neurons counted is shown in the parentheses above the columns.
To confirm the cellular effect induced by v-KIND knockdown, we next investigated whether v-KIND RNAi rescued the dendritic phenotype induced by v-KIND overexpression. Hippocampal neurons were cotransfected with FLAG-tagged v-KIND plus RNAi-2 (v-KIND+RNAi-2) or with FLAG-tagged v-KIND plus Control-2 (v-KIND+Control-2) at DIV3, and were observed at DIV21 (Fig. 5 B). v-KIND+Control-2 transfected cells had simple and less branched dendritic patterns (Fig. 5 B, b), similar to those of v-KIND-overexpressing cells (Fig. 5 B, a). On the other hand, v-KIND+RNAi-2 transfected cells exhibited more complex dendritic patterns (Fig. 5 B, c). Exogenous FLAG-tagged v-KIND expression in transfected hippocampal neurons was visualized by immunostaining with anti-FLAG antibody (Fig. 5 B, d–f). In contrast to v-KIND and v-KIND+Control-2 (Fig. 5 B, d and e, respectively), v-KIND+RNAi-2 transfected cells had significantly decreased FLAG immunoreactivity (Fig. 5 B, f), indicating that RNAi effectively inhibited exogenous FLAG-tagged v-KIND. Similar results were also observed in RNAi-1 and RNAi-3 transfected cells. Statistical analysis by combining all data of these RNAi expression experiments (RNAi-1, -2, and -3) and their control experiments (Control-1, -2, and -3) indicated that the average total dendrite length per cell transfected with v-KIND, v-KIND+ Control, and v-KIND+RNAi was 1,159 ± 381, 921 ± 317, and 2,280 ± 332 μm, respectively (Fig. 5 E) and that the average number of dendritic branch tips was 15 ± 6, 14 ± 4, and 31 ± 8, respectively (Fig. 5 F). These data indicated that cotransfection with RNAi rescued retarded dendrite growth by v-KIND overexpression with regard to both length (Fig. 5 E, P < 0.001) and branching (Fig. 5 F, P < 0.001). Collectively, these results suggest that v-KIND controls the development of dendritic arborization, probably by limiting dendrite growth, such as dendritic branching and extension.
v-KIND interacts with MAP2
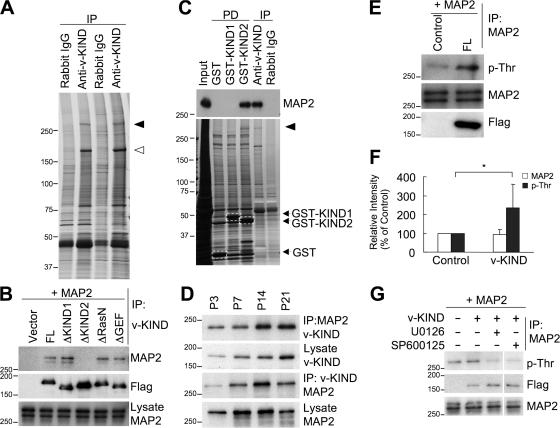
To clarify the molecular mechanism underlying v-KIND-mediated control of dendrite growth, we analyzed v-KIND associated proteins by coimmunoprecipitation from brain lysates with anti- v-KIND antibody followed by MASS spectrometry analysis (Fig. 6 A). Silver staining after separating on sodium dodecyl sulfate-PAGE (SDS-PAGE) revealed a high molecular weight band of ∼280 kD specifically precipitated by anti-v-KIND antibody, but not by rabbit IgG. This band was identified as MAP2 by MASS spectrometry analysis (Fig. 6 A).
Figure 6.
v-KIND binds HMW MAP2 and induces its Thr phosphorylation. (A) Association of v-KIND with MAP2 in postnatal mouse cerebellum. Silver staining of proteins associated with v-KIND in P21 mouse cerebella which were coimmunoprecipitated by the anti-v-KIND antibody. Open arrowhead, v-KIND protein; solid arrowhead, a band of ∼280 kD. (B) Association of v-KIND mutants with MAP2. v-KIND and the deletion of the first KIND (ΔKIND1), deletion of the second KIND domain (ΔKIND2), deletion of RasN (ΔRasN), and deletion of RasGEF (ΔGEF) were coexpressed in COS cells together with MAP2. Immunoprecipitations using the anti-v-KIND antibody were immunoblotted by anti-MAP2 antibody. (C) the KIND2 domain was responsible for MAP2 binding in mouse cerebellum. Equal amounts of mouse cerebellar proteins pulled down by GST-KIND1 and GST-KIND2 were subjected to immunoblotting by anti-MAP2a/b antibody (top panel) and visualized by silver staining (bottom panel). Silver-stained bands in the rectangle as indicated by the arrows are GST fusion proteins used. Positive control, immunoprecipitations by anti-v-KIND antibody; negative control, GST. Solid arrowhead, a band of ∼280 kD (MAP2). (D) Developmental association of v-KIND with MAP2. Top two panels, quantity of MAP2 coimmunoprecipitated with v-KIND. Bottom two panels, quantity of v-KIND coimmunoprecipitated with MAP2. (E) v-KIND overexpression increased p-Thr levels of MAP2. MAP2 was coexpressed with or without FLAG-tagged v-KIND and then it was immunoprecipitated and followed by immunoblotting by the anti-p-Thr antibody. Top panel, MAP2 Thr phosphorylation; middle panel, MAP2 in immunoprecipitations; bottom panel, v-KIND in the whole-cell lysates (anti-FLAG antibody). (F) Band intensity from three independent experiments in E was quantified by densitometry analysis and statistically analyzed. Relative band intensity with v-KIND overexpression to the MAP2 alone was shown. The data are presented as mean ± SD (F). *, P < 0.05 (t test). (G) ERK and JNK are involved in v-KIND induced MAP2 Thr phosphorylation. MAP2 was overexpressed together with v-KIND, and cells were treated by inhibitors of ERK1/2 (U0126) at 10 μM or JNK-1 (SP600125) at 25 μM for 2 h. Thr phosphorylation levels of treated and untreated MAP2 were checked by anti-p-Thr antibody.
We first examined the interaction between v-KIND and MAP2 by coimmunoprecipitation experiments using COS7 cells lysates with exogenously coexpressed MAP2 and FLAG-tagged v-KIND or various deletion mutants: mutants lacking the first KIND domain (ΔKIND1), the second KIND domain (ΔKIND2), RasN (ΔRasN), or RasGEF (ΔGEF). Anti-v-KIND antibody coimmunoprecipitated MAP2 coexpressed with v-KIND, ΔKIND1, ΔRasN, and ΔGEF, but not ΔKIND2 (Fig. 6 B). These results suggest that v-KIND interacts with MAP2 via the KIND2 domain. To further confirm the MAP2 binding region of v-KIND, we performed pull down assays from mouse brain lysates using GST fusion KIND1 and KIND2 proteins. Immunoblotting and silver staining analyses showed that GST-KIND2, but not GST-KIND1, pulled down endogenous brain MAP2 (Fig. 6 C, top). These results demonstrated that the KIND2 domain is responsible for interaction with MAP2.
We analyzed changes in the interaction between v-KIND and MAP2 during mouse brain development by coimmunoprecipitation using anti-v-KIND and anti-MAP2 antibodies (Fig. 6 D; and Fig. S3, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200702036/DC1). The quantity of v-KIND that coimmunoprecipitated with MAP2 peaked at P14, and the quantity of MAP2 that coimmunoprecipitated with v-KIND sharply increased at P14 and decreased at P21. Thus, the peak stage for the association between v-KIND and MAP2 appears to parallel the mid-late developmental stage of dendrite development. In fact, colocalization of v-KIND with MAP2 in P21 mouse cerebellum was also detected in the IGL and in neurons of the deep cerebellar nuclei (Fig. S3, C–K).
v-KIND enhances Thr phosphorylation of MAP2
MAP2 is one of the most phosphorylated proteins in neurons and its phosphorylation state correlates with dendrite development (Sanchez et al., 2000). We evaluated whether serine (Ser) and Thr phosphorylation levels of MAP2 expressed in COS7 cells were affected by coexpression of v-KIND. Phosphorylated MAP2 was detected by immunoprecipitation with anti-MAP2 antibody followed by immunoblotting with anti-phospho-Ser and anti–phospho-Thr (p-Thr) antibody (Fig. 6 E). Ser phosphorylation in MAP2 did not change significantly (unpublished data). In contrast, v-KIND coexpression increased p-Thr immunoreactivity of MAP2 (Fig. 6 E, top), which was confirmed by statistical analysis of quantified immunoreactive band intensity (Fig. 6 F, P < 0.05). These results suggested that coexpressed v-KIND enhances Thr phosphorylation of MAP2.
Many protein kinases phosphorylate MAP2 during brain development (Sanchez et al., 2000). ERK and JNK-1, downstream kinases of Ras signaling pathways, are involved in Thr phosphorylation of MAP2 (Holzer et al., 2001; Bjorkblom et al., 2005). We investigated the effects of the ERK1/2 inhibitor U0126 (Favata et al., 1998) or the JNK-1 inhibitor SP600125 (Bennett et al., 2001) on v-KIND induced p-Thr immunoreactivity of MAP2 (Fig. 6 G). U0126 or SP600125 treatment reduced Thr phosphorylation levels of MAP2 augmented by v-KIND coexpression, suggesting that JNK-1 and ERK, downstream targets of Ras signaling, are involved in the v-KIND–mediated MAP2 phosphorylation.
v-KIND is anchored to MAP2 through the KIND2 domain
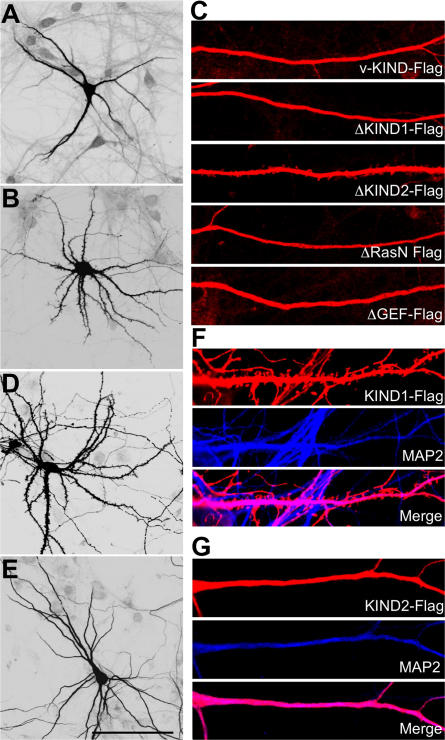
To study the protein structure and functional relationships of v-KIND in dendrite growth, we expressed various FLAG-tagged v-KIND constructs (v-KIND, ΔKIND1, ΔKIND2, ΔRasN, ΔGEF, KIND1 domain, and KIND2 domain) in hippocampal cells and immunocytochemically analyzed by anti-FLAG antibody (Fig. 7). Overexpressed v-KIND was mainly distributed in the soma and dendrite shafts where MAP2 was concentrated (Fig. 7, A and C). Similar to v-KIND, ΔKIND1, ΔRasN, and ΔGEF were also concentrated in the soma and dendritic shafts (Fig. 7 C). In contrast, ΔKIND2 was ubiquitously localized in the soma, dendrites shafts, spines, and axons (Fig. 7, B and C). Overexpressed KIND1 domain was widespread in dendrites and dendritic protrusions, including spines (Fig. 7, D and F), whereas overexpressed KIND2 domain had a similar localization as v-KIND, and was mainly localized in dendritic shafts, but not in spines (Fig. 7, E and G). Because KIND2 strongly binds with MAP2 (Fig. 6 B), these results indicate that v-KIND is anchored to MTs via the KIND2 domain.
Figure 7.
KIND2 domain anchors v-KIND to MT. Hippocampal neurons were transfected at DIV10, fixed and immunolabeled by the anti-FLAG antibody. (A and B) Confocal images of neurons overexpressing v-KIND (A), ΔKIND2 (B). (C) Localization of overexpressed v-KIND and deletion mutants in dendrites. (D and E) Neurons (DIV21) with exogenously expressed KIND1 (D) and KIND2 domains (E). (F and G) Colocalization of exogenously expressed KIND1 (F) and KIND2 (G) domains with MAP2 (blue, anti-Map2a/b). Bar, 100 μm.
MAP2 is a target in the signal of v-KIND-related regulation of dendrite development
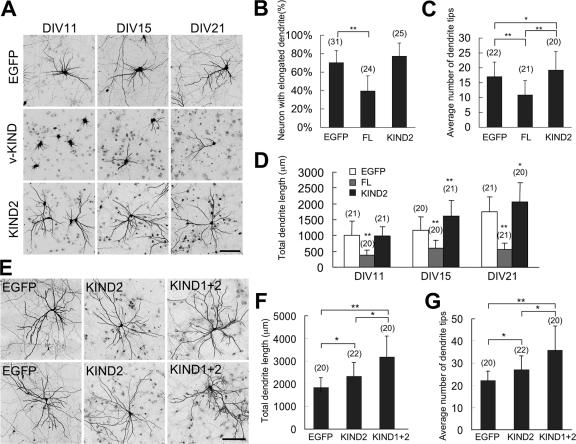
KIND2 binds MAP2 as strongly as v-KIND protein, indicating that KIND2 is sufficient to replace endogenous v-KIND for MAP2 binding (Figs. 6 and 7). We transfected the FLAG-tagged v-KIND or KIND2 domain into hippocampal neuronal primary cultures at DIV3, and observed the dendrite morphology at DIV11, DIV15, and DIV21 (Fig. 8). Expression of exogenous v-KIND and KIND2 proteins was confirmed by immunocytochemistry using anti-FLAG antibody (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200702036/DC1). In contrast to KIND2 overexpression induced significant increase in the total dendrite length (Fig. 8 A, bottom three panels and Fig. 8 D; P < 0.001) and branching (Fig. 8 C, P < 0.05) compared with no expression of v-KIND constructs. These results demonstrate that the MAP2-binding KIND2 domain acts as a dominant negative form of v-KIND in the neuron, and that MAP2 is a target for v-KIND which controls dendrite growth. Moreover, it is noted that overexpression of the construct containing both KIND1 and KIND2 domains (KIND1+2) in hippocampal neurons induced significant increases in both dendrite length (Fig. 8 F) and branching (Fig. 8 G) compared with overexpression of KIND2 alone (Fig. S4 B). These results suggest another binding partner of v-KIND via KIND1 might also be involved in v-KIND-mediated regulation of dendrite growth and complexity and/or that the functional ability of KIND2 might be enhanced in the presence of KIND1.
Figure 8.
Overexpression of the KIND2 domain promotes dendrite growth. Hippocampal neurons were transfected at DIV3, neurons were fixed and observed at DIV11, DIV15, and DIV21. Bar, 100 μm. (A) Representative images of hippocampal neurons overexpressing EGFP, v-KIND plus EGFP, or the KIND2 domain plus EGFP. (B–D) Statistical analysis of the percentage of neurons with elongated dendrites at DIV11 (B), dendritic tip numbers at DIV21 (C), and total length of dendrite branches (D) in individual neurons expressing EGFP, v-KIND, or KIND2. (E) Representative images of hippocampal neurons overexpressing the KIND2 domain, or the KIND1+2 domains of the v-KIND protein. (F and G) Statistical analysis of total dendrite length (F) and average dendritic tip number (G) of individual neurons. In statistical analysis, data from at least three independent experiments are presented as mean ± SD. *, P < 0.05, **, P < 0.001 (t test). The number of neurons counted is shown in the parentheses above the columns.
V-KIND has an important role in the control of cerebellar granule cell dendrite development
We demonstrated the fundamental properties of v-KIND in hippocampal neurons as described above. We then analyzed the role of v-KIND in dendrite growth of cerebellar granule cells was analyzed by a series of v-KIND overexpression and knockdown experiments. Overexpression of v-KIND caused stunted short dendrites (Fig. 9, A and F, total dendrite length, P < 0.001) in cerebellar granule cells, whereas that of KIND2 alone induced extended long dendrites (Fig. 9, A and G, total dendrite length, P < 0.05). These observations were very similar to those in hippocampal neurons. The effects of v-KIND knockdown on granule cells were analyzed by transfecting with each RNAi construct or each Control construct soon after cell dissociation followed by fixation for EGFP cell morphology imaging at DIV7 (Fig. 9 B). Western blot analyses showed effective knockdown of v-KIND expression levels in RNAi-2-transfected cultures at DIV7 and DIV14 in comparison with those in Control-2-transfected cultures (Fig. 9 C). In addition, v-KIND knockdown by RNAi-2 promoted dendrite growth in granule cell cultures compared with that with Control-2 (Fig. 9 G, P < 0.01), which was similar to hippocampal neurons shown in Fig. 5, A and D. These results indicated that v-KIND has an important role in the control of neuron dendrite development in both cerebellar granule cells and hippocampal neurons.
Figure 9.
v-KIND has an important role in the control of cerebellar granule cell dendrite development. (A) Confocal images of cerebellar granule cells overexpressing EGFP, v-KIND plus EGFP, or KIND2 plus EGFP. (B) Confocal images of cerebellar granule cells transfected with v-KIND RNAi (RNAi-1, -2, and -3) or the control RNA (control-1, -2, and -3). Cerebellar granule cells were transfected by the calcium phosphate method using a CellPhect Transfection kit at DIV1, and cells were fixed at DIV7. Imaging of the marker protein EGFP shows the neuron morphology. (C) Knockdown of v-KIND was confirmed by immunoblotting with anti-v-KIND antibody. RNAi constructs (Control-2 or RNAi-2) were transfected into cerebellar granule cells soon after dissociation using the Mouse Neuron Nucleofector kit and the Nucleofector device (Amaxa). Cells were lysed at DIV7 and DIV14, respectively. Immunoblotting was performed with the anti-v-KIND antibody (top panel), and anti-α-tubulin antibody was used as a quantity control (bottom panel). (D) MAP2 Thr phosphorylation was reduced by v-KIND knockdown. The data of RNAi-2 are shown. Top panel, immunoblotting with the anti-phosphorylated Thr antibody (p-Thr); middle panel, immunoprecipitated MAP2; bottom panel, immunoblotting by anti-v-KIND antibody. (E) Quantitative analysis of intensities of MAP2 Thr phosphorylation band and v-KIND band in v-KIND RNAi-2 or control-2 transfected cells from three independent experiments (see Materials and methods). Relative intensity to the control was shown (% control) as mean ± SD. *, P < 0.05 (t test) as compared with the control. (F) Statistical analysis of the total dendrite length of cerebellar granule cells overexpressing EGFP, v-KIND plus EGFP, and KIND2 plus EGFP. (G) Statistical analysis of the total dendrite length of cerebellar granule cells transfected with RNAi-2 plus EGFP (RNAi-2), or Control-2 plus EGFP (Control-2). In statistical analysis, data from at least three independent experiments are presented as mean ± SD. *, P < 0.05, **, P < 0.01, ***, P < 0.001 (t test). The number of neurons counted is shown in the parentheses above the columns.
We investigated MAP2 Thr phosphorylation in cerebellar granule cells when endogenous v-KIND expression was knocked down by RNAi. Western blotting analysis and quantitative statistical analysis showed that MAP2 Thr phosphorylation was reduced concomitantly with decreased v-KIND levels by knockdown with RNAi-2 (Fig. 9, D and E, P < 0.05). Similar data were also obtained from hippocampal neurons after v-KIND knockdown by RNAi (unpublished data). These results suggest that v-KIND is involved in MAP2 Thr phosphorylation, which is likely important for dendrite development.
Discussion
In this study, we characterized a novel function for a brain-specific, two KIND domain-containing Ras GEF, v-KIND protein. v-KIND had a widespread distribution pattern in mouse brain, and was predominantly expressed in the IGL of cerebellum from around P12 to adult stage. In cerebellum, v-KIND protein was localized in the soma of granule cells and glomeruli, where granule cell dendrites extended to make synapses with mossy fibers and Golgi cell axons, and in hippocampus, it was present in the soma and dendrites of CA1-3 neurons. v-KIND activated Ras through the RasGEF domain and interacted with MAP2 through the KIND2 domain. Overexpression of exogenous v-KIND protein suppressed dendrite growth, whereas knockdown of v-KIND by RNAi promoted dendrite growth. These findings suggest a new signaling pathway linking small GTPase Ras and MT-associated MAP2 via v-KIND to control dendrite growth (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200702036/DC1).
v-KIND links signaling of activated Ras to MAP2
v-KIND was first reported as a novel brain-specific RasGEF (Mees et al., 2005). In the present study, v-KIND bound to and activated Ras in mouse brain lysates through the C-terminal RasGEF domain, indicating that v-KIND certainly acts as a brain RasGEF. Within the GEF family, v-KIND is unique to have two KIND domains at the N-terminal region. The KIND domain is a recently identified protein domain whose functional properties remain yet unclear (Ciccarelli et al., 2003). Our results demonstrated that the KIND2 domain is functionally distinct from the KIND1 domain and specifically interacts with MAP2 in in vitro expression systems and in vivo brain lysates. In addition, overexpressed KIND2 was concentrated in dendritic shafts of neurons and largely colocalized with MAP2. These results suggest that v-KIND is anchored to the dendrites through the interaction between the KIND2 domain and MAP2.
MAP2 contains a number of Ser and Thr residues that are highly conserved in mouse, rat, and human, and many of these residues are highly phosphorylated in vivo (Sanchez et al., 2000). Both ERK and JNK-1 are downstream molecules of the Ras signaling pathways and are involved in Thr phosphorylation of MAP2 (Holzer et al., 2001; Bjorkblom et al., 2005). Our results showed that v-KIND induced MAP2 Thr phosphorylation, which was blocked by inhibitors of ERK or JNK-1. These results suggest that v-KIND links the Ras signaling cascades, including downstream signaling via ERK or JNK1, to MAP2 phosphorylation.
v-KIND controls dendrite growth
The v-KIND overexpression and RNAi knockdown experiments suggested important roles of v-KIND in controlling dendrite growth, although the underlying molecular mechanism remains elusive. One possibility might be the physical and functional association between v-KIND and MAP2, that is, v-KIND-MAP2 interaction and v-KIND-induced MAP2 Thr phosphorylation. During dendrite development, normal MT structures are required for both dendritic elongation at early stages and dendritic maturation at later stages. MAP2 phosphorylation levels are developmentally controlled along with dendrite growth (Riederer et al., 1995; Quinlan and Halpain, 1996). Increased phosphorylation of MAP2 causes MAP2 to detach from MTs, resulting in decreased MT stability and attenuated actin-bundling activity (Jameson and Caplow, 1981; Murthy and Flavin, 1983; Selden and Pollard, 1983; Hoshi et al., 1992; Sanchez et al., 2000). Our results showed that v-KIND overexpression promotes MAP2 Thr phosphorylation and causes impaired dendrite growth, whereas v-KIND RNAi knockdown reduces MAP2 Thr phosphorylation and induces increased dendrite growth. Moreover, we showed that the MAP2-binding domain KIND2 alone acts as a dominant negative and its overexpression eventually increases dendrite length and complexity. These data suggest a possibility that v-KIND functions through controlling of MAP2 phosphorylation levels during dendrite development.
The present study showed that v-KIND might associate with other cytoskeletal element(s) via the KIND1 domain except for the interaction with MAP2 via the KIND2 domain. In this regard, it is notable that overexpression of the construct containing both KIND domains (KIND1+2) induced more branching and longer dendritic patterns than that of the KIND2 domain alone. Moreover, although v-KIND prominently colocalized with MAP2 in dendritic shafts, it was also observed in dendritic terminals where it partially colocalized with F-actin in developing hippocampal neurons. Altogether, these results suggest that apart from MAP2, other binding partner(s) may also cooperate with v-KIND in controlling complex dendrite development.
Functional role of v-KIND in cerebellar development
Dendrite growth and complexity appear to be regulated by a combination of positively and negatively acting factors in dendrite growth (Scott and Luo, 2001). Hippocampal neurons and cerebellar granule cells have different complexities in dendritic patterns, and hippocampal neurons have much longer and more complicated dendrite patterns than cerebellar granule cells. Cerebellar granule cells possess very short, claw-like dendrites around the mossy fiber terminals within the glomerular rosettes. These dendrites not only make excitatory synapses with large mossy terminals, but also form inhibitory synapses with Golgi cell axon terminals within the synaptic glomeruli (Ito, 1997). Moreover, in addition to synaptic contacts, these dendrites form numerous dendrodendritic attachment plaques or puncta adherentia in glomeruli. Whether the differences in the dendrite complexity between hippocampal neurons and cerebellar granule cells are, at least in part, caused by differential levels of protein expression is not known.
We initially isolated v-KIND as a gene predominantly expressed in cerebellar granule cells of the IGL during mid-late developmental stages of postnatal mouse brain. v-KIND immunoreactivity was enriched in the somatodendritic regions of granule cells and especially the glomeruli. Moreover, we demonstrated that v-KIND overexpression suppresses the dendrite growth of cerebellar granule cells, whereas v-KIND knockdown promotes it. Thus, we suggest that v-KIND acts as a signaling molecule in controlling or limiting dendrite growth to form short but unique structures that extend within very narrow spaces and are surrounded by densely packed granule cell soma in the IGL during the mid-late developmental period. Slight differences observed in the extent of the effects of v-KIND overexpression and knockdown between cerebellar granule cells and hippocampal neurons might be due to differences in the expression levels of v-KIND, which negatively controls dendrite growth, as well as any positively acting factors in dendrite growth in these two cell-types.
Materials and methods
RT-PCR analysis
A series of first-strand cDNAs was produced by reverse-transcription (RT) from 20 ng of total cerebellar RNAs at varying developmental stages, using an oligo-dT primer. The cDNA sequence corresponding to the nucleotide positions 583 to 1182 (a.a 175–394) of v-KIND (GenBank/EMBL/DDBJ accession no. AJ580324) was amplified using the primers 5′-GCAGGTGGGGAAAACTCC-3′ (forward primer) and 5′-CTGGCGTCTCCAGCAAAT-3′ (reverse primer). The cycling conditions were as follows: denaturing at 94°C for 3 min, amplification by 25 cycles at 94°C (15 s), 55°C (30 s), and 72°C (1 min), and extension at 72°C for 5 min. To analyze tissue distribution, total RNAs prepared from various tissues of P7 or P21 mice were used for RT-polymerase chain reaction (PCR). The RT-PCR of glyceraldehyde- 3-phosphate dehydrogenase with primers 5′-GCCATCAACGACCCCTTCATTGACCTC-3′ (forward primer) and 5′-GCCATGTAGGCCATGAGGTCCACCAC-3′ (reverse primer) were used as internal controls.
In situ hybridization
In situ hybridization (ISH) brain histochemistry was basically performed as described previously (Huang et al., 2006). The cDNA sequence corresponding to nucleotide positions 1962 to 2561 (a.a 654–857) of the v-KIND cDNA was used as a template to prepare the digoxigenin-labeled antisense riboprobes using a digoxigenin-dUTP labeling kit (Roche Diagnostics). Paraffin sections of mouse brain (10 μm thick) were fixed in 4% paraformaldehyde for 5 min, washed twice in phosphate buffered saline (PBS), and treated with freshly prepared 10 μg/ml proteinase K (Invitrogen) at room temperature. After acetylation, the sections were subjected to digoxigenin-based hybridization procedures. In brief, the sections were incubated in hybridization buffer containing 0.2 μg/ml digoxigenin-labeled riboprobes at 60°C overnight in a humid chamber. The hybridized sections were washed by successive immersion in 1 × SSC (150 mM NaCl and 15 mM sodium citrate, pH 7.0; 60°C, 10 min, twice), 2 × SSC (37°C, 10 min), 2 × SSC containing 20 μg/ml RNase A (37°C, 30 min), 2 × SSC (37°C, 10 min), and 0.2 × SSC (60°C, 30 min, twice). The hybridization signals were detected using a digoxigenin detection kit (Roche Diagnostics). ISH images (Fig. 1) were captured with a microscope (Olympus BX51) equipped with a 2×/NA 0.08 (PlanApo) or 20x/NA 0.7 (UPlanApo) objective and a charged-coupled device camera (ProgRes C14; JENOPTIK) and processed with Photoshop (Adobe).
Cell culture
COS-7 cells were grown in culture dishes in DME/10% fetal bovine serum at 37°C and 10% CO2 in a humidified incubator. Hippocampal dissociated primary cultures were prepared from embryonic day 17 Wistar rats (Nippon SLC) as described previously (Shiraishi et al., 2003). In brief, excised hippocampi were treated with 45 U papain (Worthington), 0.01% DNase I (Boehringer-Mannheim), 0.02% DL-cystein, 0.02% bovine serum albumin, and 0.5% glucose in PBS for 20 min at 37°C. After adding 20% bovine serum, cells were dissociated by repeatedly passing them through a 1 ml plastic pipette tip. Dispersed cells were plated at a density of 1.104 cells/cm2 onto poly-L-lysine-coated (Sigma-Aldrich) glass coverslips (Matsunami) in neurobasal medium (GIBCO BRL, Life Technologies) containing 2% B27 supplement (Invitrogen), 500 μM l-glutamine, 0.1 mg/ml streptomycin (Meiji), and 100 U/ml penicillin (Banyu, Tokyo, Japan). Cultures were maintained in a humidified atmosphere of 5% CO2 in air at 37°C.
Antibodies
Antibodies against mouse v-KIND were generated by immunizing rabbits with a synthetic peptide, CEKHSRKIQDKLRRMKATFQ (aa1724–1742), after conjugation with keyhole limpet hemocyanin. Affinity purification of the antibody was performed using a HiTrap NHS-High Performance column (Amersham Biosciences). The antibody was used at a dilution of 1:2000 for immunoblotting and 1:1,000 for immunohistochemistry and immunocytochemistry. Antibodies against MAP2a/b (M1406) and MAP2 (M4403) were obtained from Sigma-Aldrich; the antibody against GFP (SC-9996) was from Santa Cruz Biotechnology, Inc.; the anti-p-Thr antibody (#9381) was from Cell Signaling Technology; and the antibody against FLAG (F3165) was from Sigma-Aldrich. All antibodies were used at a dilution of 1:1,000 in immunoblotting and 1:200 in immunohistochemistry and immunocytochemistry.
Plasmid construction
Mouse v-KIND was isolated by PCR from mouse cerebellum and cloned into a pCAG vector with a FLAG-tag (a gift from Dr. Junichi Miyazaki, Div. Stem Cell Regulation Research, Osaka University Graduate School of Medicine, Osaka, Japan). v-KIND mutants were prepared by PCR-based methods and inserted into pCAG vectors with a FLAG-tag. H-Ras cDNA was isolated by PCR from mouse cerebellum and cloned into a pCAG vector with a HA-tag. HMW MAP2 cDNA was a gift from Dr. N. Cowan (NYU Medical Center, New York, NY) and was constructed by PCR-based methods and cloned into pCAG vector with a HA-tag. KIND1 and KIND2 domain fragments were prepared by PCR-based methods, and were inserted into pGEX-4T-2.
Transfection and data analysis
COS-7 cells were transfected by Lipofectamine 2000 reagent (Invitrogen) 24 h after plating at a density of 2.0 × 105 cells/cm2. After an additional 6 h of incubation, cells were washed and cultured in DME/10% fetal bovine serum for 2 d. Hippocampal neurons were transfected by the Ca2+-phosphate-mediated method with some modification (Haubensak et al., 1998; Kohrmann et al., 1999). For 105 cells plating on each coverslip, 2 μg of plasmid was dissolved in 10 μl 250 mM CaCl2, mixed with 10 ml 2×BBS (280 mM NaCl, 1.5 mM Na2HPO4, 50 mM BES, pH 7.1), and added to 400 μl of the fresh culture medium. After incubation for 1 to 2 h at 37°C with 2.5% CO2, cells were washed with HBSS and cultured in conditioned medium. The calcium phosphate method using a CellPhect Transfection kit (Amersham Biosciences) was used to transfect cerebellar neurons on DIV1 in serum-free defined medium on poly- l-lysine-coated glass coverslips. To quantify the percentage of cells with elongated dendrites at DIV11, 20 fields were randomly selected from three independent experiments with v-KIND or KIND2 overexpression based on healthy morphology, then analyzed by counting the number of cells with dendrites that were longer than 20 μm. To quantify dendritic length and arbor numbers in neurons overexpressing v-KIND protein or KIND2 domain, dendritic length and the number of arbors of neurons randomly selected from three independent experiments (15 fields total) were measured. Results are presented as the mean ± SD and were analyzed with t test using the Excel 2003 software program. P values less than 0.05 were considered significant.
RNAi
Three types of v-KIND stealth RNAi were obtained from Invitrogen. Their sense sequences were as follows: CAUCCAGGAGGAAUUUGCCUUUGAU (control sequence CAUGGAGGAUAAGUUUCCUUCCGAU), GAGCAGCUGCUAAAGAACCUCUUCA (control sequence GAGGUCGAAUCAAGACUCCUACUCA), and GAGACGGGAGGUUUCACCAUGACUA (control sequence GAGGGAGGAUUUCACUACAGACCUA). These RNAi were transfected into COS cells using the Lipofectamine 2000 reagent according to the manufacture's directions (Invitrogen), and into hippocampal neurons together by pCAG-EGFP by Ca2+ phosphate precipitation at a concentration of 50 nM as described above. For immunoblotting experiments, RNAi was transfected into cerebellar granule cells soon after dissociation using the Mouse Neuron Nucleofector kit and the Nucleofector device as previously described (Amaxa) (Huang et al., 2002; Liu et al., 2003).
Immunohistochemistry and immunocytochemistry
ICR mice were anesthetized with diethyl ether and transcardially perfused with 4% paraformaldehyde in PBS. The excised brains were immersed for 2 h in the same fixative and cryosectioned (20 μm thick). For immunohistochemistry (IHC), fixed brain sections were preincubated with 5% normal goat serum in PBS containing 0.03% Triton X-100 for 1 h and then incubated with primary antibody overnight at 4°C. After washing with PBS, immunodetection was performed using horseradish peroxidase-conjugated secondary antibody (1:500) and diaminobenzidine. IHC images (Fig. 1) were captured with a microscope (Olympus BX51) equipped with a 2x/NA 0.08 (PlanApo) or 20x/NA 0.7 (UPlanApo) objective and a charged-coupled device camera (ProgRes C14; JENOPTIK). The acquired images were processed with Photoshop (Adobe). For immunocytochemistry, cultured cells were fixed with 4% paraformaldehyde with 4% sucrose, permeabilized in 0.2% Triton X-100 in PBS for 5 min at room temperature, preincubated with 5% normal donkey serum in PBS for 1 h, and then incubated with each affinity-purified antibody for 16 h at 4°C. Multiple staining was performed using combinations of rabbit anti-v-KIND antibody (1:1,000), mouse anti-MAP2 antibody (1:200; Sigma-Aldrich), mouse anti-Flag antibody (1:200; Sigma-Aldrich), and Alexa 647-phallotoxin (1:50; AlexaFluor 647 phalloidin; Invitrogen). For fluorescent immunostaining, cells were then incubated with AlexaFluor 488-, 555-, or 647-conjugated secondary antibody (Invitrogen) in 5% donkey serum/PBS for 1 h at room temperature. Immunofluorescent images were captured with a confocal microscope (LSM 510 META; Carl Zeiss MicroImaging, Inc.) equipped with a 20x/NA 0.50 (Plan-NEO FLUAR), 40x/NA 0.75 (Plan-NEO FLUAR), or 100x/NA 1.4 oil iris (Plan-APO CHROMAT) objective lens (20×, Fig. 8; 40×, Figs. 4, 5, and 7; 100×, Figs. 3 and 9) and LSM 510 META software (Carl Zeiss MicroImaging, Inc). The acquired images were processed with Photoshop (Adobe).
Protein extraction, immunoprecipitation, and immunoblotting
Protein extraction and Western blotting analysis were performed as described previously (Huang et al., 2006). In brief, COS cells or mouse cerebella were lysed and homogenized in 1% Triton X-100 buffer (50 mM Hepes, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonylfluoride). The subcellular fractionation of the P7 and P21 mouse cerebella was performed as described previously (Huang et al., 2006). In brief, the P7 and P21 mouse cerebella were first homogenized in homogenization buffer (0.32 M sucrose, 5 mM Tris·HCl, 1 mM EGTA, 1 mM dithiothreitol, 1mM pepstatin A, 1mM leupeptin, and 1mM Na3VO4). The protein lysates were centrifuged at 1,000 g for 10 min, the pellet was lysed in 1% Triton X-100 buffer (PPt1), and the supernatant was centrifuged at 105,000 g for 1 h. The pellet was lysed in 1% Triton X-100 buffer (PPt2+3), and the supernatant was used as Sup3. For immunoprecipitation, after centrifuging at 1,000 g for 10 min, protein solutions (containing ∼1 mg protein) were mixed with the primary antibody and incubated for 1 h on ice. The mixtures were rotated with Protein A sepharose or Protein G sepharose (Amersham Biosciences) for 1 h at 4°C. The sepharose was washed four times with 1% Triton X-100 buffer. After boiling in sample buffer (0.4 M Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, and 0.04% bromophenol blue) for 5 min, equal portions of protein solution were separated by SDS-PAGE and probed with diluted primary and horseradish peroxidase-conjugated secondary antibodies (1:2,000). Silver staining was performed using the silver staining kit according to the manufacturer's instructions (Daiichi Pure Chemicals Co.). The GST fusion protein pull-down assay was performed as follows. Escherichia coli expressing GST fusion proteins were lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 25% sucrose, 1% Triton X-100, 5 mM MgCl2), and the cell extracts containing ∼10 μg GST-fusion proteins were coupled to glutathione- sepharose (Amersham Biosciences) by rotating for 1 h at 4°C. After washing three times with 1% Triton-X/PBS, the sepharose was coupled to cerebellar protein lysates, which had been precleared with glutathione-sepharose for 1 h at 4°C. After rotating for 1 h at 4°C, the GST-fusion protein complex was washed 5 times with cell lysis buffer and subjected to SDS-PAGE and immunoblotting. In GTP-γ-S and GDP-β-S treatment, protein lysates were treated with 0.1 mM GTP-γ-S (Sigma Chemical Co.) or 1.0 mM GDP-β-S (Sigma-Aldrich) in the presence of 10 mM EDTA, pH 8.0 at 30°C for 15 min (to activate or inactivate Ras). Then the GST pull-down assay was performed as described above. Quantitative analysis of immunoblotting band intensity was performed by Quantity-One software (Bio-Rad Laboratories). Relative intensity to the control was shown (% control). Values are mean ± SD. *, P < 0.05 as compared with the control.
Mass spectrometry
For identification of v-KIND interacting proteins, in-gel digestion was performed following Coomassie staining, bands corresponding to MAP2 were excised, and the gel pieces were destained with 50% CH3CN in 50 mM NH4HCO3 solution. After removing the supernatant, cysteine residues were reduced with dithiothreitol, carbamido methylated with iodoacetamide, and the proteins were digested with trypsin at 37°C overnight. The tryptic peptides were recovered by sequentially adding 50% CH3CN/1% trifluoroacetic acid, 20% HCOOH/25% CH3CN/15% i-PrOH, and 80% CH3CN solutions. The supernatants were collected and pooled into one tube, and the volume was reduced in vacuo. The dried tryptic peptides were suspended in 2% CH3CN/0.1% trifluoroacetic acid and applied to the following liquid chromatography-tandem mass spectroscopy system. Chromatographic separation was accomplished with the MAGIC 2002 HPLC system (Michrom BioResources, Inc.). Peptide samples were loaded onto a Cadenza C18 custom-packed column (0.2 × 50 mm, Michrom BioResources, Inc.), and eluted using a linear gradient of 5% to 60% CH3CN in 0.1% HCOOH for 30 min with a flow of 1 ml/min. Samples were ionized with the Nanoflow-liquid chromatography electrospray ionization, and tandem mass spectrometry data were obtained with an LCQ-Deca XP ion trap mass spectrometer (Thermo Electron Corp.). The Mascot database searching software (Matrix Science Inc.) was used for the identification of acetylated proteins.
Online supplemental material
Fig. S1: subcellular distribution of v-KIND in mouse cerebellum. Fig. S2: three oligonucleotide Stealth v-KIND RNAi efficiently knocked down exogenously expressed v-KIND in COS7 cells. Fig. S3: association of v-KIND with MAP2. (A-B), Coimmunoprecipitation of v-KIND and MAP2 from brain lysate. Fig. S4: FLAG-tagged v-KIND protein and the mutant overexpressed in hippocampal neurons. Fig. S5. v-KIND links Ras signaling to MTs. v-KIND is anchored to the dendritic shaft through the KIND2 domain by its MAP2 binding affinity. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702036/DC1.
Acknowledgments
We thank Dr. N. Cowan for providing the HMW Map2 cDNA. The pCAG expression vector was a gift from Dr. Jun-ichi Miyazaki. We also thank Dr. Adrian Moore (Molecular Neuropathology Group, BSI, RIKEN) and our laboratory staff, especially Dr. Tetsushi Sadakata, for their helpful comments and advice.
J. Huang's present address is Drug Discovery Research Lab, Hanno Research Center, TAIHO Pharmaceutical Co., Ltd., 1-27, Misugidai, Hanno-shi, Saitama, 357-8527, Japan.
Abbreviations used in this paper: DIV, days in vitro; GEF, guanine nucleotide exchange factor; HMW MAP2, high molecular weight MAP2; KINDs, kinase non-catalytic C-terminal domains; MAP2, microtubule associated protein 2; MT, microtubule; v-KIND, very-KIND.
References
- Alonso, M., J.H. Medina, and L. Pozzo-Miller. 2004. ERK1/2 activation is necessary for BDNF to increase dendritic spine density in hippocampal CA1 pyramidal neurons. Learn. Mem. 11:172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, B.L., D.T. Sasaki, B.W. Murray, E.C. O'Leary, S.T. Sakata, W. Xu, J.C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, et al. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA. 98:13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, R., and A. Matus. 1984. Light and electron microscopic studies of the distribution of microtubule-associated protein 2 in rat brain: a difference between dendritic and axonal cytoskeletons. J. Comp. Neurol. 226:203–221. [DOI] [PubMed] [Google Scholar]
- Bjorkblom, B., N. Ostman, V. Hongisto, V. Komarovski, J.J. Filen, T.A. Nyman, T. Kallunki, M.J. Courtney, and E.T. Coffey. 2005. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: role of microtubule-associated protein 2 as an effector. J. Neurosci. 25:6350–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils, J., and P. Chardin. 1999. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24:306–311. [DOI] [PubMed] [Google Scholar]
- Ciccarelli, F.D., P. Bork, and E. Kerkhoff. 2003. The KIND module: a putative signalling domain evolved from the C lobe of the protein kinase fold. Trends Biochem. Sci. 28:349–352. [DOI] [PubMed] [Google Scholar]
- Cline, H.T. 2001. Dendritic arbor development and synaptogenesis. Curr. Opin. Neurobiol. 11:118–126. [DOI] [PubMed] [Google Scholar]
- Dotti, C.G., C.A. Sullivan, and G.A. Banker. 1988. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8:1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favata, M.F., K.Y. Horiuchi, E.J. Manos, A.J. Daulerio, D.A. Stradley, W.S. Feeser, D.E. Van Dyk, W.J. Pitts, R.A. Earl, F. Hobbs, et al. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623–18632. [DOI] [PubMed] [Google Scholar]
- Friedrich, P., and A. Aszodi. 1991. MAP2: a sensitive cross-linker and adjustable spacer in dendritic architecture. FEBS Lett. 295:5–9. [DOI] [PubMed] [Google Scholar]
- Gao, F.B., and B.A. Bogert. 2003. Genetic control of dendritic morphogenesis in Drosophila. Trends Neurosci. 26:262–268. [DOI] [PubMed] [Google Scholar]
- Goi, T., G. Rusanescu, T. Urano, and L.A. Feig. 1999. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol. Cell. Biol. 19:1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin, M., and M. Segal. 2003. Protein kinase C and ERK involvement in dendritic spine plasticity in cultured rodent hippocampal neurons. Eur. J. Neurosci. 17:2529–2539. [DOI] [PubMed] [Google Scholar]
- Harada, A., J. Teng, Y. Takei, K. Oguchi, and N. Hirokawa. 2002. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J. Cell Biol. 158:541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak, W., F. Narz, R. Heumann, and V. Lessmann. 1998. BDNF-GFP containing secretory granules are localized in the vicinity of synaptic junctions of cultured cortical neurons. J. Cell Sci. 111:1483–1493. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., S. Hisanaga, and Y. Shiomura. 1988. MAP2 is a component of crossbridges between microtubules and neurofilaments in the neuronal cytoskeleton: quick-freeze, deep-etch immunoelectron microscopy and reconstitution studies. J. Neurosci. 8:2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer, M., U. Gartner, F.J. Klinz, F. Narz, R. Heumann, and T. Arendt. 2001. Activation of mitogen-activated protein kinase cascade and phosphorylation of cytoskeletal proteins after neurone-specific activation of p21ras. I. Mitogen-activated protein kinase cascade. Neuroscience. 105:1031–1040. [DOI] [PubMed] [Google Scholar]
- Hoshi, M., K. Ohta, Y. Gotoh, A. Mori, H. Murofushi, H. Sakai, and E. Nishida. 1992. Mitogen-activated-protein-kinase-catalyzed phosphorylation of microtubule-associated proteins, microtubule-associated protein 2 and microtubule-associated protein 4, induces an alteration in their function. Eur. J. Biochem. 203:43–52. [DOI] [PubMed] [Google Scholar]
- Huang, J., H. Hamasaki, T. Nakamoto, H. Honda, H. Hirai, M. Saito, T. Takato, and R. Sakai. 2002. Differential regulation of cell migration, actin stress fiber organization, and cell transformation by functional domains of Crk-associated substrate. J. Biol. Chem. 277:27265–27272. [DOI] [PubMed] [Google Scholar]
- Huang, J., R. Sakai, and T. Furuichi. 2006. The docking protein Cas links tyrosine phosphorylation signaling to elongation of cerebellar granule cell axons. Mol. Biol. Cell. 17:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M. 1997. Cerebellar microcomplexes. Int. Rev. Neurobiol. 41:475–487. [DOI] [PubMed] [Google Scholar]
- Jameson, L., and M. Caplow. 1981. Modification of microtubule steady-state dynamics by phosphorylation of the microtubule-associated proteins. Proc. Natl. Acad. Sci. USA. 78:3413–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, Y.N., and L.Y. Jan. 2003. The control of dendrite development. Neuron. 40:229–242. [DOI] [PubMed] [Google Scholar]
- Jaworski, J., S. Spangler, D.P. Seeburg, C.C. Hoogenraad, and M. Sheng. 2005. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J. Neurosci. 25:11300–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, Y.H., C. Ruiz-Canada, M. Gorczyca, and V. Budnik. 2002. The Ras1-mitogen- activated protein kinase signal transduction pathway regulates synaptic plasticity through fasciclin II-mediated cell adhesion. J. Neurosci. 22:2496–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrmann, M., W. Haubensak, I. Hemraj, C. Kaether, V.J. Lessmann, and M.A. Kiebler. 1999. Fast, convenient, and effective method to transiently transfect primary hippocampal neurons. J. Neurosci. Res. 58:831–835. [PubMed] [Google Scholar]
- Kumar, V., M.X. Zhang, M.W. Swank, J. Kunz, and G.Y. Wu. 2005. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J. Neurosci. 25:11288–11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.J., J. Ding, A.S. Kowal, T. Nardine, E. Allen, J.D. Delcroix, C. Wu, W. Mobley, E. Fuchs, and Y. Yang. 2003. BPAG1n4 is essential for retrograde axonal transport in sensory neurons. J. Cell Biol. 163:223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. 2002. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18:601–635. [DOI] [PubMed] [Google Scholar]
- Mees, A., R. Rock, F.D. Ciccarelli, C.B. Leberfinger, J.M. Borawski, P. Bork, S. Wiese, M. Gessler, and E. Kerkhoff. 2005. Very-KIND is a novel nervous system specific guanine nucleotide exchange factor for Ras GTPases. Gene Expr Patterns. 6:79–85. [DOI] [PubMed] [Google Scholar]
- Miller, F.D., and D.R. Kaplan. 2003. Signaling mechanisms underlying dendrite formation. Curr. Opin. Neurobiol. 13:391–398. [DOI] [PubMed] [Google Scholar]
- Murthy, A.S., and M. Flavin. 1983. Microtubule assembly using the microtubule-associated protein MAP-2 prepared in defined states of phosphorylation with protein kinase and phosphatase. Eur. J. Biochem. 137:37–46. [DOI] [PubMed] [Google Scholar]
- Quinlan, E.M., and S. Halpain. 1996. Emergence of activity-dependent, bidirectional control of microtubule-associated protein MAP2 phosphorylation during postnatal development. J. Neurosci. 16:7627–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond, L., and A. Ghosh. 2001. The role of Notch and Rho GTPase signaling in the control of dendritic development. Curr. Opin. Neurobiol. 11:111–117. [DOI] [PubMed] [Google Scholar]
- Redmond, L., A.H. Kashani, and A. Ghosh. 2002. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 34:999–1010. [DOI] [PubMed] [Google Scholar]
- Riederer, B.M., E. Draberova, V. Viklicky, and P. Draber. 1995. Changes of MAP2 phosphorylation during brain development. J. Histochem. Cytochem. 43:1269–1284. [DOI] [PubMed] [Google Scholar]
- Sanchez, C., J. Diaz-Nido, and J. Avila. 2000. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 61:133–168. [DOI] [PubMed] [Google Scholar]
- Scott, E.K., and L. Luo. 2001. How do dendrites take their shape? Nat. Neurosci. 4:359–365. [DOI] [PubMed] [Google Scholar]
- Selden, S.C., and T.D. Pollard. 1983. Phosphorylation of microtubule-associated proteins regulates their interaction with actin filaments. J. Biol. Chem. 258:7064–7071. [PubMed] [Google Scholar]
- Shiomura, Y., and N. Hirokawa. 1987. Colocalization of microtubule-associated protein 1A and microtubule-associated protein 2 on neuronal microtubules in situ revealed with double-label immunoelectron microscopy. J. Cell Biol. 104:1575–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi, Y., A. Mizutani, K. Mikoshiba, and T. Furuichi. 2003. Coincidence in dendritic clustering and synaptic targeting of homer proteins and NMDA receptor complex proteins NR2B and PSD95 during development of cultured hippocampal neurons. Mol. Cell. Neurosci. 22:188–201. [DOI] [PubMed] [Google Scholar]
- Taylor, S.J., R.J. Resnick, and D. Shalloway. 2001. Nonradioactive determination of Ras-GTP levels using activated ras interaction assay. Methods Enzymol. 333:333–342. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., and H.T. Cline. 2004. Rho GTPases and activity-dependent dendrite development. Curr. Opin. Neurobiol. 14:297–304. [DOI] [PubMed] [Google Scholar]
- Whitford, K.L., P. Dijkhuizen, F. Polleux, and A. Ghosh. 2002. Molecular control of cortical dendrite development. Annu. Rev. Neurosci. 25:127–149. [DOI] [PubMed] [Google Scholar]
- Wu, G.Y., K. Deisseroth, and R.W. Tsien. 2001. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat. Neurosci. 4:151–158. [DOI] [PubMed] [Google Scholar]
- Yang, H., and R.R. Mattingly. 2006. The Ras-GRF1 exchange factor coordinates activation of H-Ras and Rac1 to control neuronal morphology. Mol. Biol. Cell. 17:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]