Abstract
We report here a genome-wide analysis of alternative splicing in 2 million human expressed sequence tags (ESTs), to identify splice forms that are up-regulated in tumors relative to normal tissues. We found strong evidence (P < 0.01) of cancer-specific splice variants in 316 human genes. In total, 78% of the cancer-specific splice forms we detected are confirmed by human-curated mRNA sequences, indicating that our results are not due to random mis-splicing in tumors; 73% of the genes showed the same cancer-specific splicing changes in tissue-matched tumor versus normal datasets, indicating that the vast majority of these changes are associated with tumorigenesis, not tissue specificity. We have confirmed our EST results in an independent set of experimental data provided by human-curated mRNAs (P-value 10–5.7). Moreover, the majority of the genes we detected have functions associated with cancer (P-value 0.0007), suggesting that their altered splicing may play a functional role in cancer. Analysis of the types of cancer-specific splicing shifts suggests that many of these shifts act by disrupting a tumor suppressor function. Sur prisingly, our data show that for a large number (190 in this study) of cancer-associated genes cloned originally from tumors, there exists a previously uncharacterized splice form of the gene that appears to be predominant in normal tissue.
INTRODUCTION
One of the most prevalent and successful models in cancer research is that cancer involves changes in gene expression (1). However, given recent indications that alternative splicing is a widespread mechanism of functional regulation in the human genome (2–7), it is interesting to ask whether cancer might also involve changes in mRNA splicing. Cancer-associated splice variants have been reported for genes such as EGFR (8), CD44 (9) and NER (10). In the last few months, many more alternative splice variants were discovered in cancer, for example, tyrosine hydroxylase (11), lactate dehydrogenase (12), cadherin-11 (13), fibronectin (14) and Brn-3a (15). A computational study has indicated that expressed sequence tags (ESTs) derived from tumors often show apparently different splicing patterns than the canonical RefSeq mRNA sequence (detected in 455 genes at P < 0.05) (16), suggesting that alterations of splicing might be widespread in human cancers.
In this study, we assess several key challenges for genomics-based analyses of alternative splicing’s role in cancer. Such studies inherit both advantages and disadvantages from the high-throughput datasets (such as ESTs) upon which they are based. Genomics data are large and comprehensive (e.g. up to 4 million human ESTs, representing 6900 cDNA libraries, of which 5700 can be unambiguously classified as tumor or normal tissue in origin), providing statistical power for revealing surprising patterns—in this study, a large shift in the relative frequency of alternative splice forms between normal versus tumor samples. However, unlike a traditional biology experiment, the EST data were not designed to test a specific hypothesis with detailed controls, and considerable care is required for their interpretation, for example, to evaluate possible sampling bias. For EST-based detection of cancer-specific splice forms, we see several major questions.
First, do these changes in splicing actually contribute to cancer? Simplistically, are they ‘causes’ or merely ‘symptoms’ of tumorigenesis? Some cancer-specific variants appear to make important functional contributions to the transformed state, such as inhibiting apoptosis [CD79 (17)] or blocking tumor suppressor activity [BIN1 (18)], whereas others apparently do not (19). Since the EST data provide only a statistical association for the occurrence of specific splice forms in tumors, this is a difficult question to answer, but can be addressed in part by statistical tests versus other data about gene function, as we will illustrate.
A second, related question concerns the character of the splicing change itself. Does it appear to be a regulated switch in splicing (like tissue-specific splicing, mediated by regulatory factors that turn on certain splice forms in one tissue), or simply a loss of splicing specificity in tumors? For example, pheochromocytoma tumors showed a large increase in incomplete splice products of the Ret tyrosine kinase, including failure to splice out intron 2 (19). Such a loss of splicing specificity may be indicated in several ways: (i) production of many minor variant forms in an apparently non-specific manner; (ii) incomplete splicing, e.g. intron retention; (iii) nonsense products that result in non-functional protein and/or nonsense-mediated decay (NMD) (20). Whereas in normal cells such failures of the splice site selection machinery occur rarely [e.g. for the HPRT gene, spliceosomal errors were observed in 2–3% of transcripts (21)], their frequency can be greatly elevated in tumors [e.g. 10–20% of transcripts for RET (19)]. We will refer to such failures throughout this paper as ‘loss of splicing specificity’, and assess its prevalence in cancer splice forms.
Third, apparent cancer-specificity can be difficult to distinguish from tissue-specific splicing reflecting the particular cell type which gave rise to the tumor. For example, if the normal tissue samples for a particular gene represented a variety of tissues, but the tumor samples were mostly derived from one tissue with a tissue specific variant, that variant would show a misleading statistical association with cancer. What fraction of apparent cancer-specific splice forms actually reflects tissue-specific artifacts? We seek to answer this question through analysis of a panel of tissue-matched tumor versus normal EST library sets.
MATERIALS AND METHODS
Library classification
Tissue source information for 6900 human EST libraries (UniGene release January 2002) was exhaustively examined to produce a consistent cancer/normal classification. We used histological information provided by ORESTES (Ludwig Institute for Cancer Research, http://www.ludwig.org.br/), NCI-CGAP (Cancer Genome Anatomy Project, http://cgap.nci.nih.gov/) and NIH-MGC (Mammalian Gene Collection, http://mgc.nci.nih.gov/). We also performed text searches to classify other EST libraries. All tumor types were combined into a single pool, as were all normal tissue libraries. 1160 EST libraries were excluded because they could not be clearly assigned to either cancer or normal (for example, if its histology was unclear, or pre-cancerous).
LOD score calculation
Suppose a gene G has two mutually exclusive (i.e. alternative) splices S and S′. By ‘mutually exclusive’ we mean two splices that share one splice site, but differ at the other splice site, and which thus cannot both be present in a single transcript (7). For our hidden variables, let θT and θN represent the hidden frequency of S in the tumor sample pool and normal tissue pool, respectively. Similarly, let θ′T = 1 – θT and θ′N = 1 – θN represent the hidden frequencies of S′ in tumor and normal pools. For our observations, let nT and n′T be the total number of ESTs in the tumor pool observed to have splice S or S′, respectively. Similarly, let nN and n′N be the total number of ESTs in the normal pool observed to have splice S or S′, respectively.
We calculated the confidence that θ′T is greater than θ′N as a log odd ratio:
We calculated this LOD score by two methods: Fisher’s Exact Test (22), and direct numerical integration as previously described (23,24), using
LOD scores from both methods generally agreed to within ±0.5.
Validation of cancer-specific alternative splicing
Whereas ESTs were produced by a small number of high-throughput sequencing projects from standard cDNA libraries whose tissue origins are catalogued in public databases, mRNAs are an independent dataset consisting of sequences deposited by thousands of individual investigators world-wide. We had to investigate their tissue origins manually, by reading their GenBank entries or published papers to determine whether they were from tumors or normal tissue samples. We performed validation of cancer-specificity on the 128 alternative splicing events in 89 human gene clusters (LOD3 dataset) using mRNA data in NCBI UniGene. We also visualized and validated all aspects of the genomic mapping of our clusters, exons and introns, splices sites, alternative splicing and the impacts on protein structure and function versus the literature, by examining all the features in the genomic-EST-mRNA multiple sequence alignments.
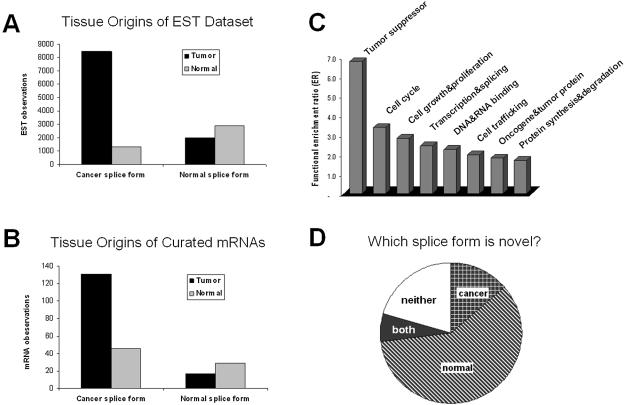
These mRNA validation results (Fig. 2B) appear to be representative of a general pattern that is valid for most or all of the individual genes in the LOD3 dataset, and are unlikely to be an anomalous result due to a small number of unusual genes. First of all, we were able to identify mRNAs specifically from tumor or normal tissue for almost all of the LOD3 genes (82/89 = 92%). On average, there were about three such mRNAs per gene. To exclude the possibility that the results in Figure 2B might be dominated by an anomalous subset of genes that had many mRNAs, we excluded all genes with more than five mRNAs. This yielded results virtually identical to Figure 2B (n′T 113, n′N 35, nT 13, nN 24).
Figure 2.
Independent validation, functional enrichment and novelty of cancer-specific alternative splice forms. (A) The tissue distribution of EST observations for both cancer and normal splice forms. (B) The tissue distribution of mRNA observations for both cancer and normal splice forms. (C) Functional categories that were enriched among the cancer-specific alternatively spliced genes, expressed as the ratio of genes observed in the LOD3 set relative to a random sample of 100 alternative spliced genes. (D) Classification of which splice forms within 316 cancer-specific alternatively spliced genes were novel. Splice forms were classified as known if they matched an existing mRNA sequence in GenBank, otherwise as novel.
Although most individual genes have too few counts (3 mRNAs or less) to yield a statistically significant confirmation of a cancer-specific shift in splicing, the counts for most individual genes match the pattern in Figure 2B, i.e. n′T > n′N and nN > nT. Of the genes with non-zero counts of the S′ form, we found n′T > n′N for 89% of the individual genes (50/56). Of the genes with non-zero counts of the S form, we found nN > nT for 69% of the individual genes (18/26). For genes with sufficient mRNAs to calculate θ′T and θ′N directly from the mRNA counts (i.e. non-zero counts for both tumor and normal samples, and non-zero counts for both S′ and S splice forms), we found θ′T > θ′N (indicating a cancer-specific shift in splicing) for 70% of the individual genes (7/10). These data indicate that the results in Figure 2B are not an artifact of an anomalous subset of genes, but are instead representative of the majority of genes in the LOD3 dataset.
We categorized the types of observed shifts as follows: (i) Switch S → S′ from normal tissue to tumor, defined as θ′T ≥ 2/3 and θ′N ≤ 1/3; (ii) Loss of S in tumors (θ′T ≥ 2/3 and θ′N > 1/3); (iii) Gain of S′ in tumors (θ′T < 2/3 and θ′N ≤ 1/3).
RESULTS
Genome-wide detection of cancer-specific alternative splicing
We based our analysis on our previously validated identification of alternative splicing from human ESTs aligned to genomic sequence (7,24). To identify changes in splicing that are characteristic of the transformed state, we pooled 4067 EST libraries from tumor samples, and compared against a separate pool of 1737 EST libraries from normal tissue. Nearly all normal samples were from primary tissue; a tiny fraction (0.4%) was from cultured cells explicitly classified as ‘normal’. By pooling many different tumors, we sought specific characteristics that are shared by many cancer types, and which are present far more frequently in tumors than normal samples.
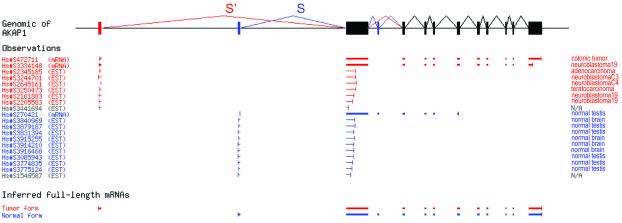
For example, in gene AKAP1 (Fig. 1), we detected a splice form S′ that was detected in six ESTs and two mRNAs representing a series of different tumors, and an alternative splice form S detected in nine ESTs and one mRNA, all obtained from normal tissue samples. These EST data produced a strong log-odds confidence score (LOD 4.29, equivalent to a P-value of 0.000051) that the ratio of splice form S′ over splice form S was much higher in the tumor pool than in the normal tissue pool.
Figure 1.
Detection of cancer-specific alternative splicing in AKAP1. Raw data showing the schematic alignment of all 17 ESTs aligning to the S/S′ region of the gene structure (including two ESTs of unclassified origin, excluded from our calculation) and 3 mRNAs. In normal tissues, splice form S was strongly preferred (9/9 ESTs and 1/1 mRNA), but in tumors, splice form S′ replaced it entirely (6/6 ESTs and 2/2 mRNA). The odds ratio for the null hypothesis that no shift from S to S′ occurs between normal and tumor samples is less than 10–4. The UniGene sequence identifier is shown for each sequence.
By this LOD score measure, we detected cancer-specific alternative splicing in 316 genes above LOD score 2 (i.e. P < 0.01), with 89 genes above LOD 3 (P < 0.001). To assess the significance of these results, we calculated the number of genes expected by random chance. For the LOD2 calculation, our database contained 4900 genes with sufficient EST data (a minimum of seven total ESTs for S and S′). Thus 4900 × 0.01 = 49 genes with a LOD score of 2 are expected by random chance (versus 316 observed). For the LOD3 calculation, only 4100 genes had sufficient data (a minimum of 11 ESTs for S and S′), indicating 4100 × 0.001 = 4 genes with a LOD score of 3 expected by random chance (versus 89 observed). Thus it is likely that the majority of our results are significant.
Representative examples are shown in Table 1, and the full list is in Appendix 1 in Supplementary Material. These genes showed a strong shift in the ratio of S′ form over S form in the tumor pool versus the normal pool. For example, among the 89 genes above LOD score 3, there was a 9.3-fold shift from splice form S to splice form S′ between normal tissue samples versus tumors (Fig. 2A).
Table 1. Examples of cancer-specific alternatively spliced genes.
| Cluster_id | Gene description | LOD | Tumor samples | Normal samples | |||
|---|---|---|---|---|---|---|---|
| S′ | S | S′ | S | ||||
| i | Hs.2384 | Tumor protein D52 | 6.18 | 18 | 0 | 10 | 20 |
| Hs.77572 | BCL2/adenovirus E1B 19 kDa interacting protein 1 (BNIP1) | 4.68 | 25 | 0 | 2 | 5 | |
| Hs.78921 | PRKA anchor protein 1 (AKAP1) | 4.29 | 6 | 0 | 0 | 9 | |
| Hs.5443 | BCL2-associated athanogene 5 (BAG5) | 3.22 | 7 | 0 | 2 | 8 | |
| Hs.99881 | Lactate dehydrogenase C (LDHC) | 2.76 | 2 | 0 | 1 | 20 | |
| ii | Hs.154718 | Tumor protein D52-like 2 (D54) | 6.66 | 76 | 0 | 9 | 7 |
| Hs.82202 | Ribosomal protein L17 (RPL17) | 4.27 | 49 | 3 | 4 | 6 | |
| Hs.110849 | Estrogen-related receptor alpha (ERRA) | 3.89 | 20 | 0 | 6 | 7 | |
| Hs.103081 | Ribosomal protein S6 kinase, 70 kDa, polypeptide 2 (S6K2) | 3.41 | 40 | 1 | 10 | 6 | |
| iii | Hs.69547 | Myelin basic protein (MBP) | 3.52 | 44 | 43 | 9 | 36 |
| Hs.334368 | Hypothetical protein MGC11257 | 3.04 | 30 | 30 | 2 | 17 | |
S′, putative cancer-specific splice form. S, putative normal splice form. i, ii, iii: three major categories of cancer-specific splicing (see text).
Validation of cancer-specific alternative splicing by independent mRNA data
Are these EST results reliable? To assess this, we analyzed a different set of experimental data: human-curated mRNA sequences from GenBank, which were not included in the original EST calculation because no tumor/normal classifications of their sources were available in a form usable by our calculation. To perform validation, we had to read the GenBank record (and in some cases the original publications) for each mRNA, to determine whether its tissue source was tumor, normal, or other/unknown. For the 89 genes above LOD 3, we were able to classify 223 mRNAs as derived from tumor or normal tissue. We examined these mRNAs for the cancer-specific splice forms observed in the ESTs, and found an almost identical result in this independent experimental data (Fig. 2B). The putative normal splice form S was observed predominantly in mRNAs from normal tissue, whereas the S′ splice form was observed predominantly (131/177 mRNAs) in mRNAs derived from tumors. These mRNA data strongly confirm the EST results (P-value 10–5.7). It should be emphasized that the mRNA experimental data are largely independent of the EST experimental data: they came from different tissue samples, different experimental methods, and different investigators. Of the mRNAs in Figure 2B, 67% were from cDNA libraries not represented by any ESTs counted in Figure 2A; even for the smaller subset of mRNAs that had some potential overlap with the EST data, only 5% of the ESTs in Figure 2A were derived from one of the same cDNA sample preparations. Thus, not only are the mRNAs independent in sequencing, but also largely independent sample preparations. This indicates a reproducible overall tendency of these genes to be spliced differently in tumor versus normal tissue.
In some cases, the tumor-specific splicing identified by our calculation has been independently reported. Table 2 lists a number of genes in which our tumor-specific splicing has been independently demonstrated by other experimental methods. For example, our EST analysis identified cancer-specific splicing in BIN1, an important tumor suppressor gene (found to be deleted in 50% of carcinoma cell lines). Our EST analysis indicated that exon 12A is included preferentially in tumors as opposed to normal tissues. This has been demonstrated independently (both by northern blots and by western blot with an antibody specific to the protein sequence coded by exon 12A), using tissue-matched normal versus tumor samples (normal fetal melanocytes versus melanomas) (18). Moreover, this splice change has critical functional consequences for tumorigenesis. Whereas normal BIN1 has strong tumor suppressor activity (blocks c-Myc or adenovirus E1A+Ras mediated focus-formation, and suppresses proliferation of HepG2 cells), the variant including exon 12A lacks tumor suppressor activity in these assays. This was confirmed in human melanoma cells using a cre-inducible adenovirus vector to introduce BIN1 or the exon 12A variant (18).
Table 2. Independent validation of cancer-specific splicing.
| Gene | LOD | Alt splice? | Splice form cancer specific? | Functional impact? |
|---|---|---|---|---|
| ACT1 (NFκB activator 1) | 2.5 | Confirmed (34) | Confirmed to be dominant in 10 cancer cell lines (‘epithelial-like’ adherent cancer cells, non-adherent cancer cells, melanomas), but did not examine same normal samples (34) | Cancer form shown to activate NFκB, but no specific change demonstrated |
| BIN1 (tumor suppressor) | 1.6 | Confirmed (18) | Confirmed in eight melanoma cell lines, compared with fetal melanocytes (normal) (18) | Eliminates tumor suppressor activity |
| CC3 (metastasis suppressor) | 0.9 | Confirmed (35) | Cancer-specific splice form (TC3) confirmed to be present in all tumor cell lines expressing CC3, versus only very low levels in normal tissues (35) | Switches CC3 activity from inducing apoptosis to inhibiting apoptosis |
| FGFR1 (fibroblast growth factor receptor 1) | 0.9 | Confirmed (36) | Confirmed in a panel of matched glioblastomas versus normal brain white matter. The level of the cancer-specific form correlated strongly with tumor malignancy (36,37) | Changes fibroblast growth factor receptor from low-affinity form (normal brain) to high affinity form (tumor) |
| LDHC (lactate dehydrogenase) | 2.8 | Confirmed (12) | Confirmed in a large panel of lung cancer and melanoma samples, versus many normal tissues (12) | Removes NAD binding |
| NABC1 (breast carcinoma amplified 1) | 3.2 | Confirmed (38) | Observed at higher levels in breast tumors (and not in breast normal) (38) | Unknown |
| NER (nuclear hormone receptor) | 0.7 | Confirmed (10) | Confirmed in 116 of 128 primary cancers and in 31 of 39 cancer cell lines, and absent in the corresponding normal tissues (10) | Removes DNA binding domain |
Distinguishing cancer-specificity from tissue-specificity
Our EST analysis screened for alternative splice forms that are characteristic of tumors in general (rather than a specific tumor type), by pooling all tumor samples as a single group. However, in genes with ESTs from only one tumor type or normal tissue type, our data might reflect a change in splicing that is characteristic of one specific tumor type, or simply a tissue-specific variant not related to cancer. By ‘tissue’, we simply mean the documented origin of the sample (e.g. ‘brain’).
To check whether our results might be an artifact of tissue-specific differences between the tumor pool versus normal pool, we repeated our analysis on tissue-matched tumor versus normal samples. Much EST sequencing (e.g. CGAP, ORESTES) has focused on matched pairs of tumor and normal samples from the same tissue (e.g. brain tumor versus brain normal), permitting us to compare the splicing patterns in tumor versus normal samples from the same tissue. These new data confirm our original conclusions. Of the 64 genes in our LOD3 set that were expressed in the tissues included in our new analysis, 47 (73%) showed the same cancer-specific splicing changes between the tissue-matched tumor versus normal datasets (at a confidence level of greater than 95%; see Appendix 3 in Supplementary Material). These new data show that most of our original dataset stands up to this more stringent tissue-matched test. It should be emphasized that since this test involves switching from the complete EST data to just ESTs from a single tissue, it always reduces the LOD score, and a 100% validation rate is by no means expected. Even though such splice forms appear to be tumor-associated, they may simply be symptoms of tumorigenesis (such as loss of differentiation), rather than significant functional contributors to cancer.
As a further test of our original results, we evaluated how broad the supporting evidence for each cancer-specific form was. In total, 97% of our results were supported by both multiple tumor libraries and multiple normal tissue libraries (e.g. AKAP1, Fig. 1). This again suggests that the fraction of our results which might be artifacts of sampling only one tissue is small.
Functional impact of cancer-specific alternative splicing
The observation of cancer-specific splicing is interesting, but raises an important question: are these splice forms cancer-specific because they actually make a functional contribution to cancer? Alternatively, these splice forms might simply be a symptom of tumorigenesis. It is known in some cases that cancer causes loss of splicing specificity, generating minor variant splice forms (19). Since these forms occur much more in tumors than in normal samples, they would appear to be tumor-specific. Such forms could be interesting as diagnostic markers of cancer, but in some cases may not contribute to tumorigenesis. To check whether the cancer-specific splice forms we detected might be due to loss of splicing specificity, we compared all our splice forms against human-curated mRNA sequences from GenBank; 78% of our cancer-specific splice variants were confirmed by known mRNA sequences. These human-curated mRNA sequences are unlikely to be non-specific splice products, because: (i) such spliceosomal error products ordinarily constitute a very small fraction of the mRNAs in a cell [from 2% in normal cells (21), up to 20% in some tumors (19)]; (ii) human-curated mRNA sequence ordinarily is checked against northern blots (or RT–PCR) to confirm a match against the major transcript bands from one or more tissues; (iii) these mRNA sequences show no signs of spliceosomal error (e.g. intron retention); (iv) these mRNAs do not show signs of nonsense products (e.g. only 3% of mRNAs in our data show nonsense-mediated decay). Thus, even if we assume that all cancer splice forms not confirmed by known mRNA are due to loss of splicing specificity, these would constitute a quarter or less of our data.
As a second test of this question, we checked whether these genes show a functional association with cancer. Whereas splice changes that make no functional contribution to cancer could occur in any gene with equal probability, splice changes that are associated with cancer because they contribute functionally to tumorigenesis are much more likely to be found in genes known to be involved in cancer. (It should be emphasized that the converse is not true: for genes with no known association with cancer, changes in their splicing might still contribute functionally to tumorigenesis.) We checked whether our LOD3 genes were associated with cancer by first checking where they were originally cloned from. For these genes, two times as many mRNA sequences were cloned from tumors (148) than from normal tissues (75), indicating that most of these genes were originally cloned and sequenced in studies of cancer. In contrast, for a random sample control set of 100 alternatively spliced genes, fewer mRNAs were cloned from tumors (108) than from normal (118). This is a statistically significant result (P-value 10–4.4).
To reveal specific functional biases, we categorized gene functions in both the LOD3 set and control set (Appendices 1 and 2 in Supplementary Material), and calculated the ratio of counts in each category (LOD3 set/control). These data show a strong, non-random association of these genes with functional categories known to be involved in cancer (Fig. 2C). For example, tumor suppressor genes were seven times more frequent in the cancer-specific splicing set than in the control set. The most enriched categories were tumor suppressors, cell cycle genes, cell growth and proliferation genes, and transcription factors; 52% of the cancer-specific splicing genes had functions associated with cancer (tumor suppressor, oncogene and tumor protein, cell cycle, cell growth and proliferation, apoptosis, transcription and splicing, protein synthesis and degradation). In contrast, only 20% of the control set fell into these categories. This is a statistically significant association (P-value 0.0007).
Discovery of novel splice forms
What fraction of the splice forms we identified is novel? In over three quarters of the genes above LOD 2 (over 190 in this study), at least one of the two splice forms (cancer-specific or normal-specific) appears to be novel (i.e. no mRNA with this splicing has been deposited in public databases; see Fig. 2D). Thus, while the set of genes we identified will be highly familiar to cancer researchers, many of the specific splice forms are novel.
A very interesting pattern emerges when we ask which splice form is novel. In most of these genes, one splice form is known and one novel. Surprisingly, it is the normal tissue splice form that tends to be novel, while it is the abnormal (cancer-specific) splice form that usually matches the known mRNA sequences for these genes (Fig. 2D). For a large number of cancer-associated genes (over 190 in this study), the sequence deposited in GenBank appears in light of our data to be an abnormal (cancer-specific) form, and there is no record of the form we observe in normal tissues.
Distinct types of cancer-specific shifts in splicing
We observed three distinct types of shifts in splicing between normal tissues and tumors. In some cases there was an almost complete switch from one form (S) in normal tissues to a different form (S′) in tumors (Group i in Table 1; see Materials and Methods). In other genes, we observed both forms present equally in normal tissue, and a loss of the S form in tumors (Group ii in Table 1). Finally, in some genes we observed a significant increase in the S′ form in tumors, but with the S form abundant in both tumors and normal tissue (Group iii in Table 1).
These categories make interesting suggestions about possible mechanisms. For example, if a cancer-specific form S′ is caused by loss of splicing specificity in tumors, then it should not be observed at high frequency in normal tissue. This would rule out the Loss of S category (ii) and favor the Gain of S′ category (iii). If we hypothesize a cancer-specific form S′ has a dominant, oncogenic effect, again we expect it to fall into the Gain of S′ category (iii). In contrast, if we hypothesize that it causes loss of function of a tumor suppressor, it should have a recessive effect that will only be revealed in the absence of the normal (S) form. This hypothesis would favor the Loss of S category (ii).
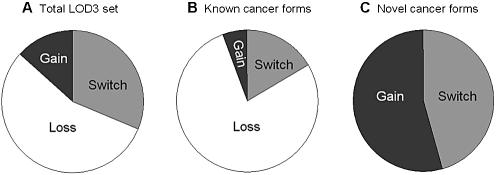
In the LOD3 dataset, 56% of the genes displayed a Loss of S shift in tumors (Group ii), while 31% showed a strong Switch (Group i), and only 13% Gain of S′ (Group iii; Fig. 3A). We obtained the same result for the larger LOD2 dataset. These data indicate first that relatively little of our data is due to loss of splicing specificity in tumors. Second, the preponderance of the Loss of S group suggests that disruption of a tumor suppressor function may be a dominant mechanism for cancer-specific splicing in these genes.
Figure 3.
Types of cancer-specific splicing shifts. (A) The relative proportions of LOD3 cancer-specific splicing that constituted a complete shift from one splice form in normal to another form in tumors (Switch); increase in one splice form in tumors (Gain); or loss of one form in tumors (Loss). (B) The same proportions, measured in genes for which the cancer-specific splice form was previously known; (C) in genes for which the cancer- specific form is novel.
Further analysis supports this interpretation. For LOD3 genes where the cancer-specific S′ form matches the known mRNA sequence, and is thus probably not non-specific splicing, the Loss of S category grew to 78%, while the Gain of S′ and Switch categories fell to 5% and 17%, respectively (Fig. 3B). In contrast, for genes where the S′ form is novel, the composition changed dramatically. The Loss of S category effectively vanished, while the Gain of S′ and Switch categories each grew to around 50% (Fig. 3C). This is a statistically significant result (P-value 0.0019). These data suggest that in this group (where S′ is novel, constituting about one-fifth of our cancer-specific splicing), tumor-induced loss of splicing specificity may indeed be a major factor.
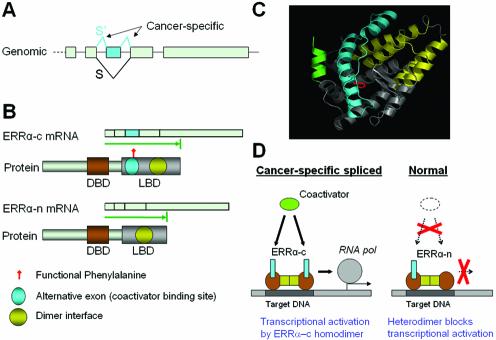
Figure 4 shows a novel cancer-specific splicing example representative of the Loss of S category, Estrogen-related receptor α (ERRα). ERRα is an orphan nuclear receptor transcription factor implicated in breast cancer, and has constitutive activity independent of hormone. This protein up-regulates expression of aromatase, which converts androgen to estrogen and stimulates breast cancer cell growth (25), and of pS2 (also known as TFF1), an estrogen-inducible breast cancer marker (26). We identified two splice forms of ERRα differing by a single exon skip (Fig. 4A). A novel splice form characteristic of normal tissue is supported by seven independent EST sequences. In normal tissue, an equal mix of both forms was observed (Table 1). In contrast, in 20 tumor ESTs, only the full-length form was observed, yielding a cancer-specificity LOD score of 3.9. The exon-skip (normal) form causes a 57 aa in-frame deletion removing ERRα’s transcriptional coactivator binding site (Fig. 4B and C), but leaving its dimerization region intact. Moreover, Phe-329 is a critical amino acid for the constitutive activity of ERRα; the mutant protein F329A forms a heterodimer with wild-type ERRα protein, inhibiting its activity, and causes a dominant negative phenotype (27). The exon skip in normal tissue also removes this functionally critical amino acid (red in Fig. 4C). These data suggest that in normal tissue the exon-skip form can dimerize with the full-length form and suppress its activity (a dominant negative effect). In tumors, in contrast, the absence of the exon-skip form may leave the full-length form constitutively active, in the absence of ligand, as has been observed experimentally (28–30).
Figure 4.
Cancer-specific alternative splicing of ERRα. (A) Genomic structure of the last 5 exons of ERRα gene. Exons are shown as gray boxes and the cancer-specific splices (S′) and exon are colored cyan. (B) The two alternative mRNA and protein isoforms of ERRα. The protein-coding region for each isoform is represented as a green arrow. The functional protein domains are marked on protein isoforms (DBD, DNA binding domain; LBD, ligand binding domain). (C) 3-D structure model of ERRα ligand-binding domain (LBD). The cancer-specific exon is colored cyan. The dimer interface is colored yellow. The functional phenylalanine is colored red. The coactivator peptide is colored green. (D) Proposed model of functional regulation of alternative splicing on ERRα. The cancer-specific spliced form ERRα-c contains complete coactivator-binding site and is a fully functional transcription factor in cancer tissue. The truncated form ERRα-n misses part of the coactivator binding site and is likely to heterodimerize with the full-length form, suppressing its activity (dominant negative effect) in normal tissue.
DISCUSSION
There are many lines of evidence for our conclusions of tumor-specific splicing: the original pooled-tumors versus pooled-normal EST results; the tissue-matched tumor versus normal EST results; the independent mRNA dataset; the evidence of functional association with cancer; independent experimental studies of a number of these genes (Table 2). There are certainly possible artifacts in each of these datasets, and statistical analysis of pre-existing data cannot prove that our results are free of artifacts. In the absence of a double-blind, randomized experiment design, one can speculate that all the results are due to hidden sample biases in each dataset. This is possible, but unlikely, as it would require a coincidence of hidden sample biases in each of these many datasets. The concurrence of these many lines of evidence indicates a broadly reproducible tendency for these genes to be spliced differently in tumors than in normal tissue. Since our results have already been confirmed by independent experimental methods for a substantial number of genes (Table 2), it is likely that a significant fraction of our results are valid.
In our view, the most difficult questions about any observed tumor specificity are the problem of tissue specificity, and the possibility of tumor-induced loss of splicing specificity. However, our data indicate fairly strong bounds on these problems. Three-quarters of our original cancer-specific splices were validated by a new analysis of tissue-matched tumor versus normal samples. Thus while tissue specific artifacts remain a concern, they appear to be only a small fraction of our data. Similarly, 78% of our cancer-specific splice forms are validated by human curated mRNA sequences, indicating that they are not due to loss of splicing specificity in tumors.
What kinds of roles might tumor-specific splicing play in cancer? Based on our research design, it seems unlikely that these cancer-specific splices are initial triggers of tumorigenesis. We have sought to detect splice forms that are characteristic of all cancers (rather than a specific type), by pooling all tumor samples as a single group. Given that there are diverse molecular events that can initially trigger cancer, it seems unlikely that the initial causes would be shared among all these tumor samples. On the other hand, many changes in cell behavior are required for successful maintenance of the transformed state, and these changes would be expected to be shared by all growing tumors. This suggests that many of the cancer-specific splice forms we detected may play a role in maintenance of the transformed state. This interpretation is supported by the types of specific gene functions in which we observed cancer-specific splicing: pro-apoptotic factors (e.g. BIN1; BAG5; BNIP1; CC3); tumor suppressors; oncogenes and tumor proteins; cell cycle and growth.
Some of the cancer-specific splice forms we detected may be medically important. For example, breast cancer often progresses from being dependent on estrogen, to hormone independence, rendering antiestrogen drug treatments useless (26). We propose that cancer-specific splicing of ERRα eliminates the dominant negative form ERRα-n in some tumors, rendering ERRα constitutively active, stimulating growth even in the absence of hormone and producing the hormone independent form of breast cancer. This hypothesis is supported by the observation that diethylstilbestrol (DES), a synthetic estrogen drug used for the treatment of advanced breast and prostrate cancer (31,32), appears to act through ERRα, completely abolishing the ERRs’ transcriptional activity on the pS2 promoter. DES has been shown to inhibit the growth even of estrogen-independent cancer cells entirely lacking the estrogen receptor (26,33).
Our data include discovery of many novel splice forms of cancer-associated genes, and suggest a significant new direction for cancer research. It should be emphasized that to obtain a statistically significant LOD score in our calculation, the evidence for both splice forms must be strong, at least three or four independent observations of each splice form, and usually much more (Table 1). Surprisingly, while most of the cancer-specific splice forms we detected match known mRNA sequences, most of the splice forms characteristic of normal tissue were novel. Given that so many of the genes identified by our analysis were first cloned from tumor samples, it is perhaps not surprising that their cancer-specific forms are already known. However, it appears that for many of these genes more sequencing of mRNAs from normal tissue is needed. It is possible that once a complete mRNA sequence for a given gene was deposited and confirmed, the philosophy of ‘one gene, one product’ may have ended the search for additional forms. Even in current, sophisticated sequencing projects (e.g. the Mammalian Gene Collection), it is common practice to halt further sequencing of a gene once a single full-length mRNA is deposited. Discovery of the normal forms of these genes as provided by our data, and comparison with the cancer-specific forms, should shed a useful new light on the action of these known cancer genes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank M. Sawaya for 3-D structure modeling, and D. Eisenberg, R. Landgraf, S. Nelson, K. Mitsouras, M. Roy, R. Wall and Y. Xing for their helpful discussions and comments on this work. C.J.L. was supported by National Institutes of Health grant MH65166 and Department of Energy grant DEFC03-87ER60615.
REFERENCES
- 1.Ross D.T., Scherf,U., Eisen,M.B., Perou,C.M., Rees,C., Spellman,P., Iyer,V., Jeffrey,S.S., Van de Rijn,M., Waltham,M. et al. (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet., 24, 227–235. [DOI] [PubMed] [Google Scholar]
- 2.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 4.Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 5.IHGS Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wikstrand C.J., Hale,L.P., Batra,S.K., Hill,M.L., Humphrey,P.A., Kurpad,S.N., McLendon,R.E., Moscatello,D., Pegram,C.N., Reist,C.J. et al. (1995) Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res., 55, 3140–3148. [PubMed] [Google Scholar]
- 9.Naor D., Nedvetzki,S., Golan,I., Melnik,L. and Faitelson,Y. (2002) CD44 in cancer. Crit. Rev. Clin. Lab. Sci., 39, 527–579. [DOI] [PubMed] [Google Scholar]
- 10.Saito H., Nakatsuru,S., Inazawa,J., Nishihira,T., Park,J.G. and Nakamura,Y. (1997) Frequent association of alternative splicing of NER, a nuclear hormone receptor gene in cancer tissues. Oncogene, 14, 617–621. [DOI] [PubMed] [Google Scholar]
- 11.Parareda A., Villaescusa,J.C., Sanchez de Toledo,J. and Gallego,S. (2003) New splicing variants for human Tyrosine Hydroxylase gene with possible implications for the detection of minimal residual disease in patients with neuroblastoma. Neurosci. Lett., 336, 29–32. [DOI] [PubMed] [Google Scholar]
- 12.Koslowski M., Tureci,O., Bell,C., Krause,P., Lehr,H.A., Brunner,J., Seitz,G., Nestle,F.O., Huber,C. and Sahin,U. (2002) Multiple splice variants of lactate dehydrogenase C selectively expressed in human cancer. Cancer Res., 62, 6750–6755. [PubMed] [Google Scholar]
- 13.Feltes C.M., Kudo,A., Blaschuk,O. and Byers,S.W. (2002) An alternatively spliced cadherin-11 enhances human breast cancer cell invasion. Cancer Res., 62, 6688–6697. [PubMed] [Google Scholar]
- 14.Castellani P., Borsi,L., Carnemolla,B., Biro,A., Dorcaratto,A., Viale,G.L., Neri,D. and Zardi,L. (2002) Differentiation between high- and low-grade astrocytoma using a human recombinant antibody to the extra domain-B of fibronectin. Am. J. Pathol., 161, 1695–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiarugi V., Del Rosso,M. and Magnelli,L. (2002) Brn-3a, a neuronal transcription factor of the POU gene family: indications for its involvement in cancer and angiogenesis. Mol. Biotechnol., 22, 123–127. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z., Lo,H.S., Yang,H., Gere,S., Hu,Y., Buetow,K.H. and Lee,M.P. (2003) Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res., 63, 655–657. [PubMed] [Google Scholar]
- 17.Cragg M.S., Chan,H.T., Fox,M.D., Tutt,A., Smith,A., Oscier,D.G., Hamblin,T.J. and Glennie,M.J. (2002) The alternative transcript of CD79b is overexpressed in B-CLL and inhibits signaling for apoptosis. Blood, 100, 3068–3076. [DOI] [PubMed] [Google Scholar]
- 18.Ge K., DuHadaway,J., Du,W., Herlyn,M., Rodeck,U. and Prendergast,G.C. (1999) Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl Acad. Sci. USA, 96, 9689–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Hir H., Charlet-Berguerand,N., de Franciscis,V. and Thermes,C. (2002) 5′-End RET splicing: absence of variants in normal tissues and intron retention in pheochromocytomas. Oncology, 63, 84–91. [DOI] [PubMed] [Google Scholar]
- 20.Lewis B.P., Green,R.E. and Brenner,S.E. (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA, 100, 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skandalis A., Ninniss,P.J., McCormac,D. and Newton,L. (2002) Spontaneous frequency of exon skipping in the human HPRT gene. Mutat. Res., 501, 37–44. [DOI] [PubMed] [Google Scholar]
- 22.Bartoszynski R. and Niewiadomska-Bugaj,M. (1996) Probability and Statistical Inference. John Wiley & Sons, New York, NY, USA. [Google Scholar]
- 23.Kan Z., States,D. and Gish,W. (2002) Selecting for functional alternative splices in ESTs. Genome Res., 12, 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q., Modrek,B. and Lee,C. (2002) Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res., 30, 3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C., Zhou,D. and Chen,S. (1998) Modulation of aromatase expression in the breast tissue by ERR alpha-1 orphan receptor. Cancer Res., 58, 5695–5700. [PubMed] [Google Scholar]
- 26.Lu D., Kiriyama,Y., Lee,K.Y. and Giguere,V. (2001) Transcriptional regulation of the estrogen-inducible pS2 breast cancer marker gene by the ERR family of orphan nuclear receptors. Cancer Res., 61, 6755–6761. [PubMed] [Google Scholar]
- 27.Chen S., Zhou,D., Yang,C. and Sherman,M. (2001) Molecular basis for the constitutive activity of estrogen-related receptor alpha-1. J. Biol. Chem., 276, 28465–28470. [DOI] [PubMed] [Google Scholar]
- 28.Xie W., Hong,H., Yang,N.N., Lin,R.J., Simon,C.M., Stallcup,M.R. and Evans,R.M. (1999) Constitutive activation of transcription and binding of coactivator by estrogen-related receptors 1 and 2. Mol. Endocrinol., 13, 2151–2162. [DOI] [PubMed] [Google Scholar]
- 29.Hong H., Yang,L. and Stallcup,M.R. (1999) Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J. Biol. Chem., 274, 22618–22626. [DOI] [PubMed] [Google Scholar]
- 30.Greschik H., Wurtz,J.M., Sanglier,S., Bourguet,W., van Dorsselaer,A., Moras,D. and Renaud,J.P. (2002) Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol. Cell, 9, 303–313. [DOI] [PubMed] [Google Scholar]
- 31.Peethambaram P.P., Ingle,J.N., Suman,V.J., Hartmann,L.C. and Loprinzi,C.L. (1999) Randomized trial of diethylstilbestrol vs. tamoxifen in postmenopausal women with metastatic breast cancer. An updated analysis. Breast Cancer Res. Treat., 54, 117–122. [DOI] [PubMed] [Google Scholar]
- 32.Malkowicz S.B. (2001) The role of diethylstilbestrol in the treatment of prostate cancer. Urology, 58, 108–113. [DOI] [PubMed] [Google Scholar]
- 33.Reddel R.R. and Sutherland,R.L. (1987) Effects of pharmacological concentrations of estrogens on proliferation and cell cycle kinetics of human breast cancer cell lines in vitro. Cancer Res., 47, 5323–5329. [PubMed] [Google Scholar]
- 34.Xia Y.F., Li,Y.D., Li,X. and Geng,J.G. (2002) Identification of alternatively spliced Act1 and implications for its roles in oncogenesis. Biochem. Biophys. Res. Commun., 296, 406–412. [DOI] [PubMed] [Google Scholar]
- 35.Whitman S., Wang,X., Shalaby,R. and Shtivelman,E. (2000) Alternatively spliced products CC3 and TC3 have opposing effects on apoptosis. Mol. Cell. Biol., 20, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi F., Saya,H., Bruner,J.M. and Morrison,R.S. (1994) Differential expression of two fibroblast growth factor-receptor genes is associated with malignant progression in human astrocytomas. Proc. Natl Acad. Sci. USA, 91, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin W., Huang,E.S., Bi,W. and Cote,G.J. (1999) Redundant intronic repressors function to inhibit fibroblast growth factor receptor-1 alpha-exon recognition in glioblastoma cells. J. Biol. Chem., 274, 28035–28041. [DOI] [PubMed] [Google Scholar]
- 38.Correa R.G., de Carvalho,A.F., Pinheiro,N.A., Simpson,A.J. and de Souza,S.J. (2000) NABC1 (BCAS1): alternative splicing and downregulation in colorectal tumors. Genomics, 65, 299–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.