Abstract
We have used cryo-electron microscopy of kinesin-decorated microtubules to resolve the structure of the motor protein kinesin's crucial nucleotide response elements, switch I and the switch II helix, in kinesin's poorly understood nucleotide-free state. Both of the switch elements undergo conformational change relative to the microtubule-free state. The changes in switch I suggest a role for it in “ejecting” adenosine diphosphate when kinesin initially binds to the microtubule. The switch II helix has an N-terminal extension, apparently stabilized by conserved microtubule contacts, implying a microtubule activation mechanism that could convey the state of the bound nucleotide to kinesin's putative force-delivering element (the “neck linker”). In deriving this structure, we have adapted an image-processing technique, single-particle reconstruction, for analyzing decorated microtubules. The resulting reconstruction visualizes the asymmetric seam present in native, 13-protofilament microtubules, and this method will provide an avenue to higher-resolution characterization of a variety of microtubule- binding proteins, as well as the microtubule itself.
Introduction
Kinesin (Brady, 1985; Vale et al., 1985) is a force-generating molecule that is probably ubiquitous and essential in organisms with microtubules. In neurons, it powers vesicular trafficking directed toward the synapse; in all cells, it is involved in the restructuring that occurs during cell division (Kreis and Vale, 1999). This exquisite molecular machine can convert up to 35% of the chemical energy released by ATP hydrolysis into mechanical energy (Nishiyama et al., 2002), as the dimeric form “walks” step-by-step along microtubule protofilaments (Block, 1998). Numerous measurements have characterized kinesin's biophysical properties (Howard, 2001; Peterman et al., 2004).
Atomic resolution structures of monomeric kinesin obtained by x-ray crystallography revealed a “switch I/switch II” nucleotide-sensing architecture, similar to that found in G proteins and myosins (Kull et al., 1996; Sablin et al., 1996). Moreover, a “relay helix” hypothesis emerged from the crystal structures and other data (Vale and Milligan, 2000). Based on the observation of two distinct conformations of the “switch II helix,” crystal structures of kinesin were designated as either “ATP-like” or “ADP-like.” In the hypothesis (presented in an adapted form below [see Fig. 5, c and d]), the switch II helix of ADP-bound and nucleotide-free kinesin (the first two nucleotide states, respectively, of microtubule-bound kinesin) would be positioned away from the nucleotide catalytic site. This conformation of the switch II helix, it was proposed, would resemble that found in the class of kinesin crystal structures regarded as ADP-like. In ATP-bound kinesin, according to the hypothesis, the switch II helix moves toward the nucleotide active site, mimicking the switch II helix conformation found in a second class of kinesin crystal structures, regarded as ATP-like. This switch II helix movement was hypothesized to control kinesin's presumed force-delivering element, the neck linker, by obstructing the neck linker's docking onto the core of kinesin in the ADP-like position and permitting docking in the ATP-like position.
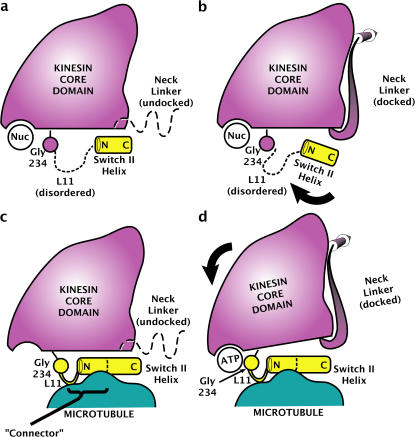
Figure 5.
Model for microtubule-induced activation of kinesin's ATP sensing mechanism by partially ordering the L11 segment. The represented view orientation is the same as in Fig. 2 c and Fig. 3 (a and b). Shown are coexisting, nucleotide-insensitive conformations of the switch II helix in the absence of the microtubule, modeled after the ADP-like (a) and ATP-like (b) crystal structures of K349. The presence of various nucleotides in the active site does not trigger any movement of Gly 234, whose position is in any case uncorrelated with that of the switch II helix because L11 is disordered. (c and d) Adapted relay helix model. In both nucleotide-free (c) and ATP-bound (d) states of kinesin, microtubule contacts order the C-terminal portion of L11 into an extension of the switch II helix plus an additional structured loop. ATP-induced movement of Gly 234 relative to the kinesin core, perhaps activated by the closing of switch I (Minehardt et al., 2001), is relayed through the structured L11 assembly (i.e., the connecter) to the switch II helix, which in turn controls the structure of the neck linker. As previously proposed (Kikkawa et al., 2001), we depict the switch II helix fixed on the surface of the microtubule, so that its movement relative to the kinesin core domain causes the core to move relative to the microtubule surface.
The first direct tests of the relay helix hypothesis have been incomplete and contradictory. A pair of newly published cryo-EM structures at a resolution of ∼1 nm of the KIF1A kinesin (Kikkawa and Hirokawa, 2006) are consistent with the relay helix theory for two of the three principal nucleotide states, finding the switch II helix ADP-like in ADP-bound, microtubule-bound kinesin and ATP-like in microtubule-bound kinesin with the nonhydrolyzable ATP analogue 5′-adenylyl-imidodiphosphate (AMPPNP). However, nucleotide-free kinesin was not included in the KIF1A study. Furthermore, a second very recent cryo-EM study of a different, unconventional kinesin (Kar3) is seemingly at odds with the relay helix theory (Hirose et al., 2006). In Kar3, the switch II helix was visualized apparently in the ADP-like position for both the ADP–kinesin–microtubule and AMPPNP–kinesin–microtubule states. Moreover, in the nucleotide-free state of Kar3, the switch II helix was only partially visible and was interpreted to have restructured as a loop. Thus, the generality of the relay helix theory is in doubt, and the role of the nucleotide-free state of kinesin in particular is highly unclear.
Another puzzle related to the relay helix theory concerns the communication between the nucleotide pocket and the switch II helix. Instead of showing clearly defined, nucleotide-dependent states for switch II, the crystal structures showed a range of positions for the switch II helix—apparently unconnected to the identity of the active site nucleotide. For example, it was possible to crystallize KIF1A with an ATP-like switch II helix conformation but with ADP in the active site (Kikkawa et al., 2001), in contrast to ATP-like switch II helix conformations found in crystal forms reported with either AMPPCP (5′-adenylyl-methylenediphosphate; Kikkawa et al., 2001) or AMPPNP (Nitta et al., 2004). On the other hand, the more recent structures of KIF2C kinesin display an ADP-like switch II helix conformation in the presence of either ADP or AMPPNP (Ogawa et al., 2004). Also, electron paramagnetic resonance and crystallography studies of the human kinesin heavy chain (KHC) confirmed that in solution kinesin freely exchanges between ATP- and ADP-like conformations in the presence of either ADP or AMPPNP (Sindelar et al., 2002). Thus, although microtubule-bound kinesin is hypothesized to have the conformation of its switch II helix controlled by the active site nucleotide, crystallography experiments to date have only identified “uncontrolled” switch II helix movements.
We have previously speculated how microtubule binding may activate kinesin's switch II nucleotide sensing mechanism by structuring kinesin's loop “L11,” N-terminally adjacent to the switch II helix (Sindelar et al., 2002). However, other subsequent reports have not addressed this aspect of kinesin's mechanism. Here, we use a new method of cryo-EM data analysis that, when combined with high-quality image data, can attain better than nanometer-resolution reconstructions of the kinesin–microtubule complex. Our resulting structure of nucleotide-free kinesin complexed to the microtubule exhibits at least three remarkable features. By directly visualizing the switch II helix, we find that the helix is in an ADP-like position, confirming one aspect of the relay helix theory for our KHC kinesin construct—in striking contrast to the nucleotide-free Kar3 structure with an apparently “melted” switch II helix. Furthermore, we show that the switch II helix is N-terminally extended by rearrangement of the loop L11 (which is unstructured in crystal structures of our construct). This structured extension, apparently stabilized by microtubule contacts, is a prime candidate for explaining the microtubule-induced activation of kinesin's switch II nucleotide sensing mechanism. Finally, we infer a possible role for switch I in disrupting kinesin's interactions with ADP upon binding to the microtubule.
Results and discussion
Identifying and reconstructing the microtubule seam
To determine the structure of the asymmetric 13-protofilament microtubule, we applied single-particle reconstruction techniques (see Materials and methods) to our set of nucleotide-free kinesin-decorated microtubule images. The essential step of the single-particle analysis was to determine the orientation of a given microtubule's symmetry-disrupting “seam.” Once the seam orientations were identified in our imaged, kinesin-decorated microtubules, we produced a 15 Å–resolution reconstruction of the entire, asymmetric microtubule volume (see Materials and methods). The reconstruction, shown in Fig. 1, clearly reveals the microtubule seam, validating our reconstruction techniques. Seams in microtubules have been imaged before (Kikkawa et al., 1994; Sosa et al., 1997), but 3D reconstructions have never been produced of the native 13-protofilament form, because of technical limitations in image processing. Here, we have applied a method capable of reconstructing the 13-protofilament microtubule form and have derived a cryo-EM reconstruction of this form, including the asymmetric seam.
Figure 1.
Isosurface view of the reconstruction of the microtubule seam. Note that this reconstruction is filtered to a resolution of 15 Å and, unlike the symmetry-averaged one presented in the following figures, is not amplitude sharpened. The vertical dashed line indicates the location of the seam; horizontal curved dashed lines indicate the helical path of the bound kinesin monomers, interrupted by the seam.
A high-resolution kinesin–microtubule complex structure
To maximize the resolution of the nucleotide-free kinesin– microtubule complex, we averaged together the 13 unique protofilaments reconstructed in Fig. 1 to produce a new motor– tubulin complex with the highest possible signal-to-noise ratio. After this step and subsequent image processing (see Materials and methods), we derived the kinesin–tubulin complex shown in Figs. 2–4 . We note that in the averaging step, we assumed that all 13 copies of the motor–tubulin complex were identical—an assumption that could in principle be violated because our structure contained an asymmetric seam. However, inspection of our nonaveraged reconstruction (Fig. 1) does not reveal obvious conformational differences between different sites (for example, differing motor occupancy or orientation), justifying our assumption at least to the first approximation.
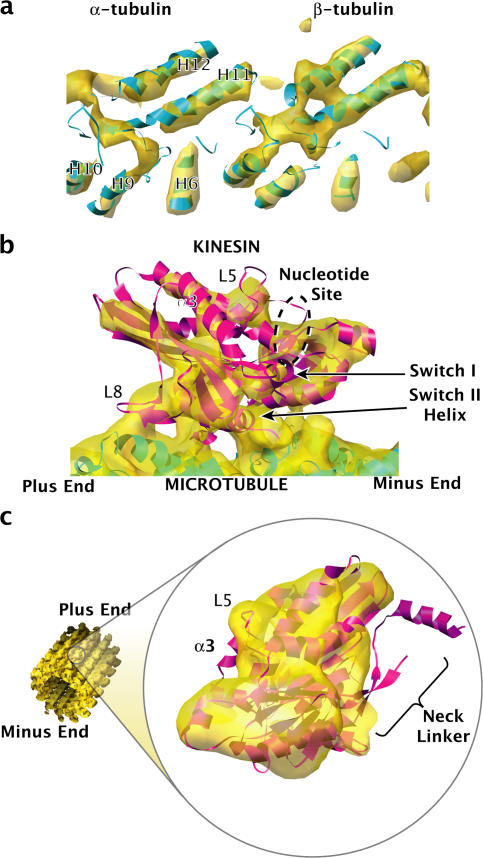
Figure 2.
Fits of the K349 kinesin crystal structures into our cryo-EM map, suggesting a disordered neck linker. (a) Representative density in the microtubule region of our cryo-EM map (gold) and the fitted crystal structure of tubulin (blue). See Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200612090/DC1). Molecules and surfaces were rendered with UCSF Chimera (Pettersen et al., 2004). (b) Fit of the ADP-like K349 crystal structure (Kull et al., 1996) into the map, indicating the location of various structural elements (Video 2). (c) Fit of the ATP-like K349 crystal structure (Sindelar et al., 2002) into the map, demonstrating that the neck linker is out of density. The C terminus of kinesin's helix α3 is also out of density, possibly correlated to a rearrangement of switch I.
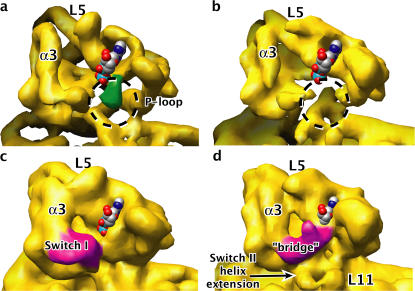
Figure 4.
Density map changes in the nucleotide pocket of the nucleotide-free kinesin–microtubule state, relative to the crystallized kinesin–ADP state. The space-filled ADP model indicates the position of the nucleotide pocket. Orientation of the view is approximately the same as in Fig. 2 b. (a and c) 9-Å–resolution synthetic maps, at two different contour levels, generated from the crystal structure of ADP-like K349 kinesin (with the ADP coordinates removed). (b and d) Our experimental map, at contour levels equivalent to panels a and c, respectively. In panel a, density for the P-loop in the crystal structure is highlighted in green; in panel b, the corresponding void in the map is circled. In panel c, density corresponding to switch I is highlighted in magenta; in panel d, density corresponding to the bridge between switch I and the nucleotide site is also magenta.
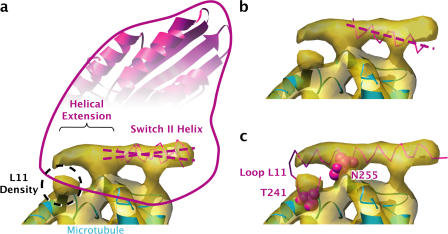
Figure 3.
Density for the switch II helix in our cryo-EM map resembles the switch II helix conformation seen in ADP-like crystal structures of kinesin and reveals an N-terminal extension. View orientation is identical to that of Fig. 2 c. (a) Density map of the switch II helix, with the fitted ADP-like K349 and KIF1A crystal structures superimposed. For clarity, the N-terminal segment of KIF1A's switch II helix, although ordered, was left out of the rendering. The KIF1A structures in panels a and b were aligned with our fitted K349 crystal structure by least-squares fitting using SwissPDBViewer. The axis of the switch II helix density is intermediate between those of the two crystal structure helices. (b) The axis of the switch II helix from the fitted ATP-like K349 and KIF1A crystal structures does not align with the axis of the helix density. (c) The crystal structure of Kar3 (mutation R598A; Yun et al., 2001) aligned with the K349 crystal structure coordinates from panel a suggests the architecture of the N-terminal switch II helix extension. The two space-filled residues, T241 and N255, are highly conserved throughout the kinesin family and apparently contact the microtubule surface.
At the resolution of the symmetry-averaged structure, ∼9-Å (see Materials and methods) helices in tubulin were distinct from one another, even the packed antiparallel helices H11 and H12 (Fig. 2 a). All helices in kinesin were resolved but were slightly less distinct, possibly because of imperfect occupancy (estimated to be ∼75%) on the microtubule lattice. However, the large β sheet at the core of kinesin appeared as a continuous twisted sheet of density, a loop (L8 in kinesin) appeared as a distinct arm connecting kinesin to the microtubule surface, and the switch II helix was clearly resolved at the microtubule binding interface.
To evaluate tubulin's secondary structure elements, the coordinates of bovine tubulin (Protein Data Bank [PDB] ID 1JFF; Lowe et al., 2001) were fitted into the cryo-EM map using cross-correlation implemented in Fourier space (see Materials and methods). The agreement between positions of helices in our map and the fitted tubulin crystal structure was excellent (Fig. 2 a and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200612090/DC1).
The switch II helix is ADP-like
Two crystal structures of monomeric human kinesin, those of the K349 construct (used in our experiments) with and without a docked neck linker (PDB IDs 1MKJ [Sindelar et al., 2002] and 1BG2 [Kull et al., 1996], respectively), were fit into our map using cross-correlation (see the previous section). The positioning of the molecules was unambiguous, with alignments between the two fitted core motor domains differing by <2°, when the principal axes of the moments of inertia were compared (see Materials and methods). The fit of the crystal structure into our molecular envelope is excellent in most regions of the protein chain (Fig. 2 b and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200612090/DC1). However, the docked neck linker present in 1MKJ was found to be out of density (Fig. 2 c). No compensating region of unoccupied density was found near the docked neck linker region. Thus, our density map strongly suggests that the neck linker is disordered in our experimental conditions. This result agrees with numerous other experiments using techniques including single molecule fluorescence (using bifunctional labels; Asenjo et al., 2006) and electron paramagnetic resonance (Rice et al., 1999).
In all crystal structures observed to date, a disordered neck linker in kinesin is accompanied by a switch II helix in the ADP-like position; furthermore, the switch II helix in the ADP–kinesin–microtubule complex (with a disordered neck linker) was likewise in the ADP-like position (Kikkawa and Hirokawa, 2006). It therefore seemed likely that the nucleotide-free kinesin–microtubule complex would have an ADP-like switch II helix conformation as well (Vale and Milligan, 2000). Our cryo-EM map allowed this prediction to be tested directly, as density corresponding to the switch II helix was clearly apparent.
We compared the position and orientation of the density corresponding to the switch II helix in our map with the switch II helix positions predicted by crystal structure fits, using ∼20 kinesin crystal structures from PDB. As shown in Fig. 3 (a and b), our helical density falls within the switch II helix orientations classified as ADP-like from K349 and KIF1A crystal structures, as well as others (not depicted). We quantified this observation by fitting a long switch II helix from Kar3 (Fig. 3 c) into our density using cross-correlation and comparing the principal axis of the moment of inertia of this fitted helix with the principal axes of the K349 and KIF1A helices. This analysis showed that the KIF1A ADP-like switch II helix was rotated ∼8° counterclockwise (in the plane of Fig. 3) compared with our helical density, whereas the K349 ADP-like helix was rotated ∼6° clockwise. Thus, ADP-like switch II helices can vary over a considerable range of orientations relative to the core motor domain, and our switch II helix density lies squarely in this range.
On the other hand, fitted ATP-like kinesin structures displayed switch II helices distinct from our helix density, as shown in Fig. 3 b. In particular, the C termini of the crystal structure helices (Fig. 3 b, right) are well out of density and far from the apparent helical axis of the density. A moment of inertia comparison showed the ATP-like switch II helices to be rotated ∼12–15° clockwise in the plane of Fig. 3, relative to the helix density. This angular difference, substantially greater than the estimated error of our orientation determination (∼5°), demonstrates that the switch II helix orientation in our structure is measurably distinct from that found in the ATP-like crystal structures. Collectively, these results constitute direct evidence to support the “relay helix” prediction of kinesin's switch II helix ADP-like conformation in the nucleotide-free, microtubule-bound state.
A microtubule-stabilized extension of the switch II helix
An N-terminal extension of the switch II helix, relative to the crystal structures of our K349 construct, is evident in our cryo-EM density map in Fig. 3. The visible density extends the helix by several turns. Furthermore, the extension in our map is accompanied by an extra “sphere” of density, closely associated with the microtubule surface, in the vicinity of the N terminus of the helix. Intriguingly, these features correspond to the architecture of switch II seen in several crystal structures (Kikkawa et al., 2001; Nitta et al., 2004; Shipley et al., 2004), including that of Kar3 (mutation R598A; Yun et al., 2001), as shown in Fig. 3 c. The structural element at the N terminus of the switch II helix, loop L11, is often disordered in crystal structures but visible in that of Kar3 and projects through the sphere of density in our alignment.
The structural alignment in Fig. 3 c serves as a useful indicator of residues likely to be involved in the kinesin–microtubule interaction for kinesin's nucleotide-free state. This alignment indicates that the two space-filled residues shown in Fig. 3 c (corresponding to Asn 255 on the K349 switch II helix and Thr 241 on K349 loop L11), highly conserved in the kinesin family (Sablin et al., 1996), account for the two microtubule contact points of the switch II helix extension–L11 assembly. Both of these residues are involved in the microtubule-induced restructuring of kinesin seen in our cryo-EM structure. In the fitted ADP-like K349 crystal structure in Fig. 3 a, not only does the sidechain of Asn 255 face away from the microtubule surface (not depicted) but the backbone departs from the α-helical structure of subsequent residues 256–271 of the switch II helix. In contrast, the equivalent residue Asn 650 contained within the α-helical extension of Kar3 (Fig. 3 c) has swiveled ∼180° to point its sidechain at microtubule helix H11′ and/or the subsequent loop on the microtubule surface. Residue Thr 241 is not present in the K349 crystal structure and is presumably disordered, whereas the equivalent residue Ser 636 in the Kar3 model faces helix H3′ on the microtubule surface. Thus, our modeling suggests that rearrangement of critical conserved microtubule-binding residues in kinesin supports the restructuring of switch II and L11 seen in our nucleotide-free kinesin–microtubule complex.
ADP release does not strongly reorient kinesin on the microtubule
We compared the microtubule-bound orientation of our fitted K349 core domain with that of ADP- and AMPPNP-bound KIF1A reported by Kikkawa and Hirokawa (2006) (PDB IDs 2HXF and 2HXH). A least-squares superposition and moment-of-inertia analysis showed only a 5° rotation of the core domain of our structure compared with ADP-bound KIF1A. On the other hand, AMPPNP-bound KIF1A was reoriented by 15° relative to our structure. Thus, our nucleotide-free K349 core domain orientation is much more similar to ADP-bound, microtubule-complexed KIF1A. Because kinesin's orientation on the microtubule is hypothesized to depend on whether the switch II helix is ATP- or ADP-like, the similarity between our structure and ADP- bound KIF1A is further confirmation that the switch II helix is ADP-like in both structures.
Restructuring of the nucleotide pocket
To compare the nucleotide pocket in our experimental map with that expected from crystal structures of K349–ADP, we generated a 9-Å–resolution synthetic map using the fitted PDB coordinates of ADP-like K349 (but with ADP omitted) and tubulin from Fig. 2 b. At this resolution, several structured loops are visible as lobes of density in both synthetic and experimental maps. Examples include kinesin's loops L5 and L8 (Figs. 2 and 4), as well as the loop between tubulin's β strands B9 and B10 (not depicted). This observation suggests that it is feasible to detect, in our maps, conformational change of kinesin's nucleotide binding pocket, which contains two functionally important loops: the ADP-coordinating “P-loop” and the switch I nucleotide response element.
When viewed at an isocontour level of ∼2 σ, as shown in Fig. 4 (a and b), the synthetic map displays a lobe of density marking the location of the P-loop. In the experimental density map, however, this lobe does not appear even at lower contour levels (Fig. 4, b and d). This difference, which was consistently observed in reconstructions using two independent half datasets (unpublished data) suggests a rearrangement of the P-loop in kinesin's nucleotide-free state relative to crystal structures.
Although no alternative density site for the P-loop is apparent at the 2 σ contour level in our map of nucleotide-free kinesin, at a lower threshold (1.2 σ) a “bridge” is revealed in our experimental map between the density for switch I and the P-loop site, as shown in Fig. 4 d. The bridge, which features a distinctive protuberance, was consistently observed in two independent half dataset reconstructions (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200612090/DC1). The appearance of this bridge is accompanied by a rotation and shortening of helix α3, N-terminal to the switch I loop, and a movement of the switch I density itself toward the nucleotide site as it forms the bridge (Fig. 4 d).
These observations suggest a model for ADP release by kinesin after microtubule binding (Hackney, 1988). In guanine nucleotide-binding proteins, whose mechanism of nucleotide release is somewhat better understood, a consistent feature of the nucleotide-free state is disruption of the P-loop—evidently contributing to nucleotide release (Vetter and Wittinghofer, 2001). This disruption appears to be caused by any of a variety of mechanisms, including direct interactions between the P-loop and residues of the guanine-nucleotide exchange factor (GEF), or an indirect pathway such as GEF-modulated conformational change of switch II that in turn disrupts the P-loop. As shown in Fig. 4 d, our data are consistent with a disruption of the P-loop but suggest that interactions by switch I may contribute to the disruption.
A probable trigger for the apparent restructuring of switch I in our structure is the microtubule-induced restructuring of L11 and the N-terminal extension of switch II as described (Fig. 3). As seen in Fig. 4 b, switch I in our structure approaches the extended switch II N terminus, possibly attracted by new contacts made available by the L11/switch II restructuring. Indeed, based on the Kar3 model of Fig. 3 c, absolutely conserved residue Glu 250 in the switch II helix extension would be positioned at the interface, where our density for switch I meets the helical extension (not depicted). Furthermore, in a KIF1A ADP-like crystal structure—with an extended switch II helix—the equivalent of K349's Glu 250 forms a salt bridge with the equivalent of K349's Arg 203 from switch I. Thus, given the proximity between the switch II helix and switch I in our structure, it is likely that this conserved interaction forms there as well.
This potential chain of communication, from switch II to switch I to the nucleotide pocket, is supported by experiments that showed that mutations in either switch I (equivalent to R203A in our K349 construct) or switch II (equivalent to N255K or E236A in K349) decoupled microtubule binding from kinesin's ADP affinity (Yun et al., 2001). Our structure suggests a more detailed explanation for the decoupling mechanism, particularly in the case of the Asn mutation. If the N255K mutation destabilizes the helical extension by changing the residue's microtubule interactions, then in our mechanism, the extension would no longer be present to attract switch I toward the nucleotide site while kinesin is bound to the microtubule. Thus, microtubule binding would no longer eject ADP and would be uncoupled from ADP affinity.
Mechanism of plus end–directed motors appears distinct from that of Kar3
Together with the ADP and AMPPNP kinesin–microtubule complexes of Kikkawa and Hirokawa (2006), our nucleotide-free structure completes a nanometer-resolution picture of the nucleotide binding and hydrolysis cycle for the plus end–directed kinesins. Notably, the position of the switch II helix was identified for all three of these nucleotide states and in each case was found to agree with the predictions of the relay helix theory.
Previously reported structures of Kar3 complexed to the microtubule (Hirose et al., 2006) do not appear to follow the relay helix paradigm, as mentioned in the Introduction. Although it is hard to reconcile these results with ours and those of Kikkawa and Hirokawa (2006), we note that Kar3 moves toward the minus end of the microtubule, whereas our KHC construct and KIFA are a plus end–directed motors. As such, Kar3 possesses a different force-delivering element, the helical “neck” (Sablin et al., 1998; as opposed to conventional kinesin's neck linker). Thus, it is at least conceivable that minus end–directed kinesins have evolved a switch II mechanism distinct from the “relay helix.”
Model for microtubule activation of the relay helix
The mechanism that communicates the state of the nucleotide to the switch II helix is a crucial, unknown component of the proposed relay helix mechanism for kinesin. This mechanism presumably involves the formation of a hydrogen bond between switch II's absolutely conserved Gly 234 and ATP's γ-phosphate—a critical feature conserved as far as is known across proteins with functional switch II domains (Sablin and Fletterick, 2001). One possibility for this mechanism is our “connecter” hypothesis, depicted in Fig. 5 (c and d).
We predict that this connecter, consisting of our observed N-terminal extension of the switch II helix as well as the C-terminal, microtubule-contacting segment of L11, forms upon microtubule binding and persists after ATP binds in kinesin's active site (Fig. 5 d). In this process, the microtubule contacts would act as a kind of glue, stabilizing the connecter so that ATP-induced movement of Gly234 and the adjacent, N-terminal segment of L11 would be transmitted through the relay helix to the neck linker. The apparent microtubule interactions of conserved residues Thr 241 and Asn 255 in our structure, sufficient to stabilize and restructure a portion of L11 relative to crystal structures, reinforce this idea. The fact that some crystal structures of kinesin show N-terminal extensions of the switch II helix relative to our construct show that such an extension is possible, if not necessarily stable, in the absence of microtubules. Furthermore, density consistent with the helical extension is evident in both ADP- and AMPPNP-bound forms of the KIF1A–microtubule complex (Kikkawa and Hirokawa, 2006).
Though an N-terminal extension of the switch II helix is not seen in the AMPPNP–KIF1A crystal structure (Nitta et al., 2004), it is unlikely in any case that the conformations of L11 and the adjoining Gly 234 are the same in the ATP–kinesin–microtubule complex as in the crystal structure. This is because the AMPPNP–KIF1A crystal structure lacks a hydrogen bond between the nucleotide's γ-phosphate and KIF1A's Gly 251 (the equivalent of Gly 234 in K349). The switch II helix extension we propose in the ATP state of the kinesin–microtubule complex could help lead to the formation of such a hydrogen bond. Further work to extend the resolution of the kinesin–microtubule complex will be required to fully elaborate the role of L11 and Gly 234 in kinesin's ATP-sensing mechanism.
Summary
Our structure has allowed us to propose further details of how microtubule binding affects kinesin's interactions with bound nucleotide—in particular, how it may release bound nucleotide upon microtubule binding and how microtubule binding may permit the switch II helix to respond to the identity of kinesin's active-site nucleotide. Furthermore, our structure determination methods open up a route to high-resolution characterization of lateral interactions of the microtubule protofilaments—even those that occur at the microtubule seam. Future work will extend the resolution of both kinesin and the microtubule, allowing us to test the various aspects of the structural mechanisms of these proteins.
Materials and methods
Sample preparation
Human monomeric kinesin K349 cys-lite, derived from the conventional human KHC, was expressed from a plasmid (a gift from N. Nabor, University of California, San Francisco, San Francisco, CA) and prepared as described previously (Rice et al., 1999). We chose the mutant form 220C, which has motility and hydrolysis properties close to wild-type kinesin (Rice et al., 1999), for its high purification yield (∼100 mg per 2-liter culture). After elution from the final column (mono Q), ∼1-ml aliquots of ∼2 mg/ml were frozen in elution buffer + 20% sucrose. For use in EM experiments, protein was thawed on ice and dialyzed overnight against binding buffer (25 mM Pipes, pH 6.8, 25 mM NaCl, 1 mM EGTA, and 2 mM MgCl2). The resulting protein solution was concentrated to 15–20 mg/ml using centricon concentrators (Millipore). After this treatment, our kinesin preparation remained stable with substantial microtubule binding activity after several months at +4°C, making it an especially useful construct for extended experimental study.
Glycerol-free tubulin was purchased from Cytoskeleton, Inc., and polymerized according to a modified protocol to minimize aggregated, nonpolymerized tubulin. 25 μl of frozen tubulin aliquots was thawed on ice and ultracentrifuged at maximum speed for 10 min (100,000 rpm; TLA 120.2 rotor [Beckman Coulter]). The supernatant was added to a chilled glass vial, 1 mM GTP was added, and the vial was placed in a 37° incubator for 15 min. At this point, 1.25 μl taxol (2 mM in DMSO), diluted in 11.25 μl polymerization buffer (80 mM Pipes, pH 6.8, 1 mM EGTA, and 2 mM MgCl2), was slowly and carefully stirred into the polymerization vial. After 15–30 more minutes, 12.5 μl of the polymerized microtubules were mixed with ∼25 μl of above-prepared kinesin solution and 5 μl apyrase (10 mg/ml in deionized water). After 5 min, this mixture was layered onto a room-temperature glycerol cushion (50% glycerol + binding buffer + 200 μM taxol) and ultracentrifuged at 40,000 rpm for 20 min. The resulting kinesin–microtubule pellet was briefly washed 2× with binding buffer + 200 μM taxol and resuspended in 15 μl of the same buffer.
Maintaining the kinesin decoration on the microtubules during grid preparation for cryo-EM proved to be extremely difficult. Any of the following three factors apparently caused the kinesin to dissociate before the grids could be frozen: (1) glow-discharging the grids, (2) the presence of undiluted salts and buffer molecules from the binding buffer, and (3) blotting for more than a fraction of a second. To overcome these difficulties, we arrived at the following protocol: 0.3 μl of the kinesin–microtubule mixture was added to 3 μl of deionized water and mixed by pipetting. The resulting mixture was applied to a homemade holey carbon grid, and excess fluid was wicked away by touching the edge of the grid to a piece of filter paper repeatedly, until the meniscus was barely visible. The grid was then blotted and quickly (<0.5 s later) plunged into liquid ethane. Because our grids were not subjected to glow discharge, the ice was of poor quality and each grid contained relatively few areas suitable for imaging.
EM
Approximately 350 images were collected on film using a microscope (JEM-4000; JEOL) operating at 400 kV and 60 kx magnification, with defocus values ranging from 0.8 to 2 μm. Developed films were scanned using a robotic system incorporating a scanner (CoolPix; Nikon) operating at 6.3 μm/pixel (Typke et al., 2005). The final digitized images had a sampling of ∼1 Å per pixel.
Single-particle image analysis
13-protofilament microtubules were selected manually using the boxer program of the EMAN package (Ludtke et al., 1999), whose interactive functions facilitated the division of each microtubule into short (750- × 750-pixel) overlapping segments. Subsequent single-particle image processing was performed using customized scripts written for the SPIDER package (Frank et al., 1996), roughly following the methodology described by Li et al. (2002).
To generate an initial model for reference-based alignment, the atomic microtubule model of Li et al. (2002) was “decorated” using the crystal structure of human monomeric kinesin in orientations reflecting the AMPPNP-bound kinesin–microtubule complex of Kikkawa et al. (2001). Reference projections of this volume (filtered to ∼1-nm resolution) were then calculated at 0.3° intervals around the microtubule axis, including up to ±25° out-of-plane tilt. Although we did not expect our final map to exactly resemble this initial atomic model, the differences between our final map and the initial reference model served as a control to ensure that our reconstruction methods were not simply reproducing the model with which we started.
Finding the seam
Reference-based analysis of kinesin-decorated 13-protofilament microtubules (Figs. S1 and S2, available at http://www.jcb.org/cgi/content/full/jcb.200612090/DC1) was complicated by the presence of a seam that disrupts the quasi-helical symmetry (Mandelkow et al., 1986). The practical implication of the seam was that a complete 360° axial rotation of the decorated reference microtubule produced 13 fairly close matches against an experimentally imaged microtubule segment, of which only one gave the correct seam alignment. However, small differences in the magnitude of the image-reference cross-correlation indicated the true position of the seam (Fig. S2). Furthermore, seam determinations along consecutive microtubule segments typically agreed, amplifying our certainty that we had correctly identified the seam position. The low signal-to-noise ratio in our images caused some fraction (frequently 25% or more) of boxed segments to yield incorrect seam identifications that were one or more protofilaments away from the correct seam, identified by the majority of segments. However, this type of error decreased when increasingly accurate reference models were used in subsequent rounds of refinement.
Occasional microtubules produced extremely poor seam alignments, which appeared to jump randomly or cyclically between seam orientations with each segment. We identified these microtubules as 12- or 14-protofilament ones that had eluded the initial visual screening process. Thus, our reference alignment procedure also served as a backup screen for 13-protofilament microtubules.
3D volume reconstruction
After reference-based alignments were derived for the microtubule segments (Figs. S1 and S2), a 3D volume was obtained by weighted back projection. To increase the quality of our final map, the back-projection process was modified to directly integrate contrast transfer function (CTF) correction via a customized C program. The goal of the modification was to implement CTF correction with CTF-squared weighting of the Fourier components of the images, resembling the helical reconstruction methods presented, for example, by Yonekura and Toyoshima (2000). Within the program, the Fourier transform of each microtubule segment was first multiplied by its experimentally determined CTF (which included astigmatism). The CTF was derived by straightening entire microtubules using previously determined reference alignment parameters; the entire microtubule was then Fourier transformed to find the position and shape of the layer line absences indicating CTF minima. Subsequently in the program (but before back projection), the Fourier transform of each image was multiplied by a general, exponential weighting function derived from all other image segment transforms overlapping in Fourier space (Frank et al., 1996) and divided by the sum of the squares of the CTFs for the overlapping Fourier space measurements. The general exponential weighting function used was identical to that used in SPIDER's bp 3d command. The resulting Fourier space object was inverse Fourier transformed into an image and back projected through the target volume; this process was repeated for all microtubule segments to produce a final volume.
Another feature of our volume reconstruction methods took advantage of the pseudosymmetry of the 13-protofilament microtubules used here. As mentioned above, the seam disrupted true helical symmetry in the microtubules. However, it was possible to average the 13 pseudosymmetry mates in Fourier space, during back projection, to improve the resolution of the basic kinesin–tubulin subunit along one protofilament. The most straightforward way to describe our symmetry-averaging method is to describe the real-space analogue. Rotating a microtubule volume by n × 360°/13 and translating by n/13 of the pseudohelical repeat distance of 12 nm (where n is an integer) results in a new volume that superimposes on the original volume except for superimposing α tubulin on β tubulin in n of the protofilaments. Applying this process for n = 0–12 and superimposing the 13 resultant volumes will reveal one protofilament where kinesin and α and β tubulin superimpose for all 13 volumes. Thus, averaging the 13 volumes produced this way results in a strange volume that has one correctly averaged protofilament. To apply this idea in Fourier space before back projection, we created 13 copies of each segment image, corresponding to n = 0–12. We then added n × 360°/13 to the reference-determined axial orientation parameter of segment and translated the segment in the axial direction of the microtubule by n/13 of the repeat distance of 12 nm. Thus, the input to our back-projection algorithm consisted not only of the original segment images but also 12 additional copies related by symmetry.
The final volume incorporated data from 131 micrographs, representing 188 microtubules for a total of 2,460 unique image segments. Each image segment contained approximately nine tubulin dimer repeats multiplied by 13 protofilaments. However, each image segment also overlapped neighboring image segments by ∼50%. Thus, our reconstruction contained information from 2,460 × 9 × 13 × 0.5 tubulin dimer repeats = ∼150,000 tubulin dimers.
After the first round of refinement, the reconstructed microtubule volume was filtered to 1.6-nm resolution and used to generate a new set of reference images. The reference-alignment process was then repeated with the new reference set. The second reference alignment did not change most of the initially discovered orientation parameters by >1° when compared with the first reference alignment. This agreement indicated that our initial reference model, though it represented the wrong nucleotide state of kinesin, was sufficient to yield relatively high-quality orientation parameters in the initial reference alignment.
To compensate for amplitude attenuation at higher resolutions, the reconstruction was sharpened by applying a B-factor of −100, which was derived by comparing resolution-dependent amplitudes of our map to a synthetic map generated from PDB coordinates. The resolution of our reconstructions was estimated by two independent methods. First, the program RMEASURE (Sousa and Grigorieff, 2007) reported a resolution of 15.0 Å for the asymmetric reconstruction and 9.0 Å for the symmetry-averaged reconstruction (using the program's conservative output, with an estimated Fourier shell correlation [FSC] cutoff of 0.5). Second, the FSC (van Heel, 1987) was compared between two reconstructions: one using odd-numbered image segments and the other using even-numbered image segments. The resulting curve descended below FSC = 0.5 at 15.1-Å resolution for the asymmetric reconstruction and at 9.1-Å resolution for the symmetry-averaged reconstruction.
PDB model fitting and analysis
X-ray crystal structures of tubulin and kinesin were fit into our map using the COLORES program from the SITUS package (Wriggers and Birmanns, 2001). Principal axes of the moments of inertia of fitted crystal structures were computed using the program GEM (Browner et al., 1992).
Public data deposition
The coordinates of kinesin and tubulin, fitted into our density map, are available online from PDB (www.rcsb.org; ID 2P4N). The density map has been deposited in EMDB and is available online (http://www.ebi.ac.uk/msd-srv/emsearch/index.html; accession no. 1340).
Online supplemental material
Figs. S1 and S2 describe details of the single-particle analysis and 3D reconstruction procedures. Fig. S3 shows two independent half dataset reconstructions of kinesin's nucleotide pocket. Videos 1 and 2 are 3D reconstructions of tubulin and kinesin, respectively, from our density map, with superimposed ribbon diagrams of fitted PDB structures. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200612090/DC1.
Supplementary Material
Acknowledgments
We gratefully acknowledge R. Milligan (The Scripps Research Institute, La Jolla, CA) for providing micrographs for initial algorithm testing, H. Sui (Lawrence Berkeley National Laboratory, Berkeley, CA) for providing initial SPIDER scripts, M. Kikkawa (University of Texas Southwestern Medical Center, Dallas, TX) for communicating PDB entries 2HXF and 2HXH before their public release, N. Nabor for assistance with protein purification, and E. Sablin (University of California, San Francisco) and R. Milligan for thoughtful critique of the manuscript.
This work is supported by National Institutes of Health grants GM46033 and GM51487 and by the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
Abbreviations used in this paper: AMPPNP, 5′-adenylyl-imidodiphosphate; KHC, kinesin heavy chain.
References
- Asenjo, A.B., Y. Weinberg, and H. Sosa. 2006. Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat. Struct. Mol. Biol. 13:648–654. [DOI] [PubMed] [Google Scholar]
- Block, S.M. 1998. Leading the procession: new insights into kinesin motors. J. Cell Biol. 140:1281–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, S.T. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 317:73–75. [DOI] [PubMed] [Google Scholar]
- Browner, M.F., E.B. Fauman, and R.J. Fletterick. 1992. Tracking conformational states in allosteric transitions of phosphorylase. Biochemistry. 31:11297–11304. [DOI] [PubMed] [Google Scholar]
- Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199. [DOI] [PubMed] [Google Scholar]
- Hackney, D.D. 1988. Kinesin ATPase: rate-limiting ADP release. Proc. Natl. Acad. Sci. USA. 85:6314–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, K., E. Akimaru, T. Akiba, S.A. Endow, and L.A. Amos. 2006. Large conformational changes in a Kinesin motor catalyzed by interaction with microtubules. Mol. Cell. 23:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer, Sunderland, MA. 367 pp.
- Kikkawa, M., and N. Hirokawa. 2006. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 25:4187–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa, M., T. Ishikawa, T. Nakata, T. Wakabayashi, and N. Hirokawa. 1994. Direct visualization of the microtubule lattice seam both in vitro and in vivo. J. Cell Biol. 127:1965–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa, M., E.P. Sablin, Y. Okada, H. Yajima, R.J. Fletterick, and N. Hirokawa. 2001. Switch-based mechanism of kinesin motors. Nature. 411:439–445. [DOI] [PubMed] [Google Scholar]
- Kreis, T., and R. Vale. 1999. Guidebook to the Cytoskeletal and Motor Proteins. Oxford University Press, New York. 551 pp.
- Kull, F.J., E.P. Sablin, R. Lau, R.J. Fletterick, and R.D. Vale. 1996. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 380:550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., D.J. DeRosier, W.V. Nicholson, E. Nogales, and K.H. Downing. 2002. Microtubule structure at 8 Å resolution. Structure. 10:1317–1328. [DOI] [PubMed] [Google Scholar]
- Lowe, J., H. Li, K.H. Downing, and E. Nogales. 2001. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J. Mol. Biol. 313:1045–1057. [DOI] [PubMed] [Google Scholar]
- Ludtke, S.J., P.R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. [DOI] [PubMed] [Google Scholar]
- Mandelkow, E.M., R. Schultheiss, R. Rapp, M. Muller, and E. Mandelkow. 1986. On the surface lattice of microtubules: helix starts, protofilament number, seam, and handedness. J. Cell Biol. 102:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehardt, T.J., R. Cooke, E. Pate, and P.A. Kollman. 2001. Molecular dynamics study of the energetic, mechanistic, and structural implications of a closed phosphate tube in ncd. Biophys. J. 80:1151–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, M., H. Higuchi, and T. Yanagida. 2002. Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat. Cell Biol. 4:790–797. [DOI] [PubMed] [Google Scholar]
- Nitta, R., M. Kikkawa, Y. Okada, and N. Hirokawa. 2004. KIF1A alternately uses two loops to bind microtubules. Science. 305:678–683. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., R. Nitta, Y. Okada, and N. Hirokawa. 2004. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 116:591–602. [DOI] [PubMed] [Google Scholar]
- Peterman, E.J., H. Sosa, and W.E. Moerner. 2004. Single-molecule fluorescence spectroscopy and microscopy of biomolecular motors. Annu. Rev. Phys. Chem. 55:79–96. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., T.D. Goddard, C.C. Huang, G.S. Couch, D.M. Greenblatt, E.C. Meng, and T.E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Rice, S., A.W. Lin, D. Safer, C.L. Hart, N. Naber, B.O. Carragher, S.M. Cain, E. Pechatnikova, E.M. Wilson-Kubalek, M. Whittaker, et al. 1999. A structural change in the kinesin motor protein that drives motility. Nature. 402:778–784. [DOI] [PubMed] [Google Scholar]
- Sablin, E.P., and R.J. Fletterick. 2001. Nucleotide switches in molecular motors: structural analysis of kinesins and myosins. Curr. Opin. Struct. Biol. 11:716–724. [DOI] [PubMed] [Google Scholar]
- Sablin, E.P., F.J. Kull, R. Cooke, R.D. Vale, and R.J. Fletterick. 1996. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 380:555–559. [DOI] [PubMed] [Google Scholar]
- Sablin, E.P., R.B. Case, S.C. Dai, C.L. Hart, A. Ruby, R.D. Vale, and R.J. Fletterick. 1998. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 395:813–816. [DOI] [PubMed] [Google Scholar]
- Shipley, K., M. Hekmat-Nejad, J. Turner, C. Moores, R. Anderson, R. Milligan, R. Sakowicz, and R. Fletterick. 2004. Structure of a kinesin microtubule depolymerization machine. EMBO J. 23:1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar, C.V., M.J. Budny, S. Rice, N. Naber, R. Fletterick, and R. Cooke. 2002. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat. Struct. Biol. 9:844–848. [DOI] [PubMed] [Google Scholar]
- Sosa, H., A. Hoenger, and R.A. Milligan. 1997. Three different approaches for calculating the three-dimensional structure of microtubules decorated with kinesin motor domains. J. Struct. Biol. 118:149–158. [DOI] [PubMed] [Google Scholar]
- Sousa, D., and N. Grigorieff. 2007. Ab initio resolution measurement for single particle structures. J. Struct. Biol. 157:201–210. [DOI] [PubMed] [Google Scholar]
- Typke, D., R.A. Nordmeyer, A. Jones, J. Lee, A. Avila-Sakar, K.H. Downing, and R.M. Glaeser. 2005. High-throughput film-densitometry: an efficient approach to generate large data sets. J. Struct. Biol. 149:17–29. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and R.A. Milligan. 2000. The way things move: looking under the hood of molecular motor proteins. Science. 288:88–95. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., T.S. Reese, and M.P. Sheetz. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 42:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel, M. 1987. Similarity measures between images. Ultramicroscopy. 21:95–100. [Google Scholar]
- Vetter, I.R., and A. Wittinghofer. 2001. The guanine nucleotide-binding switch in three dimensions. Science. 294:1299–1304. [DOI] [PubMed] [Google Scholar]
- Wriggers, W., and S. Birmanns. 2001. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 133:193–202. [DOI] [PubMed] [Google Scholar]
- Yonekura, K., and C. Toyoshima. 2000. Structure determination of tubular crystals of membrane proteins. III. Solvent flattening. Ultramicroscopy. 84:29–45. [DOI] [PubMed] [Google Scholar]
- Yun, M., X. Zhang, C.G. Park, H.W. Park, and S.A. Endow. 2001. A structural pathway for activation of the kinesin motor ATPase. EMBO J. 20:2611–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.