Abstract
Recent evidence suggests that low oxygen tension (hypoxia) may control fetal development and differentiation. A crucial mediator of the adaptive response of cells to hypoxia is the transcription factor Hif-1α. In this study, we provide evidence that mesenchymal condensations that give origin to endochondral bones are hypoxic during fetal development, and we demonstrate that Hif-1α is expressed and transcriptionally active in limb bud mesenchyme and in mesenchymal condensations. To investigate the role of Hif-1α in mesenchymal condensations and in early chondrogenesis, we conditionally inactivated Hif-1α in limb bud mesenchyme using a Prx1 promoter-driven Cre transgenic mouse. Conditional knockout of Hif-1α in limb bud mesenchyme does not impair mesenchyme condensation, but alters the formation of the cartilaginous primordia. Late hypertrophic differentiation is also affected as a result of the delay in early chondrogenesis. In addition, mutant mice show a striking impairment of joint development. Our study demonstrates a crucial, and previously unrecognized, role of Hif-1α in early chondrogenesis and joint formation.
Introduction
Low oxygen tension is not only a pathophysiological component of many human disorders, including cancer, heart attack, and stroke, but it is also critically important in normal fetal development and cell differentiation (Chen et al., 1999; Giaccia et al., 2004). The transcription factor hypoxia-inducible factor-1 (Hif-1) has emerged as the central regulator of hypoxic gene expression (Bunn and Poyton, 1996; Kaelin, 2002; Giaccia et al., 2003; Semenza, 2003; Liu and Simon, 2004). Hif-1 is a heterodimer consisting of two subunits, Hif-1α and Hif-1β, both of which are basic helix-loop-helix/Per-Arnt-Sim domain proteins (Kaelin, 2002). Transcriptional activation by Hif-1 occurs upon its binding to the hypoxia response element (HRE) within its target genes. Whereas Hif-1β protein is constitutively expressed, Hif-1α protein is subject to rapid degradation by oxygen-dependent proteolysis (Ohh and Kaelin, 1999; Ivan et al., 2001; Jaakkola et al., 2001; Chan et al., 2002; Min et al., 2002). Under hypoxic conditions, Hif-1α protein is stabilized, initiating a multistep pathway of activation that includes nuclear translocation, dimerization with its partner Hif-1β, recruitment of transcriptional coactivators, and binding to the HREs of Hif-1 target genes (Kallio et al., 1998).
Endochondral bone formation is a two-stage mechanism; chondrocytes first shape a template, the “cartilage anlage,” in which osteoblasts then differentiate to form bone (Erlebacher et al., 1995; Karsenty, 2003; Kronenberg, 2003; Provot and Schipani, 2005).
The chondrocytic fetal growth plate is virtually avascular, but it requires blood vessel invasion to be substituted by bone (Vu et al., 1998; Zelzer et al., 2002). We previously showed that the fetal growth plate has an out–in gradient of oxygenation. More importantly, by using a Cre-lox strategy with a Col2a1 promoter-driven Cre (Col2a1-Cre) and a floxed Hif-1α allele, we provided evidence that Hif-1α is essential for cell growth and survival of growth plate chondrocytes in vivo, as chondrocytes lacking functional Hif-1α undergo massive cell death in the center of the growth plate (Schipani et al., 2001; Pfander et al., 2003). However, this genetic model did not allow us to address the role of Hif-1α in early chondrogenesis, as deletion of Hif-1α occurred in cells that were already committed to become chondrocytes.
An essential and specific function of differentiated chondrocytes is matrix synthesis. We have also recently reported that hypoxia and Hif-1α support cartilaginous matrix formation (Pfander et al., 2003, 2004). Thus, we speculated that hypoxia and Hif-1α may be permissive factors in chondrocyte differentiation.
The goal of this study was to investigate the roles of Hif-1α in the formation of mesenchyme condensations, in the commitment of mesenchymal cells toward chondrocytes, and in early stages of chondrocyte differentiation.
Results
Limb bud mesenchyme and mesenchymal condensations express Hif-1α
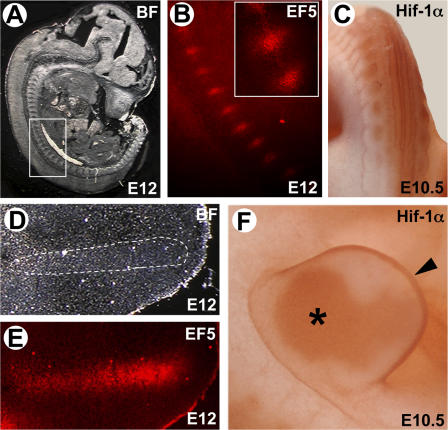
To evaluate the role of Hif-1α in limb bud mesenchyme, we first ascertained the presence of hypoxia in precartilaginous condensations by injecting the hypoxia marker EF5 into pregnant female mice at embryonic day (E) 12, a stage at which precartilaginous condensations are well formed and chondrocytes are just starting to differentiate (Lee et al., 1996). EF5 bound mesenchymal condensations that give origin to both the axial (Fig. 1, A and B) and the appendicular skeleton (Fig. 1, D and E), whereas, with the exception of the skin, no significant binding was detected in the surrounding soft tissues. These data demonstrate that mesenchymal condensations that give origin to the endochondral skeleton are hypoxic during development.
Figure 1.
Mesenchymal condensations are hypoxic, and Hif-1α is expressed in limb bud mesenchyme. The EF5 staining (B and E) and corresponding bright-field (BF) views (A and D) of developing axial skeleton (A and B) and forelimb (D and E) of E12 mouse embryo show the hypoxic tissues. The boxed area (developing axial skeleton) and the area outlined by a dashed line (presumptive digital ray) indicate the regions shown for the EF5 staining. A blow-up of two hypoxic mesenchymal condensations is shown in B. Whole mount immunohistochemistry for Hif-1α protein (C and F), on E10.5 mouse embryo shows that Hif-1α protein is detected in the somites, giving rise to the axial skeleton (C) in limb bud mesenchyme (F, asterisk) and in the apical ectoderm (F, arrowhead).
Consistent with the EF5 detection, whole mount in situ hybridization showed detectable Hif-1α mRNA in the limb bud mesenchyme and in axial condensations as early as E10.5 (unpublished data). This expression persisted in limb bud mesenchymal condensations and in axial condensations at E11.5 and E12.5 (unpublished data). A similar pattern of Hif-1α protein was detected by whole mount immunohistochemistry at E10.5 (Fig. 1, C and F) and by immunohistochemistry at E12 (Fig. S1, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200612023/DC1). Notably, Hif-1α protein was also highly expressed in the apical ectoderm of the limb bud at E13.5 (Fig. 1 F). No staining was seen in embryos treated with secondary antibody alone (unpublished data).
In summary, mesenchymal condensations in both limb bud and forming axial skeleton are highly hypoxic and express Hif-1α.
5XHRE-LacZ reporter mice express LacZ in limb bud mesenchyme and in mesenchymal condensations
We then evaluated the transcriptional activity of Hif-1α in limb bud mesenchyme by generating two types of hypoxia-inducible reporter mice. Five HREs were placed in front of either a retroviral E1b or murine hsp68 promoter fragment, driving the LacZ reporter gene to create the 5XHRE-E1b/LacZ and 5XHRE-hsp/LacZ transgenes, respectively (Fig. 2 A). The 5XHRE-E1b fragment has previously been used to successfully direct a hypoxia-specific induction of luciferase in cell culture (Shibata et al., 1998). The hsp68 promoter fragment is a well-characterized promoter capable of being activated in distinct patterns by defined heterologous enhancer elements in transgenic mice (Sumiyama and Ruddle, 2003). Neither the hsp68 nor E16 fragment alone is activated by hypoxia.
Figure 2.
HREs are activated in condensed mesenchyme. Five HREs were placed before a minimal promoter (mp) fragment driving the LacZ reporter gene to generate a hypoxia-inducible (5XHRE-mp/LacZ) transgenic reporter mouse (A). X-gal staining of E8.5 5XHRE-mp/LacZ transgenic embryos cultured ex vivo under normoxic (21% oxygen) or anoxic (0% oxygen) conditions for 24 h indicates a good response of the construct to hypoxia (B; h, heart; ys, yolk sac). Whole mount X-gal staining on 5XHRE-mp/LacZ transgenic embryos of the indicated stage shows the transcriptional activation of HREs in somites (C), developing axial skeleton (D), and limb bud mesenchyme (F), and shows a particularly strong activation in condensed limb mesenchyme (G). The pink circles in F define the margins of the limb bud. Sections of the developing axial skeleton (E) and condensed limb mesenchyme (H) show that the X-gal staining closely resembles that obtained with EF5 (Figure 1).
To confirm that the 5XHRE enhancer fragment used in our transgenic models was also hypoxia inducible in vivo, E8.5 5XHRE-E1b/LacZ embryos were cultured under normoxia or anoxic conditions for 24 h, and then analyzed for LacZ expression by X-gal staining (Fig. 2 B). In contrast to embryos cultured at 21% oxygen, a high level of LacZ expression was observed in embryos exposed to 0% oxygen for 24 h. This finding demonstrates that the 5XHRE fragment used in our experiments is induced by hypoxia to drive reporter expression in vivo. Notably, the embryos kept in normoxic conditions did not show any detectable signal. Although it is conceivable that E8.5 embryos are mildly hypoxic in utero, the short half-life of β-galactosidase (Jacobsen and Willumsen, 1995) combined with the absence of stimulation of the reporter in the presence of oxygen likely explains this result. In addition, it is important to note that both the 5XHRE-E1b/LacZ and 5XHRE-hsp/LacZ are reporter constructs for both Hif-1α and Hif-2α; therefore, stimulation of these constructs in hypoxic conditions might also result in part from Hif-2α activity.
For either transgenic line, numerous founders were generated. For the purpose of our study, we have analyzed one 5XHRE-hsp/LacZ founder line in detail. Consistent with the EF5 findings and the expression of Hif-1α mRNA and protein, whole mount β-galactosidase staining at E10.5 showed a high level of LacZ expression in the limb bud mesenchyme (Fig. 2 F). This expression was even more evident at E12.5, and at this age it overlapped with regions of mesenchyme condensations (Fig. 2, G and H). Similar to the expression of Hif-1α mRNA and protein, β-galactosidase staining was also detectable in axial condensations (Fig. 2, C–E).
Collectively, these findings support the model that Hif-1α is transcriptionally active in limb bud mesenchyme, in mesenchymal condensations of the limb bud, and in axial condensations.
Conditional knockout (CKO) of Hif-1α in limb bud mesenchyme alters the development of cartilaginous primordia
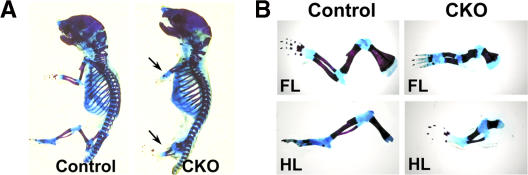
To dissect the role of Hif-1α in early chondrocyte differentiation, we conditionally inactivated Hif-1α in limb bud mesenchyme, using a Prx1 promoter-driven Cre transgenic mouse (Logan et al., 2002), and a mouse homozygote for a floxed Hif-1α allele (Hif-1αf/f; Schipani et al., 2001). The Prx1-Cre transgenic line expresses Cre recombinase prevalently in limb bud mesenchyme starting from E9.5 (Logan et al., 2002). Both Hif-1αf/f and Prx-1 Cre mice were indistinguishable from wild-type animals (unpublished data). Newborn Hif-1αf/f; Prx-1 Cre (CKO) mice, were viable, but had a characteristic shortening of both forelimbs and hindlimbs (Fig. 3 A, arrows; the length of newborn tibia is 3.4 ± 0.3 mm in control vs. 1.5 ± 0.2 mm in mutant; newborn ulna 3.9 ± 0.2 mm in control vs. 1.2 ± 0.5 mm in mutant). Real-time PCR analysis of genomic DNA extracted from newborn control and CKO paws, after removal of the skin, showed that efficiency of deletion of the Hif-1α gene was 75 ± 2.5% at this age. The result is particularly substantial, especially in light of the tissue heterogeneity of the specimens. In addition, at E14.5, accumulation of Hif-1α protein was severely decreased in CKO forelimb autopod when compared with control (Fig. 6 A). Given the nature of the immunohistochemistry, it is difficult to establish whether the remaining signal in the CKO autopod is background, or if it reflects some residual Hif-1α protein.
Figure 3.
Conditional removal of Hif-1α in limb bud mesenchyme leads to limb deformities. (A) Skeletal preparations of newborn control and CKO mice show the characteristic shortening of CKO limbs (arrows). (A) Skeletal preparations of forelimbs (FL) and hindlimbs (HL) of control and CKO newborn littermates.
Figure 6.
Joints are hypoxic and express Hif-1α. (A) Immunohistochemistry for Hif-1α on E14.5 forelimb autopod shows a particularly strong expression in the joints of the digits (arrows) and in the articular region of the wrist (arrowheads), in addition to the hypertrophic chondrocytes (asterisk). This expression is significantly reduced in CKO littermates (A′). In situ hybridization with a VEGF probe (B) closely resembles the Hif-1α protein expression pattern. EF5 staining indicates that the forming joints in the digits (C) and the articular region in the wrist (D) are highly hypoxic in E13.5 autopods. H&E staining indicates that the perichondrium surrounding the joints is particularly thick (E). Chondrocytes present at the articular surface remain hypoxic after joint cavitation, as indicated by EF5 staining of metacarpus (F) and elbow (G) of E15.5 forelimbs.
Despite the report of Prx1-Cre expression in the skull and, to some degree, in the axial skeleton, but consistent with a robust expression of Cre in limb bud mesenchyme (Logan et al., 2002), no obvious abnormalities could be observed elsewhere than in the limbs of mutant mice. Mutant limbs were severely shorter and misshapen in comparison to controls (Fig. 3 B). This finding suggests that in the absence of Hif-1α, the process of endochondral bone development was severely impaired. Histological analysis of the limb proximal bones, i.e., of the stylopod and zeugopod, at birth confirmed their extreme shortening, their severe deformities, and the massive central cell death phenotype concomitant to aberrant proliferation of viable chondrocytes (Fig. S2, C–F, available at http://www.jcb.org/cgi/content/full/jcb.200612023/DC1; and not depicted) that we previously reported in growth plates of mice in which Hif-1α had been conditionally inactivated in chondrocytes using a Col2a1 promoter-driven Cre (Schipani et al., 2001). The central cell death phenotype was already massive at E13.5 in the stylopod and zeugopod of CKO mice (Fig. S2 B; and Fig. S4, F and H).
To address the role of Hif-1α in early chondrogenesis, we then carefully analyzed the phenotype of CKO mice during early limb development (Fig. 4). The online version of this article contains supplemental material. Surprisingly, no obvious histological evidence of spatially localized loss of cell viability could be histologically observed in the distal portion of the mutant limbs, i.e., in the autopod (see below). This result, i.e., lack of the central cell death phenotype in the CKO autopod, was clearly different from what we had observed in the CKO stylopod and zeugopod. We decided to take advantage of this finding, and in our subsequent analysis, we focused exclusively on the autopod. Normally, undifferentiated mesenchymal cells in the limb bud start to condense around E11.5, and cells differentiate into chondrocytes soon after (Bi et al., 2001). The precartilaginous condensations of E12.5 forelimbs appeared similar in CKO and control (Fig. 4, C and D), as further confirmed by both PNA staining (Fig. 4, A and B) and Sox9 mRNA expression (Fig. 4, E and F). Thus, our data suggest that Hif-1α is not required for the formation of precartilaginous condensations. Consistent with the undifferentiated state of the cells at this stage, Col2a1 mRNA was expressed at very low levels in both CKO and control limbs (Fig. 4, G and H).
Figure 4.
Hif-1α is dispensable for condensation of limb bud mesenchyme. PNA staining (A and B), hematoxylin and eosin (H&E) staining (C and D), and in situ hybridization with the indicated probes (E–H) on the autopod of E12.5 forelimbs indicate that limb mesenchymal condensations form normally in the absence of Hif-1α.
At E13.5, however, the CKO autopod presented a remarkable delay in cartilage formation compared with controls (Fig. 5, A–H, and not depicted). Cells in the cartilaginous elements of control limbs showed typical chondrocyte morphology, whereas cells in CKO condensations resembled undifferentiated mesenchymal cells, with no evidence of hyaline matrix in between (Fig. 5, E and F). Alcian blue staining confirmed the paucity of proteoglycan accumulation in the mutant autopod in comparison to the control element (Fig. 5, C and D). Lastly, Col2a1 mRNA expression at this stage was slightly lower in mutant autopods than in controls (Fig. 5, G and H).
Figure 5.
Hif-1α is required for early chondrocyte differentiation. H&E staining (A and B) of E13.5 forelimb autopod, and Alcian blue staining (C and D) of E13.5 distal portion of forelimb autopod, indicate an abnormal histology of the cartilage and an abnormal cartilaginous matrix production, respectively. At this stage, a blow up of cells present in the cartilaginous primordia indicates the presence of differentiated cuboidal chondrocytes in control (E) and undifferentiated mesenchymal cells in CKO autopod (F). In situ hybridization reveals that E13.5 CKO metacarpals (H) express slightly less Col2a1 mRNA at this stage than control (G). In situ hybridization with the indicated markers performed on E14.5 forelimb control and CKO autopods confirms the dramatic delay in chondrocyte differentiation in the absence of Hif-1α (I–P).
At E14.5, histology and in situ hybridization analysis confirmed a severe delay in chondrocyte differentiation in the forelimb autopod (Fig. 5, I–P, and not depicted). Col2a1 mRNA expression was significantly lower in CKO compared with control, and, paradoxically, more intense in the distal, rather than in the proximal, portion of the mutant digital ray, whereas expression of Sox9, L-Sox5, and Sox6 mRNAs was similar in both mutants and controls (Fig. 5, I–L, and not depicted). Similar results were obtained in the E14.5 hindlimb autopod (unpublished data).
Collectively, these data indicate that Hif-1α is required for chondrogenic differentiation of mesenchymal cells, as lack of this transcription factor clearly delays the formation of cartilaginous mold.
CKO of Hif-1α in limb bud mesenchyme alters chondrocyte hypertrophy
The impairment in early chondrogenesis observed in CKO autopod could affect later steps of chondrocyte maturation, including hypertrophic differentiation. Consistent with this hypothesis, the autopod of E14.5 CKO forelimbs showed fewer regions of Indian hedgehog (Ihh) and Col10a1-expressing chondrocytes (Fig. 5, M–P). In addition, the pattern of expression of Col10a1 mRNA was clearly abnormal in the mutant versus control, as Col10a1 mRNA was detected in the distal, but not in the proximal portion of the mutant digital ray (Fig. 5 P). We do not have a good explanation for this abnormal distribution at the moment. Similar findings were also observed in the autopod of E14.5 hindlimbs (unpublished data). The massive early loss of cell viability precluded a meaningful analysis of the effect of the loss of Hif-1α in early chondrogenesis in stylopod and zeugopod. However, we previously described that cell death is restricted to the core of the cartilaginous elements (Schipani et al., 2001), and we took advantage of this to assess the degree of hypertrophic differentiation (late chondrogenesis) of chondrocytes located away from the core. In situ hybridizations analysis of superficial sections obtained from E14.5 zeugopod and stylopod confirmed a marked delay of hypertrophic differentiation in the mutant specimens (Fig. S2, K–P).
The delay of chondrocyte hypertrophy persisted at later stages of development
Histological analysis at birth of the autopod of both forelimbs and hindlimbs showed a severe reduction of hypertrophic chondrocytes and bony trabeculae in the metacarpals, metatarsals, and phalangeal elements of the mutant mice when compared with control (Fig. 7, C and D; and Fig. S3, C and D, available at http://www.jcb.org/cgi/content/full/jcb.200612023/DC1). Consistent with these data, at P9, bony trabeculae were present in control, but not in CKO, tarsal bones (Fig. S3, K and L; see the absence of pink staining in L), and no mineralized secondary ossification center was detectable in the radius and ulna of CKO animals (Fig. 8, E–F). In addition, talus and calcaneus in CKO mice showed extensive cartilaginous remnants that were absent in control bones (Fig. S3, M and N, blue staining).
Figure 7.
Hif-1α is required for joint development. H&E staining of E14.5 (A and B) and newborn (NB; C and D) forelimb autopod shows a delay in joint development in absence of Hif-1α. In situ hybridization with a GDF5 probe on E13 (E and F) and E14.5 (G and H) forelimb autopods indicates the absence of joint specification (F) and the subsequent abnormal joint development (H) in the absence of Hif-1α. The absence of some metacarpals in D results from an artifact of sectioning.
Figure 8.
CKO of Hif-1α in limb bud mesenchyme alters the formation of wrists. H&E staining of newborn (NB; A and B) and P9 (C and D) wrists and skeletal preparation of P9 wrists (E and F) show that bones present in the CKO wrist are misshapen, dislocated, and partially fused together for some of them. Wrist bones are indicated on each section. 1–3 and 4/5, distal row of carpal bones; c, central carpal bone; r and u, radial and ulnar bones; Ra and Ul, radius and ulna. Slashes between bone designators (i.e., 4/5) indicate a fusion of the indicated bones. 4/5, but never c/3, are always fused in wild-type wrist.
In situ hybridization analysis of chondrogenic markers in the autopod of newborn hindlimbs further proved that removal of Hif-1α in limb bud mesenchyme retards hypertrophy (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200612023/DC1). Expression of Ihh, Col10a1, and OP were completely absent from the region corresponding to the future phalangeal elements (Fig. S4, E–J), indicating that no hypertrophic chondrocytes were present in these areas, as also shown by histological analysis (Fig. S3, C and D). Furthermore, the distance between either the two Ihh or Col10a1 expression domains, as well as the extension of OP expression were significantly reduced in the prospective metacarpals (Fig. S4, E–H and I–J, respectively), suggesting that the overall replacement of cartilage by bone was significantly delayed. Conversely, we observed an increase in Sox9 expression in CKO bones compared with controls (Fig. S4, C and D). Because during endochondral bone development, Sox9 mRNA expression decreases over time, this observation further demonstrates that the CKO bones were, overall, younger than controls.
Collectively, our results show that Hif-1α is required for both early chondrogenesis and hypertrophic differentiation. Moreover, although we cannot exclude that the lack of Hif-1α directly affects hypertrophic differentiation, it is more likely that the impairment of hypertrophy is the indirect consequence of the initial delay in early chondrogenesis.
The joints are hypoxic, and Hif-1α regulates their development
Immunohistochemistry analysis revealed that accumulation of Hif-1α protein is particularly abundant in the prospective joints of E13.5 and E14.5 forelimb autopods, both in the digital rays and in the wrist (Fig. 6 A; and Fig. S1, C and D). Consistent with this finding, in situ hybridization showed detectable VEGF mRNA expression, which is a classical target of Hif-1α, in the same regions, at a similar stage (Fig. 6 B). In addition, analysis of EF5 staining demonstrated that the developing joints at E13.5 are highly hypoxic (Fig. 6, C and D). Notably, careful histological analysis of the autopod at E13.5 revealed a remarkable thickening of an avascular perichondrium around the area of the future joints (Fig. 6 E) that persisted at E14.5 (Fig. 7 A and Fig. S3 A). Moreover, at E15.5, after the joint space had formed, articular chondrocytes showed a significantly higher degree of hypoxia than the rest of the cartilaginous element (Fig. 6, F and G).
Collectively, these findings support the working hypothesis that hypoxia and Hif-1α could be crucial for proper joint formation, particularly in the autopod, and that the thickening of the perichondrium could play a significant role in contributing to the hypoxic status of interzone, and consequently, in modulating joint development.
Consistent with this model, the digital ray was not yet segmented into metacarpal and phalange elements in CKO forelimbs at E13.5 and E14.5, whereas this segmentation had already occurred in control limbs (Fig. 5, A and B; and Fig. 7, A and B, respectively). Distal joints, however, eventually developed at the right location in newborn CKO forelimbs (Fig. 7, C and D). Similar results were also observed in the digital ray of the hindlimbs (Fig. S3, A–D). No segmentation of the digital ray into metatarsal and phalange elements was yet evident at birth in CKO hindlimbs (Fig. S3 D), whereas a definition of metatarsals and phalanges had already occurred at E14.5 in control elements (Fig. S3 A). Also in this case, metatarsals and phalanges were eventually all properly segmented in 9-d-old (postnatal day [P] 9) CKO animals (unpublished data). Of note, thickening of the perichondrium was not affected in E13.5 and E14.5 mutant autopods (Fig. 5 B; Fig. 7 B; and Fig. S3 B), further supporting the hypothesis that this event precedes specification of the joints and can, thus, have a critical role in joint development. We also looked at joints in stylopod and zeugopod of mutant mice and observed that, in contrast to control, the interzone that normally forms between the scapula and the humerus was still absent in E13.5 mutant limbs (Fig. S3, E and F), suggesting that the role of Hif-1α in joint development is not limited to the autopod. At other sites, such as the elbow, the analysis was not conclusive, as a consequence of both the massive cell death and the severe deformities of the mutant elements (Fig. S3, G and H).
The aforementioned joint phenotype was even more severe in the ankle and wrist, which are extremely hypoxic during development (Fig. 6 D and not depicted). Consistent with the previously described impairment of early chondrogenesis, E14.5 CKO wrist had no defined cartilaginous elements, which was different from control (unpublished data). In newborn animals, the CKO wrists contained dislocated and extremely misshapen elements (Fig. 8, A and B). The malformations persisted postnatally. At P9, despite an identical number of bones in mutant and control, many bones were still misshapen, and some only partially segmented (Fig. 8, A–F, c and 3) and/or dislocated in the wrist of CKO animals (Fig. 8, C–F). Similar, but more severe, abnormalities were observed in CKO ankles (Fig. S3, I–N). Notably, the ankle of CKO mice was a single skeletal element at birth, whereas individual elements were present in control ankles (Fig. S3, I and J). At P9, all the expected bones were present in CKO ankle, but some were still only partially segmented (Fig. S3, K and N; bones 2 and 3 are fused, and c is partially fused with 4/5).
Collectively, our data indicate that lack of Hif-1α severely affects joint development, but, at this stage, they do not allow us to distinguish whether delay of joint specification or rather of joint cavitation was the cause of the phenotype. To address this issue, we looked at the expression of GDF5, a marker of joint specification that is detected even before interzone regions can be recognized histologically, and that represents one of the earliest known markers for joint formation (Storm and Kingsley, 1996; Merino et al., 1999). In E12.5 forelimbs, GDF5 mRNA appeared to be similarly expressed in the interdigital tissues of both CKO and control limbs (unpublished data). At E13.5, expression of GDF5 mRNA was already detectable in the control early prospective joints, but not in the CKO autopods (Fig. 7, E and F), indicating a delay in joint specification. At E14.5, control autopods presented strong, sharp stripes of GDF5 expression in the regions of prospective joints (Fig. 7 G), whereas in CKO, GDF5 expression was weaker and diffuse (Fig. 7 H), confirming the requirement of Hif-1α for joint development. In addition, consistent with a severe delay of segmentation, cells occupying the future joint regions were still present in the autopod of newborn CKO hindlimbs and, although they did not have detectable levels of Col2a1 mRNA, they did express Sox9, and also, weakly and with a rather diffuse pattern, GDF5 mRNA (Fig. S4, B, D, and L, respectively).
Collectively, these findings indicate that the delay in joint segmentation observed in CKO digital ray was secondary to a delay in joint specification. Furthermore, they demonstrate that GDF5 is epistatic to Hif-1α during joint formation.
Global effect of hypoxia on gene regulation in cartilage
To define the functional consequences of hypoxia in cartilage at the molecular level, and to get some insights into the molecular mechanism by which Hif-1α regulates early chondrogenesis and joint formation, metatarsals were isolated from E15.5 wild-type embryos and, after a few days in culture, exposed to 21 or 1% O2 for 8 h.
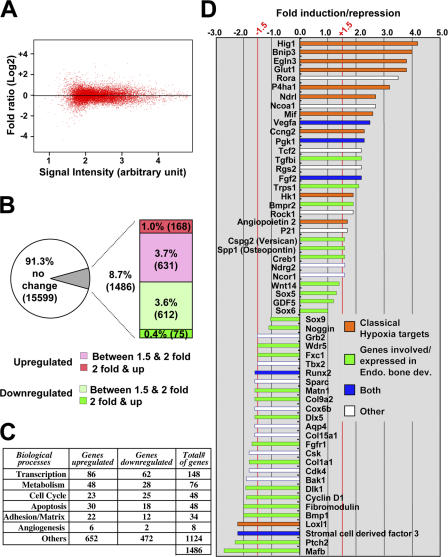
At 1% O2, Hif-1α protein is stabilized and transcriptionally active (Bishop et al., 2004; Leo et al., 2004; Greijer et al., 2005). Approximately 90% of the genes examined did not show any significant difference in response to hypoxia, as genes displaying less than −1.5- to +1.5-fold change were considered within normal range (Fig. 9, A and B). The genes modulated by hypoxia in chondrocytes belonged to a variety of biological categories, including transcription, cell cycle, apoptosis, adhesion, and angiogenesis (Fig. 9 C). For a complete list of the hypoxia-regulated genes, see Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200612023/DC1).
Figure 9.
Global effect of hypoxia on gene expression in cartilage. (A) A microarray analysis of the genes expressed in metatarsal explants and regulated upon 8 h of hypoxic treatment shows a symmetrical distribution of up- versus down-regulated genes. (B) The relative percentages of genes regulated in function of their fold induction or repression is indicated. Only the genes found to be at least 1.5-fold induced or repressed with a good P-value were considered to be regulated (see Materials and methods for all of the parameters used to filter the genes). (C) Genes regulated upon 8 h of hypoxia are involved in many diverse biological processes. A selection of a few regulated genes and their respective fold induction or repression is given in D. The genes located between −1.5- and +1.5-fold are not considered significantly regulated.
In addition to classical hypoxia-regulated genes, numerous novel targets of hypoxia, some of which had been previously involved in endochondral bone development, were identified (Fig. 9 D). Interestingly, expression of the master transcription factors of chondrogenesis, Sox9, L-Sox5, and Sox6, which play a particularly important role in early chondrogenesis (Lefebvre et al., 1998; de Crombrugghe et al., 2001; Smits et al., 2001, 2004), was virtually identical in hypoxic and normoxic specimens. The same results were obtained using primary chondrocytes cultured in hypoxic conditions for 8 h (unpublished data). These data confirm recently published findings showing that the Sox family of transcription factors is not a target of hypoxia (Hirao et al., 2006).
GDF5, Wnt14, and Noggin are main regulators of joint development (Brunet et al., 1998; Storm and Kingsley, 1999; Hartmann and Tabin, 2001; Kingsley, 2001; Guo et al., 2004). The abnormal GDF5 expression pattern observed in mutant mice raises the possibility that lack of Hif-1α may delay joint segmentation by interfering with GDF5 expression. We thus investigated whether the expression of these factors could be directly regulated by hypoxia. Microarray experiments showed that exposure to 1% hypoxia for 8 h did not significantly induce GDF5 mRNA expression in metatarsal explants (Fig. 9 D), suggesting that this factor is not a direct, transcriptional target of Hif-1α. In the same microarray assay, we also searched for levels of expression of Wnt14 and Noggin. As for GDF5, we did not observe any significant difference in Wnt14 and Noggin mRNA expression in hypoxic versus normoxic conditions (Fig. 9 D). The same results were obtained with primary chondrocytes cultured in hypoxic conditions for 8 h (unpublished data).
In the same assays, Col2a1, aggrecan, and hyaluronan synthases mRNAs were also not differentially regulated by hypoxia (unpublished data). Notably, however, expression of prolyl-4-hydroxylase α (I) (P4haI) mRNA was increased by approximately threefold after 8 h of exposure to hypoxia in both metatarsal explants (Fig. 9 D) and primary chondrocytes (not depicted). P4haI is a very well-documented gene of the hypoxia signature that is directly regulated by Hif-1α (Takahashi et al., 2000), and it plays an essential role in matrix accumulation by controlling posttranslational modifications of the α-I chain of collagen molecules.
Collectively, our results demonstrate the complexity of the hypoxia response in cartilage, and show that genes involved in posttranslational modifications of collagen, such as P4haI are up-regulated by hypoxia in chondrocytes.
Hif-1α controls chondrogenesis and joint development independently of cell death and angiogenesis
In contrast to the stylopod and zeugopod, and consistent with our histological findings, no detectable TUNEL-positive cells were found in the cartilaginous elements of either control or CKO autopod, whereas positive cells were observed, as expected, in the interdigital space of both (Fig. 10, A and B, and not depicted). These results indicate that the abnormal chondrogenesis in the autopod of CKO limbs cannot be attributed to cell death.
Figure 10.
Hif-1α regulates early chondrogenesis and joint development. TUNEL assay (A and B) performed on E14.5 forelimb autopods does not reveal any significant cell death in the absence of Hif-1α (B) in this tissue and at this stage. (C–F) In situ hybridization using a VEGFR2 probe (which marks endothelial cells) on E12.5 (C and D) and E14.5 (E and F) forelimbs does not indicate any detectable angiogenic problem in CKO autopod (G). Lack of Hif-1α alters differentiation in the absence of cell death and without impairing angiogenesis. A model for Hif-1α regulation of early chondrocyte differentiation and joint development.
Hif-1α controls angiogenesis, at least in part, by regulating expression of VEGF (Kotch et al., 1999). In the growth plate, VEGF is important for both chondrocyte survival and blood vessel invasion (Gerber et al., 1999; Maes et al., 2002; Maes et al., 2004; Zelzer et al., 2004). Thus, we looked at VEGF mRNA expression by in situ hybridization in E14.5 forelimbs. Consistent with previous studies (Zelzer et al., 2004), we observed that at this stage, VEGF mRNA was not detectable in chondrocytes forming the digital ray of control specimens, despite accumulation of Hif-1α protein (Fig. 6 B). This finding argues against a role of VEGF downstream of Hif-1α in early chondrogenesis.
To investigate whether removal of Hif-1α in limb bud mesenchyme would alter angiogenesis with yet unknown mechanisms, we then analyzed the expression of VEGF-receptor2 (VEGFR2), which is a marker of endothelial cells (Ferrara et al., 2003). At both E12.5 and E14.5, VEGFR2 mRNA expression was comparable in the soft tissue surrounding the precartilaginous and cartilaginous elements, respectively, of control and CKO (Fig. 10, C and D and E and F). Notably, at E14.5, this marker was also detectable in the bone collar of control elements (Fig. 10 E), whereas CKO forelimbs lacked the bone collar structures at this age because of their delay in hypertrophic differentiation (Fig. 10 F and not depicted).
These data indicate that lack of Hif-1α does not significantly impair angiogenesis in CKO limbs. Thus, the abnormal endochondral bone development observed in CKO bones is not caused by a global reduction of angiogenesis in the mutant limbs, but by direct effects on differentiation.
Discussion
This is the first study reporting the novel finding that Hif-1α regulates differentiation of limb bud mesenchyme. Lack of Hif-1α in limb bud mesenchyme impairs both early and late stages of chondrogenesis and joint development (Fig. 10 G). This critical role of Hif-1α in differentiation during embryonic development occurs without significant loss of cell viability and is not secondary to impairment of angiogenesis.
Limb bud mesenchyme and mesenchymal condensations express Hif-1α
Our study demonstrates that limb bud mesenchyme and mesenchymal condensations specifically express Hif-1α, as detected by in situ hybridization, and immunohistochemistry. Moreover, analyses of 5XHRE-LacZ reporter mice suggest that Hif-1α (and perhaps Hif-2α, which also binds to HRE) is transcriptionally active in this tissue. Although hypoxia is not the only regulator of Hif-1α transcriptional activity (Zelzer et al., 1998; Zundel et al., 2000), the hypoxic status of the mesenchymal condensations in the limb bud suggests, however, that hypoxia is likely a critical modulator of Hif-1α at this level.
Interestingly, at E10.5, Hif-1α mRNA and protein are also detected in the adjacent ectodermal area, particularly in the apical ectodermal ridge, which has an essential role in patterning (Mariani and Martin, 2003). Further studies will be needed to investigate the role of Hif-1α in the apical ectodermal ridge.
Lack of Hif-1α affects both early and late chondrogenesis
Hypoxia and Hif-1α support cartilaginous matrix accumulation (Pfander et al., 2003; Pfander et al., 2004) and are permissive factors in chondrocyte differentiation. The novel CKO model described in this study, demonstrates that Hif-1α is dispensable for mesenchyme condensations, but is important for early chondrogenesis. The absence of phenotype in E12.5 CKO limbs, despite Hif-1α expression, indicates that, at this stage, either this factor does not have a significant biological role, or that other redundant factors compensate for the lack of Hif-1α. Given the possibility that part of the X-gal staining observed using 5XHRE-LacZ reporter mice might be generated by Hif-2α, a putative role of this factor in endochondral bone development could be an interesting area of future research.
Our working model is that condensation of the mesenchyme leads to local hypoxia, and this stabilizes Hif-1α and increases its transcriptional activity. The transcription factors Sox9, L-Sox5, and Sox6 (the Sox trio) are required for chondrogenesis by regulating expression of cartilaginous matrix proteins, such as collagen II and aggrecan, at the transcriptional level (Lefebvre et al., 1998; Bi et al., 1999, 2001; Smits et al., 2001, 2004; Akiyama et al., 2002). Consistent with previous findings (Robin et al., 2005; Hirao et al., 2006), expression of Sox9, L-Sox5, and Sox6 mRNAs was not significantly affected by the lack of Hif-1α or hypoxia in our study. The discrepancy with the study by Robin et al. (2005) could be related to the different experimental model, as they analyze mesenchymal cell lines in vitro.
We had previously reported that hypoxia leads to increased accumulation of collagen II protein in primary chondrocytes in vitro, and that this protein accumulation is Hif-1α–dependent (Pfander et al., 2003). In contrast, expression of Col2a1 mRNA is not modulated by exposure to hypoxia for either 8 or 22 h (Pfander et al., 2003; this study). Thus, Col2a1 is likely not a target gene of Hif-1α. Interestingly, hypoxia and Hif-1α up-regulate the expression of P4haI (Fig. 9; Takahashi et al., 2000), which is a critical enzyme in posttranslational modifications of collagens. This suggests that, in the absence of Hif-1α, abnormalities in matrix accumulation could be caused, at least in part, by an impairment of posttranslational modifications of collagens. In our study, we observed very low levels of Col2a1 mRNA expression at E14.5 in mutant skeletal elements compared with control; the difference was much more subtle at E13.5, and no difference was noticeable at E12.5. These findings are consistent with the observation that hypoxia does not control transcription of the Col2a1 gene, and they suggest that the reduced levels of Col2a1 mRNA observed, particularly at E14.5, could be the indirect consequence of the initial delay in the formation of the cartilaginous mold. An abnormal matrix accumulation, which is a result, at least in part, of a defect of posttranslational modifications of collagens and, consequently, an impaired cell–matrix interaction could be the at the origin of this initial delay.
It is thus tempting to speculate that, whereas the Sox trio regulates chondrogenesis by up-regulating transcription of matrix proteins such as collagen II, hypoxia and Hif-1α may have a permissive role in chondrogenesis, at least in part, through modulation of posttranslational modifications of collagen II. This would result in a final “cooperative” effect of these transcription factors on chondrogenesis.
Consistent with our previous data (Zelzer et al., 2004), lack of Hif-1α delays not only early chondrogenesis, but also hypertrophy of mature chondrocytes. The delay of hypertrophic differentiation that we observe in the CKO limbs is likely the consequence of the impairment of early chondrogenesis.
Lack of Hif-1α alters joint development
In this study, we provide evidence that the joint space is highly hypoxic during embryonic development, and that Hif-1α regulates joint development. Lack of Hif-1α severely delays segmentation of the digital rays, and results in partial fusion of small carpal and tarsal bones. Notably, except for a few tarsal and carpal bones, our CKO mice do form synovial joints of normal appearance, after the initial developmental delay. It is still unclear whether carpal and tarsal bones originate from one single precursor or multiples. In light of both several publications (Shubin and Alberch, 1986; Settle et al., 2003) and our data showing that some carpal bones that are fused early on (Fig. 8 A) are fully segmented later (Fig. 8 C), it is possible that several carpal and tarsal bones are formed through the segmentation of bigger precursors. Thus, it is plausible that a common deficiency of segmentation is at the origin of the defects observed both in the wrist/ankle and in the digits of CKO mice.
This impairment of segmentation is secondary to a delay of joint specification, as indicated by the lack of histological and molecular evidence of the interzone in E14.5 CKO distal bones. Interestingly, however, whereas GDF5 is genetically downstream of Hif-1α in the prospective joints, mRNA expression of GDF5, Wnt14, and Noggin, which are critical regulators of joint development, are not modulated by hypoxia. This indicates that other factors are involved downstream of Hif-1α in joint development (Fig. 10 G). Notably, thickening of the perichondrium is not affected in E13.5 and E14.5 mutant autopods, further supporting the hypothesis that this event precedes specification of the joints and can thus have a critical role in joint development.
No abnormalities in joint segmentation were noted in CKO of Hif-1α in chondrocytes using a Col2a1 promoter (Schipani et al., 2001). This observation is consistent with the hypothesis that lack of Hif-1α affects very early stages of joint development. As chondrogenesis and joint formation are tightly coupled (Kornak and Mundlos, 2003), it is possible that the impairment of joint formation observed in CKO mice is secondary to the delay of early chondrogenesis. However, the high level of expression of Hif-1α in the hypoxic interzone, the severity of the phenotype, and the lack of joint problems in numerous other in vivo models of delayed endochondral bone development indicate that, in our CKO mice, the delay in joint formation is probably not only the consequence of the delay of early chondrogenesis.
Lack of Hif-1α alters differentiation in absence of cell death and without impairing angiogenesis
In contrast to what we observed in the stylopod and zeugopod of both Col2a1-Cre (Schipani et al., 2001) and Prx-1-Cre-Hif-1α CKO, the Prx-1-Cre-Hif-1α autopod does not show signs of cell death, at least not early in development. This finding allows us to conclude that the altered chondrogenic differentiation observed at these sites in absence of Hif-1α is not caused by cell death. We do not understand why Hif-1α is not a critical survival factor for chondrocytes in the autopod, but a potential explanation could rest on the geometry and thickness of these specimens. Nevertheless, these experiments allow us to distinguish for the first time the effect of Hif-1α on differentiation from its effects on survival.
An important question was whether the effect of Hif-1α on chondrocyte differentiation is cell autonomous, or rather non–cell autonomous, which we analyzed through modulation, for example, of angiogenesis in the surrounding soft tissue. Indeed, because the Prx-1 promoter is uniformly active in the limb bud mesenchyme before any condensation occurs, it was formally possible that a uniform knockout of Hif-1α in limb bud mesenchyme could affect angiogenesis, and that this, in turn, would impair chondrogenesis. In our study, we provide evidence that apparent distribution and number of blood vessels is not substantially different in CKO distal limbs versus controls. This observation indicates that impairment of angiogenesis is not the cause of the altered chondrocyte differentiation observed in CKO mutant mice.
Collectively, we propose that mesenchymal condensation or thickening of the avascular perichondrium creates a hypoxic environment, which in turn leads to Hif-1α protein stabilization that then positively regulates early chondrocyte differentiation and joint development (Fig. 10 G). This critical and previously unrecognized role of Hif-1α in early chondrogenesis and joint formation should spur future experiments to determine the critical effectors of Hif-1α that regulate cell differentiation.
Materials and methods
Analysis of EF-5 distribution
Pregnant females were injected (i.p.) at the appropriate stage with 10 mM EF5 at 1% of body weight; staining was performed using a Cy3-conjugated antibody, as previously described (Schipani et al., 2001).
Whole mount immunohistochemistry
Embryos were processed and immunohistochemistry was performed as previously described (Streit and Stern, 2001). Embryos were incubated with a primary antibody against Hif-1α (R&D Systems) diluted 1:100.
Whole mount in situ hybridization
Wholemount in situ hybridization using DIG-labeled RNA probes was performed essentially as previously described (Streit and Stern, 2001). The hybridization was performed at 65°C overnight, and the signal was detected using the colorimetric BM-Purple substrate (Roche).
Generation and screening of 5X-HRE transgenic mice
The 5XHRE-E1b/LacZ transgene was constructed by blunt-ending a 4.26-kb NcoI–PstI nuclear localization signal/LacZ–containing fragment from pIRESLacZ (a gift from A. Nagy, Samuel Lunenfeld Research Institute, Toronto, Canada) into the HindIII–XbaI sites (luciferase removed) of p5XHRE-Luciferase (Shibata et al., 1998). The 5XHRE-hsp/LacZ transgene was constructed by blunt-ending a 0.3-kb KpnI–HindIII 5XHRE- containing fragment from p5XHRE-Luciferase into the HindIII site of pHspLacZpA (compliments of J. Rossant, Samuel Lunenfeld Research Institute, Toronto, Canada). Transgenic founder mice were identified by Southern blot analysis of AccI–EcoRI–digested tail DNA probed with a 1.1-kb LacZ-containing fragment from SphI-digested mb-STOP-LacZ (Zinyk et al., 1998). Genotyping was performed on genomic DNA by PCR. Yolk sac or tail DNA was amplified for 40 cycles (1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C) on a thermal cycler. The LacZ primers 5′-GGTGATTTTGGCGATACGC-3′ and 5′-TGCAGGAGCTCGTTATCGC-3′ produce a 191-bp product.
Culture of mouse embryos
E8.5 embryos were dissected individually into cold sterile L15 media, staged for somite number, and transferred to 1 ml DME media containing 20% fetal calf serum and 1× nonessential amino acids. Embryos were cultured either in normoxic (21% O2 or 5% CO2 at 37°C) or in anoxic (hypoxia chamber; 0% O2 or 5% CO2 at 37°C) conditions for 24 h. Embryos were then processed for X-Gal staining.
Detection of β-galactosidase activity
Whole mount X-Gal staining of freshly dissected mouse embryos was performed as previously described (Zinyk et al., 1998). Cultured embryos were stained with X-Gal solution at 37°C overnight.
Generation of conditional Hif-1α knockout
Male hemizygous Prx-1 Cre transgenic mice in the Swiss-Webster background (Logan et al., 2002) were bred with animals that were homozygous for a floxed Hif-1α allele (Hiff/f) in the FVB/N background (Schipani et al., 2001). Males heterozygous for the floxed Hif-1α and homozygous for the Prx-1 transgene were crossed with female mice homozygous for the floxed Hif-1α allele to generate Hif-1αf/f; Prx-1 Cre mutant mice, which were genotyped as previously described (Schipani et al., 2001; Logan et al., 2002).
Alizarin red S staining, histology, in situ hybridization analysis, TUNEL assay, and Peanut agglutinin (PNA) staining
Alizarin red S staining was performed as previously described (Schipani et al., 1997). For light microscopy, tissues from E12.5, E13.5, and E14.5 mice (delivered by caesarean section), as well as newborn and p9 mice, were fixed in 10% formalin/PBS, pH 7.4, and stored in fixative at 4°C. Paraffin blocks, sections, and H&E staining were realized by standard histological procedures. In situ hybridizations were performed using complementary 35S-labeled riboprobes, as previously described (Schipani et al., 1997). For TUNEL assay, paraffin sections from hindlimbs of newborn mice were permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate. TUNEL assay was performed using an In Situ Cell Death Detection kit (Roche) according to the manufacturer's conditions. For PNA staining, sections were dewaxed, rehydrated, and pretreated for 20 min with 0.5% H2O2 in methanol, and washed in PBS-T (PBS + 0.05% Tween-20). Slides were then incubated with biotinylated PNA (Vector Laboratories) at 100 μg/ml in PBS-T + 0.1 mM CaCl2 for 45 min at RT, and then washed extensively with PBS-T. Bound PNA was detected using the TSA- Biotin system (Perkin Elmer) and DAB substrate (Vector Laboratories) according to the manufacturer's instructions.
Immunohistochemistry
Frozen sections of E12 and paraffin sections of E13.5 and E14.5 mouse autopods were heated at 80°C in 0.1 M citrate buffer, pH 6.0, for 2 h. After quenching of the endogenous peroxidase by incubation in 3% H2O2/PBS for 10 min at room temperature, blocking was performed with a specific solution provided by a TSA kit (NEN). Sections were incubated with a primary antibody against Hif-1α (R&D Systems) at a dilution of 1:500 at 4°C overnight. After incubation with the appropriate biotinylated secondary antibody, detection of the binding was carried out using the Streptavidin-HRP system provided by the TSA kit, following the manufacturer's instructions.
Image acquisition
All images were acquired with a microscope (Eclipse E800; Nikon) using Plan Apo 4×/0.2 NA, Plan Apo 10×/0.45 NA, Plan Apo 20×/0.75 NA, and Plan Apo 40×/0.95 NA lenses (Nikon), at ambient temperature, with air imaging medium. Cy3 fluorochrome was used for the EF5 detection. Photographs were taken using a SPOT camera (model 1.30) and SPOT software version 3.5.5 for Macintosh (both from Diagnostic Instruments). Images were assembled and legends were added using PowerPoint software (Microsoft). Figures were transferred into Photoshop software (Adobe) and brightness and contrast were modified by applying brightness/contrast adjustments to the whole image, with the strict intent of not obscuring, eliminating, or misrepresenting any information present in the original, including background.
Chondrocyte isolation and metatarsal explants
Chondrocytes were isolated from newborn wild-type mice as previously described (Pfander et al., 2004). Chondrocytes were plated at a density of 4 × 105 cells per well of 6-well plates and grown in monolayer cultures in high-glucose DME (Invitrogen) supplemented with 10% FBS (HyClone) and 1% penicillin/streptomycin. At days 5, 10, and 20 after plating, respectively, cells were exposed to 21% (normoxic) or 1% (hypoxic) O2 for 8 h; then total RNA was isolated, as previously described (Pfander et al., 2004).
Metatarsal explants were obtained from E15.5 mouse hindlimbs and cultured as previously described (Haaijman et al., 1999). After 3 d in culture, metatarsals were exposed to exposed to 21% (normoxic) or 1% (hypoxic) O2 for 8 h, and total RNA was isolated as previously described (Pfander et al., 2004). For these experiments, biological triplicates were used.
Microarray assay and real-time PCR
For microarray assay analysis, purified total RNA was subjected to quality control analysis using a Bioanalyzer (Agilent). Total RNA was reverse transcribed into cDNA labeled with Cy3 or Cy5 dye (dye swap was performed). Total RNA extracted from metatarsal explants was appropriately amplified. Detailed protocols are available online at the Massachusetts General Hospital Microarray Core website (dnacore.mgh.harvard.edu/microarray/index.shtml). Control and mutant cDNAs were equally mixed and spotted on mouse 70-mer oligonucleotide arrays. The arrays contain 19,549 oligos and provide complete coverage of the 2002 mouse genome database. Hybridization was performed in duplicate. Data were obtained from biological triplicates and analyzed using the BioArray Software Environment (BASE). Approximately 9,000 oligonucleotides provided signals statistically above background. The microarray assay analysis was performed in collaboration with the Genomic Core at Massachusetts General Hospital.
Deletion of Hif-1α was confirmed by real-time quantitative PCR analysis of genomic DNA extracted from five independent controls and mutant forelimb paws at birth after removal of the skin. Real-time PCR was performed as previously described (Pfander et al., 2003). Sequences of primers are available upon request. The Vhlh gene was amplified as an internal control. Cycle threshold (Ct) values were measured and calculated by the sequence detector software. Relative amounts of mRNA were normalized to Vhlh and calculated with the software program Excel (Microsoft). Relative genomic DNA contents were calculated as x = 2−ΔΔCt, in which ΔΔCt = ΔE − ΔC, ΔE = CtHif null − CtVhlh, and ΔC = Ctcontrol − CtVhlh. A fourfold difference in efficiency of amplification was calculated between mutants and controls, indicating that the efficiency of deletion in the mutant samples was ∼75%.
Online supplemental material
Fig. S1 shows Hif-1α protein expression in early mesenchymal condensations and in prospective joints. Fig. S2 describes the phenotype of the stylopod and zeugopod of CKO animals characterized by early cell death, disorganized growth plate chondrocytes, and delayed hypertrophy. Fig. S3 presents the abnormal joint development in CKO hindlimb autopods/ankles and in CKO stylopods (shoulder) and zeugopods (elbow). Fig. S4 describes the abnormal distribution of chondrogenic and joint-specific markers in CKO hindlimb paws. Table S1 lists the genes regulated in metatarsal explants exposed to hypoxia for 8 h. The online version of this article is available at http://www.jcb.org/cgi/content/full/jcb.200612023/DC1.
Supplementary Material
Acknowledgments
The authors thank the Histology Core at the Endocrine Unit of Massachusetts General Hospital for excellent technical assistance.
This work was supported by National Institutes of Health grants AR048191 (to E. Schipani) and CA 088480 and CA 67166 (to A.J. Giaccia).
S. Provot and D. Zinyk contributed equally to this paper.
Abbreviations used in this paper: CKO, conditional knockout; E, embryonic day; Hif-1α, hypoxia-inducible factor 1α; HRE, hypoxia response element; H&E, hematoxylin and eosin; Ihh, Indian hedgehog; P, postnatal day; p4haI, prolyl-4-hydroxylase α (I); PNA, Peanut agglutinin; VEGFR2, VEGF-receptor2.
References
- Akiyama, H., M.C. Chaboissier, J.F. Martin, A. Schedl, and B. de Crombrugghe. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16:2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, W., J.M. Deng, Z. Zhang, R.R. Behringer, and B. de Crombrugghe. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22:85–89. [DOI] [PubMed] [Google Scholar]
- Bi, W., W. Huang, D.J. Whitworth, J.M. Deng, Z. Zhang, R.R. Behringer, and B. de Crombrugghe. 2001. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. USA. 98:6698–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, T., K. Lau, A. Epstein, S. Kim, M. Jiang, D. O'Rourke, C. Pugh, J. Gleadle, M. Taylor, J. Hodgkin, and P. Ratcliffe. 2004. Genetic analysis of pathways regulated by the von hippel-lindau tumor suppressor in Caenorhabditis elegans. PLoS Biol. 2:e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet, L.J., A. McMahon, and R.M. Harland. 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 280:1455–1457. [DOI] [PubMed] [Google Scholar]
- Bunn, H., and R. Poyton. 1996. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 76:839–885. [DOI] [PubMed] [Google Scholar]
- Chan, D., P. Suthphin, N. Denko, and A. Giaccia. 2002. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J. Biol. Chem. 277:40112–40117. [DOI] [PubMed] [Google Scholar]
- Chen, E., M. Fujinaga, and A. Giaccia. 1999. Hypoxic microenvironment within an embryo induces apoptosis and is essential for proper morphological development. Teratology. 60:215–225. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe, B., V. Lefebvre, and K. Nakashima. 2001. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell Biol. 13:721–727. [DOI] [PubMed] [Google Scholar]
- Erlebacher, A., E.H. Filvaroff, S.E. Gitelman, and R. Derynck. 1995. Toward a molecular understanding of skeletal development. Cell. 80:371–378. [DOI] [PubMed] [Google Scholar]
- Ferrara, N., H. Gerber, and J. LeCouter. 2003. The biology of VEGF and its receptors. Nat. Med. 9:669–676. [DOI] [PubMed] [Google Scholar]
- Gerber, H., T. Vu, A. Ryan, J. Kowalski, Z. Werb, and N. Ferrara. 1999. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 5:623–628. [DOI] [PubMed] [Google Scholar]
- Giaccia, A., B. Siim, and R. Johnson. 2003. HIF-1 as a target for drug development. Nat. Rev. Drug Discov. 2:803–811. [DOI] [PubMed] [Google Scholar]
- Giaccia, A., M. Simon, and R. Johnson. 2004. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 18:2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greijer, A.E., P. van der Groep, D. Kemming, A. Shvarts, G.L. Semenza, G.A. Meijer, M.A. van de Wiel, J.A. Belien, P.J. van Diest, and E. van der Wall. 2005. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 206:291–304. [DOI] [PubMed] [Google Scholar]
- Guo, X., T.F. Day, X. Jiang, L. Garrett-Beal, L. Topol, and Y. Yang. 2004. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 18:2404–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaijman, A., M. Karperien, B. Lanske, J. Hendriks, C.W. Lowik, A.L. Bronckers, and E.H. Burger. 1999. Inhibition of terminal chondrocyte differentiation by bone morphogenetic protein 7 (OP-1) in vitro depends on the periarticular region but is independent of parathyroid hormone-related peptide. Bone. 25:397–404. [DOI] [PubMed] [Google Scholar]
- Hartmann, C., and C.J. Tabin. 2001. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 104:341–351. [DOI] [PubMed] [Google Scholar]
- Hirao, M., N. Tamai, N. Tsumaki, H. Yoshikawa, and A. Myoui. 2006. Oxygen tension regulates chondrocyte differentiation and function during endochondral ossification. J. Biol. Chem. 281:31079–31092. [DOI] [PubMed] [Google Scholar]
- Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. Asara, W. Lane, and W. Kaelin. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 292:464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P., D. Mole, Y. Tian, M. Wilson, J. Gielbert, S. Gakell, A. Kriegsheim, H. Heberstreit, M. Mukherji, C. Schofield, et al. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 292:468–472. [DOI] [PubMed] [Google Scholar]
- Jacobsen, K.D., and B.M. Willumsen. 1995. Kinetics of expression of inducible beta-galactosidase in murine fibroblasts: high initial rate compared to steady-state expression. J. Mol. Biol. 252:289–295. [DOI] [PubMed] [Google Scholar]
- Kaelin, W. 2002. How oxygen makes its presence felt. Genes Dev. 16:1441–1445. [DOI] [PubMed] [Google Scholar]
- Kallio, P.J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makimo, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 17:6573–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty, G. 2003. The complexities of skeletal biology. Nature. 423:316–318. [DOI] [PubMed] [Google Scholar]
- Kingsley, D. 2001. Genetic control of bone and joint formation. Novartis Found. Symp. 232:213–222. [DOI] [PubMed] [Google Scholar]
- Kornak, U., and S. Mundlos. 2003. Genetic disorders of the skeleton: a developmental approach. Am. J. Hum. Genet. 73:447–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotch, L., N. Iyer, E. Laughner, and G. Semenza. 1999. Defective vascularization of HIF 1 alpha null embryos is not associated with VEGF deficiency but with mesenchymal cell death. Dev. Biol. 209:254–267. [DOI] [PubMed] [Google Scholar]
- Kronenberg, H. 2003. Developmental regulation of the growth plate. Nature. 423:332–336. [DOI] [PubMed] [Google Scholar]
- Lee, J., D. Siemann, C. Koch, and E. Lord. 1996. Direct relationship between radiobiological hypoxia in tumors and monoclonal antibody detection of EF5 cellular adducts. Int. J. Cancer. 67:372–378. [DOI] [PubMed] [Google Scholar]
- Lefebvre, V., P. Li, and B. de Crombrugghe. 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17:5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo, C., A. Giaccia, and N. Denko. 2004. The hypoxic tumor microenvironment and gene expression. Semin. Radiat. Oncol. 14:207–214. [DOI] [PubMed] [Google Scholar]
- Liu, L., and M. Simon. 2004. Regulation of transcription and translation by hypoxia. Cancer Biol. Ther. 3:492–497. [DOI] [PubMed] [Google Scholar]
- Logan, M., J. Martin, A. Nagy, C. Lobe, E. Olsen, and C. Tabin. 2002. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 33:77–80. [DOI] [PubMed] [Google Scholar]
- Maes, C., P. Carmeliet, K. Moermans, I. Stockmans, N. Smets, D. Collen, R. Bouillon, and G. Carmeliet. 2002. Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Mech. Dev. 111:61–73. [DOI] [PubMed] [Google Scholar]
- Maes, C., I. Stockmans, K. Moermans, R.V. Looveren, N. Smets, P. Carmeliet, R. Bouillon, and G. Carmeliet. 2004. Soluble VEGF isoforms are essential for establishing epiphyseal vascularization and regulating chondrocyte development and survival. J. Clin. Invest. 113:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani, F., and G. Martin. 2003. Deciphering skeletal patterning: clues from the limb. Nature. 423:319–325. [DOI] [PubMed] [Google Scholar]
- Merino, R., D. Macias, Y. Ganan, A.N. Economides, X. Wang, Q. Wu, N. Stahl, K.T. Sampath, P. Varona, and J.M. Hurle. 1999. Expression and function of Gdf-5 during digit skeletogenesis in the embryonic chick leg bud. Dev. Biol. 206:33–45. [DOI] [PubMed] [Google Scholar]
- Min, J., H. Yang, M. Ivan, F. Gertler, W.K. Jr, and N. Pavletich. 2002. Structure of an HIF-1alpha-pVHL complex: hydroxyproline recognition in signaling. Science. 296:1886–1889. [DOI] [PubMed] [Google Scholar]
- Ohh, M., and W. Kaelin. 1999. The von Hippel-Lindau tumor suppressor protein: new perspectives. Mol. Med. Today. 5:257–263. [DOI] [PubMed] [Google Scholar]
- Pfander, D., T. Cramer, E. Schipani, and R. Johnson. 2003. HIF-1alpha controls extracellular matrix synthesis by epiphyseal chondrocytes. J. Cell Sci. 116:1819–1826. [DOI] [PubMed] [Google Scholar]
- Pfander, D., T. Kobayashi, M. Knight, E. Zelzer, D. Chan, B. Olsen, A. Giaccia, R. Johnson, V. Haase, and E. Schipani. 2004. Deletion of Vhlh in chondrocytes reduces cell proliferation and increases matrix deposition during growth plate development. Development. 131:2497–2508. [DOI] [PubMed] [Google Scholar]
- Provot, S., and E. Schipani. 2005. Molecular mechanisms of endochondral bone development. Biochem. Biophys. Res. Commun. 328:658–665. [DOI] [PubMed] [Google Scholar]
- Robin, J.C., N. Akeno, A. Mukherjee, R. Dalal, B.J. Aronow, P. Koopman, and T.L. Clemens. 2005. Hypoxia induces chondrocyte-specific gene expression in mesenchymal cells in association with transcriptional activation of Sox9. Bone. 37:313–322. [DOI] [PubMed] [Google Scholar]
- Schipani, E., B. Lanske, J. Hunzelman, A. Luz, C.S. Kovacs, K. Lee, A. Pirro, H.M. Kronenberg, and H. Jüppner. 1997. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc. Natl. Acad. Sci. USA. 94:13689–13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipani, E., H. Ryan, S. Didrickson, T. Kobayashi, M. Knight, and R. Johnson. 2001. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 15:2865–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, G. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 3:721–732. [DOI] [PubMed] [Google Scholar]
- Settle, S.H., Jr., R.B. Rountree, A. Sinha, A. Thacker, K. Higgins, and D.M. Kingsley. 2003. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 254:116–130. [DOI] [PubMed] [Google Scholar]
- Shibata, T., N. Akiyama, M. Noda, K. Sasai, and M. Hiraoka. 1998. Enhancement of gene expression under hypoxic conditions using fragments of the human vascular endothelial growth factor and the erythropoietin genes. Int J Radiat Oncol Biol Phys. 42:913–916. [DOI] [PubMed] [Google Scholar]
- Shubin, N.H., and P. Alberch. 1986. A morphogenetic approach to the origin and the basic organization of the tetrapod limb. Evol. Biol. 20:319–387. [Google Scholar]
- Smits, P., P. Li, J. Mandel, Z. Zhang, J.M. Deng, R.R. Behringer, B. de Crombrugghe, and V. Lefebvre. 2001. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell. 1:277–290. [DOI] [PubMed] [Google Scholar]
- Smits, P., P. Dy, S. Mitra, and V. Lefebvre. 2004. Sox5 and Sox6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J. Cell Biol. 164:747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm, E., and D. Kingsley. 1999. GDF5 coordinates bone and joint formation during digit development. Dev. Biol. 209:11–27. [DOI] [PubMed] [Google Scholar]
- Storm, E.E., and D.M. Kingsley. 1996. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 122:3969–3979. [DOI] [PubMed] [Google Scholar]
- Streit, A., and C. Stern. 2001. Combined whole-mount in situ hybridization and immunohistochemistry in avian embryos. Methods. 23:339–344. [DOI] [PubMed] [Google Scholar]
- Sumiyama, K., and F.H. Ruddle. 2003. Regulation of Dlx3 gene expression in visceral arches by evolutionarily conserved enhancer elements. Proc. Natl. Acad. Sci. USA. 100:4030–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., S. Takahashi, Y. Shiga, T. Yoshimi, and T. Miura. 2000. Hypoxic induction of prolyl 4-hydroxylase alpha (I) in cultured cells. J. Biol. Chem. 275:14139–14146. [DOI] [PubMed] [Google Scholar]
- Vu, T., J. Shipley, G. Bergers, J. Berger, J. Helms, D. Hanahan, S. Shapiro, R. Senior, and Z. Werb. 1998. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 93:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer, E., Y. Levy, C. Kahana, B.Z. Shilo, M. Rubinstein, and B. Cohen. 1998. Insulin induces transcription of target genes through the hypoxia-inducible factor 1 alpha. EMBO J. 17:5085–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer, E., W. McLean, Y. Ng, N. Fukai, A. Reginato, S. Lovejoy, P. D'Amore, and B. Olsen. 2002. Skeletal defects in VEGF120/120 mice reveal multiple roles for VEGF in skeletogenesis. Development. 129:1893–1904. [DOI] [PubMed] [Google Scholar]
- Zelzer, E., R. Mamluk, N. Ferrara, R. Johnson, E. Schipani, and B. Olsen. 2004. VEGFA is necessary for chondrocyte survival during bone development. Development. 131:2161–2171. [DOI] [PubMed] [Google Scholar]
- Zinyk, D.L., E.H. Mercer, E. Harris, D.J. Anderson, and A.L. Joyner. 1998. Fate mapping of the mouse midbrain-hindbrain constriction using a site-specific recombination system. Curr. Biol. 8:665–668. [DOI] [PubMed] [Google Scholar]
- Zundel, W., C. Schindler, D. Haas-Kogan, A. Koong, F. Kaper, E. Chen, A. Gottschalk, H. Ryan, R. Johnson, A. Jefferson, et al. 2000. Loss of PTEN facilitates HIF-1 mediated gene expression. Genes Dev. 14:391–396. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.