Abstract
Lafora disease (LD) is a progressive myoclonic epilepsy resulting in severe neurodegeneration followed by death. A hallmark of LD is the accumulation of insoluble polyglucosans called Lafora bodies (LBs). LD is caused by mutations in the gene encoding the phosphatase laforin, which reportedly exists solely in vertebrates. We utilized a bioinformatics screen to identify laforin orthologues in five protists. These protists evolved from a progenitor red alga and synthesize an insoluble carbohydrate whose composition closely resembles LBs. Furthermore, we show that the kingdom Plantae, which lacks laforin, possesses a protein with laforin-like properties called starch excess 4 (SEX4). Mutations in the Arabidopsis thaliana SEX4 gene results in a starch excess phenotype reminiscent of LD. We demonstrate that Homo sapiens laforin complements the sex4 phenotype and propose that laforin and SEX4 are functional equivalents. Finally, we show that laforins and SEX4 dephosphorylate a complex carbohydrate and form the only family of phosphatases with this activity. These results provide a molecular explanation for the etiology of LD.
Introduction
Lafora disease (LD; OMIM #254780) is an autosomal recessive neurodegenerative disorder resulting in severe epilepsy and death (Lafora and Gluck, 1911; Van Hoof and Hageman-Bal, 1967). It is one of five major progressive myoclonus epilepsies. LD presents itself as a single seizure in the second decade of the patient's life (Schwarz and Yanoff, 1965); this single event is followed by progressive central nervous system degeneration, culminating in death within 10 yr of the first seizure (Van Heycop Ten Ham, 1975). A hallmark of LD is the accumulation of polyglucosan inclusion bodies called Lafora bodies (LBs; Lafora, 1911; Collins et al., 1968) that are located in the cytoplasm of cells in most organs (Harriman et al., 1955; Schwarz and Yanoff, 1965; Carpenter and Karpati, 1981). LB accumulation coincides with increased neuronal nonapoptotic cell death and a number of seizures in LD patients. Thus, it is hypothesized that LBs are responsible for these symptoms and, ultimately, for the death of the patient (Yokoi et al., 1968).
Recessive mutations in EPM2B (epilepsy of progressive myoclonus type 2 gene B)/NHLRC1 (Chan et al., 2003), which encodes the E3 ubiquitin ligase malin (Chan et al., 2003; Gentry et al., 2005), are responsible for ∼40% of LD cases (Ianzano et al., 2005). Of the LD cases not attributed to mutations in EPM2B, ∼48% result from recessive mutations in EPM2A (epilepsy of progressive myoclonus type 2 gene A; Minassian et al., 1998; Serratosa et al., 1999; Ianzano et al., 2005). EPM2A encodes laforin, which contains a carbohydrate-binding module (CBM) family 20 (CBM20; Wang et al., 2002) domain followed by the canonical dual specificity phosphatase (DSP) active site motif HCXXGXXRS/T (Cx5R) (Fig. 1 A; Denu et al., 1996; Yuvaniyama et al., 1996; Minassian et al., 1998). The CBM of laforin binds complex carbohydrates in vivo and in vitro (Wang et al., 2002), and the DSP motif can hydrolyze phosphotyrosine and phosphoserine/threonine substrates in vitro (Ganesh et al., 2000; Wang et al., 2002). However, no group has detected endogenous laforin localization in tissue culture cells or in wild-type tissues likely as a result of low levels of accumulation (Chan et al., 2004; our unpublished data).
Figure 1.
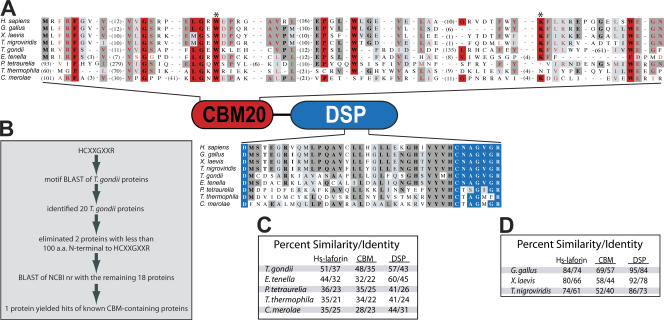
Laforin orthologues. (A) An alignment of the vertebrate and protist laforin orthologues. Residues highlighted in red are highly conserved CBM20 residues as defined by the CBM20 family (Wang et al., 2002), residues boxed in red are invariant CBM20 residues (Wang et al., 2002), and residues boxed in blue are part of the DSP catalytic site. Residues boxed in dark gray are identical, and those boxed in light gray are conserved substitutions. Asterisks mark residues that are necessary for binding to carbohydrates (Wang et al., 2002). Accession numbers are listed in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). (B) Strategy to identify laforin orthologues in T. gondii. (C) Percent similarity and identity of full-length Hs-laforin and both domains compared with those of protists. (D) Percent similarity and identity of full-length Hs-laforin and both domains compared with those of other vertebrates.
Of the 128 human phosphatases (Zolnierowicz, 2000; Alonso et al., 2004), only laforin possesses a CBM. CBM domains are predominantly found in glucosylhydrolases and glucotransferases of bacterial, fungal, or plant origin (Coutinho and Henrissat, 1999; Boraston et al., 2004; Rodriguez-Sanoja et al., 2005). The vast majority of enzymes containing a CBM use the domain to bind a specific type of carbohydrate and then enzymatically act on the sugar (Boraston et al., 2004). Accordingly, we recently showed that laforin liberates phosphate from the complex carbohydrate amylopectin, whereas other phosphatases lack this activity (Worby et al., 2006).
Ganesh et al. (2002) disrupted the EPM2A locus in a mouse model. Although this model faithfully recapitulated the disease, it yielded no molecular explanation for LD. Similarly, Chan et al. (2004) generated a transgenic mouse overexpressing inactivated laforin, and this mouse model also developed LD. Despite the availability of these two LD mouse models, the molecular etiology of LD remained unexplained. These limitations demonstrate the need to develop alternative model systems to elucidate the biology of LD. Although a molecular mechanism to explain LD has remained elusive, data cumulatively place laforin in the context of being intimately, if not directly, involved in regulating glycogen metabolism. Therefore, we focused on this indisputable aspect of LD for clues to its molecular etiology.
Insoluble glycogen, starch, and floridean starch
Glycogen is produced in the cytoplasm of the majority of archaebacterial, bacterial, fungal, and animal species. It is a water-soluble polymer composed of α-1,4-glycosidic linkages between glucose residues, with branches occurring in a continuous pattern every 12–14 residues via α-1,6-glycosidic linkages. Almost every study on LD refers to LBs as insoluble glycogen. However, definitive biochemical studies on LBs found that the arrangement and pattern of branching in LBs most closely resemble amylopectin (Yokoi et al., 1967, 1968; Sakai et al., 1970).
Amylopectin, like glycogen, is composed of α-1,4-glycosidic linkages with α-1,6-glycosidic branches but with branches arranged in a discontinuous pattern every 12–20 residues. This discontinuous and decreased amount of branching renders amylopectin insoluble. Amylopectin is one of the two components of starch, which is produced in the plastid of green plants (Viridaeplantae). Starch is an insoluble, semicrystalline heterogeneous mixture of 10–25% amylose and 75–90% amylopectin. Plants synthesize starch in chloroplasts during daylight as a transient carbon store that is used during the dark cycle to generate a usable reduced form of carbon in the absence of photosynthesis.
Floridean starch is another insoluble carbohydrate that has similar biochemical properties to amylopectin (Peat et al., 1959; Coppin et al., 2005). Floridean starch is synthesized in the cytoplasm of a variety of protists (i.e., unicellular eukaryotes) and is used as an energy source during specific stages of their life cycle. Floridean starch, like LBs and amylopectin, is made of glucose polymers with branches every 12–20+ residues in a discontinuous pattern (Coppin et al., 2005). Thus, floridean starch, amylopectin, and LBs have been described as possessing similar characteristics.
Results
Discovery of laforin orthologues
One protist that accumulates floridean starch (also called amylopectin granules) in its cytoplasm is Toxoplasma gondii (Dubey et al., 1998; Coppin et al., 2005; Guérardel et al., 2005; for review see Coppin et al., 2003). T. gondii is an obligate intracellular parasite that can infect nearly any nucleated cell from a warm-blooded animal. Like most members of Apicomplexa, T. gondii has a complex life cycle: in its intermediate hosts, it exists as a rapidly dividing tachyzoite or an encysted bradyzoite, depending on the host immune response. The bradyzoite forms floridean starch in its cytoplasm that is used as an energy source (for review see Coppin et al., 2003). Recent studies characterized the biochemical composition of T. gondii floridean starch (Coppin et al., 2005; Guérardel et al., 2005). We noted that the biochemical composition of T. gondii floridean starch was remarkably similar to that of LBs described nearly 40 yr ago (Yokoi et al., 1967, 1968; Sakai et al., 1970). Although EPM2A has been reported to be present only in vertebrates (Ganesh et al., 2001, 2004), the similarity between T. gondii floridean starch and LBs led us to explore the partially completed T. gondii genome for a laforin orthologue.
The sequence of the T. gondii genome, like the genome of many protists, was not accessible via GenBank when we initiated this study. Therefore, we searched the T. gondii database (ToxoDB; Kissinger et al., 2003) for a laforin orthologue. We used the criteria that a laforin orthologue must contain both an amino-terminal CBM and a carboxy-terminal DSP domain (Fig. 1 A). DSP domains are readily recognized by the protein families database (pfam; Bateman et al., 2004) and the National Center for Biotechnology Information's (NCBI) conserved domain database (CDD; Marchler-Bauer et al., 2005). However, CBMs are very degenerate at the primary amino acid level, and neither database consistently recognizes any of the 45 CBM families. Because CDD and pfam do not reliably recognize CBMs, we devised a multitiered search strategy to identify laforin orthologues (Fig. 1 B). First, we performed BLASTp (Altschul et al., 1997) searches using the DSP motif HCXXGXXR as an index sequence and identified 20 T. gondii proteins containing this motif. Because laforin contains an amino-terminal CBM and CBMs contain 80–100 amino acids, we eliminated two of these proteins because their HCXXGXXR motif was within the first 80 amino acids. We next performed a secondary BLAST using the NCBI nonredundant (nr) database with each of the remaining 18 proteins minus their DSP domain. If the protein contained a CBM, the BLAST identified other CBM-containing proteins. Using this strategy, we identified one protein, which we refer to as T. gondii laforin (Tg-laforin), that met the aforementioned criteria. Tg-laforin and Homo sapiens laforin (Hs-laforin) are 37% identical (Fig. 1 C). Importantly, Tg-laforin contains all of the residues important for carbohydrate binding as well as the signature residues of a DSP (Fig. 1 A).
With the discovery of a putative laforin orthologue in T. gondii, we extended our search methods to identify additional orthologues using a variety of genome databases (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). Using this strategy, we identified laforin orthologues in the four classes of vertebrates with sequenced genomes (mammals, aves, amphibians, and osteichthyes; Fig. 1, A and D). In addition, we identified putative laforin orthologues in four additional protists: Eimeria tenella, Tetrahymena thermophila, Paramecium tetraurelia, and Cyanidioschyzon merolae (Fig. 1, A and C). Although Hs-laforin contains 331 amino acids, the putative protist orthologues varied in predicted size from 323 to 727 amino acids. However, each putative orthologue contained the signature amino acids of a CBM20 and DSP; that is, four invariant aromatic amino acids (Hs-laforin F5, W32, W60, and W99) as well as DX30CX2GX2R, respectively (Fig. 1 A). Despite exhaustive efforts (we searched ∼170 eukaryotic genomes and ∼670 bacterial and archaeal genomes), we did not identify any other putative laforin orthologues. Thus, laforin is absent in all traditional nonvertebrate model organisms (e.g., yeast, fly, and worms). Laforin orthologues exist in all classes of vertebrates in which sequence data is available and in the five protists that we identified (Fig. 1, A, C, and D).
Biochemical properties and subcellular localization of laforin orthologues
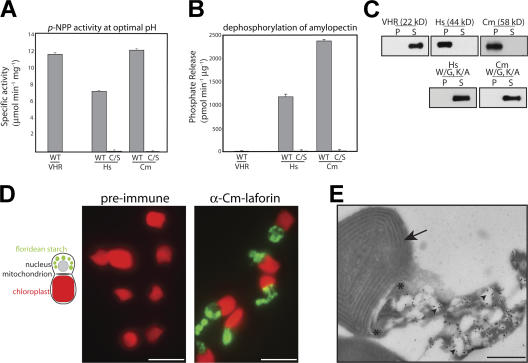
C. merolae laforin (Cm-laforin) shares the least identity with Hs-laforin (Fig. 1 C). As such, we reasoned that if it exhibited similar in vitro characteristics as Hs-laforin, the other putative orthologues were likely to as well. To test whether the identified protist proteins had similar biochemical characteristics as Hs-laforin and were thus laforin orthologues, we cloned the putative orthologue from C. merolae (Cm-laforin) and purified recombinant protein from bacteria (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). Characteristic of all DSPs, Hs-laforin exhibits phosphatase activity against the artificial substrate para-nitrophenylphosphate (p-NPP; Fig. 2 A; Ganesh et al., 2000). Cm-laforin also used p-NPP as an artificial substrate with similar kinetics as Hs-laforin (Table I) and displayed a similar specific activity (Fig. 2 A). In addition to activity against p-NPP, we recently showed that recombinant Hs-laforin releases phosphate from amylopectin (Worby et al., 2006) and that this activity is unique to laforin (Worby et al., 2006). Additionally, we fused the CBM of laforin to the DSP VH1 related (VHR) and demonstrated that although this fusion protein was an active phosphatase, it did not liberate phosphate from amylopectin (Worby et al., 2006). Fig. 2 B shows that like Hs-laforin, Cm-laforin displays a robust ability to release phosphate from amylopectin, whereas VHR does not hydrolyze phosphate from amylopectin. As predicted, the catalytically inactive Cm-laforin–C/S mutant displayed no activity against either substrate (Fig. 2, A and B). Additionally, Tg-laforin also displayed activity against both p-NPP and amylopectin (unpublished data).
Figure 2.
Biochemical characterization of laforin orthologues. (A) Specific activity of VHR, Hs-laforin, Hs-laforin–C/S, Cm-laforin, and Cm-laforin–C/S against p-NPP at their respective optimal pH. WT, wild type. (B) Phosphate release measured by malachite green assays using VHR, Hs-laforin, Hs-laforin–C/S, Cm-laforin, and Cm-laforin–C/S against amylopectin at their respective optimal pH. (A and B) Error bars indicate mean ± SD. (C) Recombinant histidine-tagged proteins were incubated with 5 mg/ml amylopectin, amylopectin was pelleted by ultracentrifugation, and proteins in the pellet (P) and supernatant (S) were visualized by Western analysis as described. VHR, H. sapiens VHR; Hs, Hs-laforin; Cm, Cm-laforin. Mutated residues are marked with an asterisk in Fig. 1 A. (D) A schematic of a nondividing C. merolae cell, defining the position of the chloroplast, mitochondrion, nucleus, and floridean starch. Cells were fixed, incubated with preimmune serum (left) or α–Cm-laforin antibody (right), and probed with a FITC-conjugated α–rabbit secondary antibody. Chloroplasts were visualized via their autofluorescence. (E) C. merolae cells were sectioned, probed with α–Cm-laforin and α–rabbit 10 nM gold-conjugated secondary antibodies, and visualized at 6,000× magnification. The arrow defines the chloroplast, asterisks mark the distal ends of a mitochondrion, and arrowheads mark three (of the many) floridean starch granules. Bars (D), 3 μm; (E) 500 nm.
Table I.
Kinetics of laforin orthologues and SEX4 utilizing p-NPP
| Vmax | kcat | km | kcat/km | |
|---|---|---|---|---|
| Hs-laforin | 3.79 μmol min−1 mg−1 | 2.41 s−1 | 0.323 mM | 7451 s−1 M−1 |
| Cm-laforin | 13.05 μmol min−1 mg−1 | 12.62 s−1 | 1.165 mM | 10832 s−1 M−1 |
| SEX4 | 2.41 μmol min−1 mg−1 | 1.54 s−1 | 0.410 mM | 3756 s−1 M−1 |
Kinetics of Hs-laforin, Cm-laforin, and A. thaliana Δ52-SEX4 (SEX4) utilizing p-NPP at pH 5.5, pH 6.0, and pH 6.0, respectively. Vmax is the maximum velocity of phosphate release from the substrate. Kcat is the catalytic constant of the phosphate-releasing reaction (equal to the turnover number). Km is the apparent equilibrium constant (the substrate concentration when the rate is at half of Vmax).
Hs-laforin is the only phosphatase in the human genome that contains a CBM and, as such, is predicted to be the only phosphatase that binds carbohydrates. Cm-laforin and Tg-laforin bound amylopectin to the same extent as Hs-laforin (Fig. 2 C and not depicted). Conversely, VHR did not bind amylopectin (Fig. 2 C). Wang et al. (2002) previously demonstrated that conserved tryptophan and lysine residues (Fig. 1 A) that participate in binding to the sugar are necessary for Hs-laforin to bind amylopectin (Fig. 2 C). Accordingly, mutation of these corresponding residues in Cm-laforin also abolished its ability to bind amylopectin (Fig. 2 C). These mutations also considerably reduced the ability of Cm-laforin to release phosphate from amylopectin (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1) while only minimally affecting its p-NPP activity (Fig. S2 B). These data suggest that Cm-laforin must be positioned correctly via the CBM in order for the DSP domain to dephosphorylate amylopectin or that the CBM binding to the carbohydrate is needed to activate the DSP.
While laforin from all three species binds α-glucans in vitro, this result may not reflect the biological localization of laforin. Moreover, the localization of Hs-laforin has never been determined in wild-type cells or tissues (Chan et al., 2004; our unpublished data). Because we identified multiple new systems to study laforin, we investigated laforin's localization in C. merolae. A C. merolae cell contains a chloroplast, mitochondrion, and nucleus and, when grown in continuous light, accumulates vast storages of floridean starch (Fig. 2 D, schematic; Viola et al., 2001). We fixed C. merolae cells and probed them with an affinity-purified α–Cm-laforin antibody. We found that endogenous Cm-laforin localized in punctate accumulations throughout the cytoplasm of cells (Fig. 2 D). To further define the localization of Cm-laforin, we performed immunogold electron microscopy staining. Ultra-thin sections of C. merolae cells were probed with the affinity-purified α–Cm-laforin antibody and a 10-nm gold particle–conjugated goat α–rabbit secondary antibody. Positive staining was observed surrounding the floridean starch granules (Fig. 2 E, arrowheads). No Cm-laforin was observed within the granules because before sectioning, no protein would have access to this region. In addition, no background staining was observed with the secondary antibody alone (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). Thus, as we hypothesized, endogenous laforin binds the outer region of insoluble carbohydrates.
Cm-laforin and Tg-laforin possess the same three in vitro properties as Hs-laforin: both use p-NPP as an artificial substrate, bind amylopectin, and release phosphate from amylopectin. Accordingly, the laforin orthologues in vertebrates and the five mentioned protists contain the critical signature primary amino acid structure of a CBM20 and DSP. Thus, our integrated bioinformatics searches for combined CBM and DSP domains correctly predicted the biochemical properties of Cm-laforin. Because the laforin orthologues are the only proteins in any of these genomes that contain a CBM and DSP, we hypothesized that these organisms may have acquired laforin from a common ancestor.
Evolutionary lineage of laforin
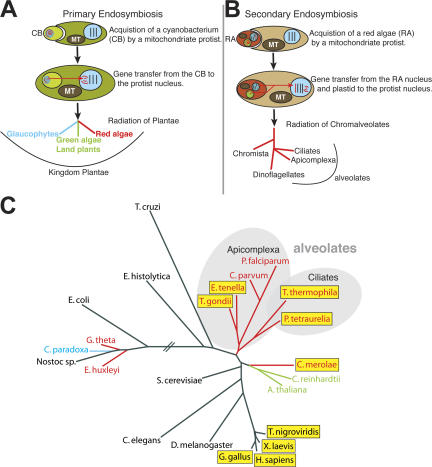
The key to the evolutionary lineage of laforin lies in the origin of the aforementioned five protists. The chromalveolate hypothesis postulates (Cavalier-Smith, 1999) that a distinct sequence of events led to the evolution of the kingdom Plantae and to subsequent progeny, including the five aforementioned protists. As illustrated in Fig. 3 A, a mitochondriate protist engulfed a cyanobacterium (Cavalier-Smith, 1982; Bhattacharya and Medlin, 1998) and eventually gave rise to the kingdom Plantae (Cavalier-Smith, 2004). Once Plantae was established, a second endosymbiosis involving red algae (Gillott and Gibbs, 1980) gave rise to the chromalveolates (Fig. 3 B; Cavalier-Smith, 1999). These engulfments were accompanied with the coevolution of “various manifestations of mitochondria” (Embley and Martin, 2006) and various forms of carbohydrate storage (Viola et al., 2001). These combined evolutionary events resulted in organisms possessing a mitosome, a hydrogenosome, or a true mitochondrion, and some organisms evolved floridean starch as their storage carbohydrate. We hypothesized that interspersed within these evolutionary events, organisms maintained, gained, or lost laforin.
Figure 3.
Evolutionary lineage of laforin. (A) Primary endosymbiosis hypothesized by the chromalveolate hypothesis (Cavalier-Smith, 1999). A cyanobacterium (CB) was engulfed by a mitochondriate protist (Cavalier-Smith, 1982; Bhattacharya and Medlin, 1998). MT, mitochondrion. Over generations, gene transfer occurred between the engulfed cyanobacterium and protist, the cyanobacterium was reduced to a plastid bound by two membranes, and the plastid-containing protist radiated into the founding members of the kingdom Plantae (Cavalier-Smith, 2004). (B) Secondary endosymbiosis hypothesized by the chromalveolate hypothesis (Cavalier-Smith, 1999). A red alga (RA) was engulfed by a mitochondriate protist (Gillott and Gibbs, 1980). Over generations, gene transfer occurred from the red alga nucleus and plastid to the nucleus of the protist, the red alga was reduced to a plastid bound by three or four membranes, and the new protist radiated into the kingdom Chromista and the alveolates, which are collectively referred to as the chromalveolates (Cavalier-Smith, 1999). The figure expands on the work of Weber et al. (2006). (C) Conservation of laforin orthologues. A phylogeny of the small subunit ribosomal RNA sequences was generated as described in Materials and methods, and accession numbers are listed in Table S3 (available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). Organisms containing laforin are boxed in yellow. Organisms from green algal descent are in green, organisms from glaucophyte descent are in blue, and organisms from red algal descent are in red. Alveolates are shaded with a gray background.
To trace the lineage of laforin, we generated a phylogeny derived from the small subunit ribosomal RNA gene of organisms belonging to diverse evolutionary niches and highlighted the organisms whose genome contains laforin (Fig. 3 C). This phylogenetic analysis revealed that each of the five protists containing a laforin orthologue is of red algal descent. However, the genome of some organisms of red algal descent lack laforin (Fig. 3 C). To determine why some organisms of red algal descent lack laforin, we analyzed the biology of each of the organisms in Fig. 3 C. We discovered that each organism of red algal descent that contained laforin also contained a true mitochondrion and produced floridean starch. Conversely, organisms of red algal descent lacking laforin either lacked a true mitochondrion or did not produce floridean starch. For example, Plasmodium falciparum is of red algal descent and possesses mitochondria; however, it does not produce floridean starch and, thus, lacks laforin (Fig. 3 C). Similarly, Cryptosporidium parvum is of red algal descent and produces floridean starch, but it has mitosomes instead of mitochondria and, thus, lacks laforin (Fig. 3 C). Conversely, C. merolae is a red alga that produces floridean starch and contains a single mitochondrion and, in agreement with our established criteria, contains laforin. Additionally, glaucophytes and green algae/land plants lack a laforin orthologue because they evolved as contemporaries of red algae and not as descendents (Fig. 3 A). Thus, our analyses generated three criteria to predict whether a protist's genome possesses laforin: the organism must (1) be of red algal descent, (2) possess a true mitochondrion, and (3) produce floridean starch. To determine whether our criteria correctly predicted the presence of laforin, we investigated the biology of each organism from the 168 eukaryotic genomes we probed. We found that in each case, our criteria correctly predicted the presence or absence of laforin (Table S5, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1).
A laforin-like protein in plants
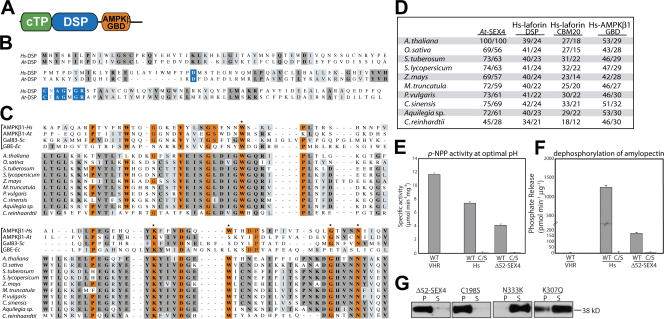
Protists such as T. gondii use insoluble floridean starch as an energy source when transitioning from inactive/hibernating life cycle stages to active/replicative stages (for review see Coppin et al., 2003). Likewise, C. merolae, a red alga that contains laforin, synthesizes insoluble floridean starch during the day and uses it as a source of energy at night. Plants have a similar diurnal cycle, producing insoluble carbohydrate in the form of starch during the day and catabolizing it during the night. Because Hs-laforin has been implicated in carbohydrate metabolism and we show that Cm-laforin binds and releases phosphate from amylopectin, we hypothesized that laforin plays a vital role in insoluble carbohydrate metabolism. Thus, we predicted that plants would also have a laforin-like activity; however, we were unable to identify a laforin orthologue in plants. Recently, several starch excess mutants that accumulate starch have been described in plants (Blennow et al., 2002; Smith et al., 2005; Zeeman et al., 2007); one of these is attributed to mutations in the starch excess 4 (SEX4) gene (At3g52180; Niittyla et al., 2006). Niittyla et al. (2006) and Kerk et al. (2006) demonstrated that the Arabidopsis thaliana SEX4 gene (previously identified as a phosphatase and called AtPTPKIS1; Fordham-Skelton et al., 2002) encodes a protein containing a chloroplast-targeting peptide (cTP) and DSP domain at its amino terminus followed by a CBM-like domain at its carboxy terminus (Fig. 4 A), suggesting that SEX4 might be a laforin-like phosphatase (Niittyla et al., 2006).
Figure 4.
Conservation and biochemical properties of SEX4. (A) Domain topography of SEX4. cTP, chloroplast-targeting peptide; DSP, dual specific phosphatase; GBD, glycogen-binding domain. (B) Alignment of the DSP of Hs-laforin (Hs-DSP) and the DSP of SEX4 (At-DSP). Residues boxed in blue are part of the DSP catalytic site, those boxed in dark gray are identical, and those boxed in light gray are conserved substitutions. (C) Alignment of the AMPKβ-GBD of four founding members of the family (top four, marked by a bracket; Polekhina et al., 2003) and the AMPKβ-GBD of SEX4 orthologues. Residues boxed in orange are highly conserved amino acids among multiple AMPKβ-GBD (Polekhina et al., 2003) proteins. Residues boxed in dark gray are identical, and those boxed in light gray are conserved substitutions. Asterisks mark residues necessary for carbohydrate binding (Polekhina et al., 2003, 2005). Accession numbers are listed in Table S5 (available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). (D) Percent similarity and identity of full-length At-SEX4 compared with SEX4 orthologues, the DSP of Hs-laforin to the DSP of each SEX4 orthologue, and the CBM20 of Hs-laforin to the glycogen-binding domain (GBD) of each SEX4 orthologue as well as the percent similarity of the GBD of Hs-AMPKβ1 to the glycogen-binding domain of each SEX4 orthologue. (E) Specific activity of VHR, Hs-laforin, Hs-laforin–C/S, Δ52-SEX4, and Δ52-SEX4-C/S against p-NPP at their respective optimal pH. (F) Phosphate release measured by malachite green assays using VHR, Hs-laforin, Hs-laforin–C/S, Δ52-SEX4, and Δ52-SEX4-C/S against amylopectin at their respective optimal pH. (E and F) Error bars indicate mean ± SD. (G) At-SEX4 binding to amylopectin. Recombinant histidine-tagged proteins were incubated with 5 mg/ml amylopectin, amylopectin was pelleted by ultracentrifugation, and proteins in the pellet (P) and supernatant (S) were visualized by Western analysis as described. Δ52-SEX4, amino-terminal truncation of SEX4; C198S, Δ52-SEX4-C198S; N333K, Δ52-SEX4-N333K; K307Q, Δ52-SEX4-K307Q.
The DSP of SEX4 shares the key DX30CX2GX2R catalytic residues with the DSP of Hs-laforin and is 24% identical to Hs-laforin (Fig. 4, B and D). Conversely, the CBM of SEX4 lacks many of the invariant CBM20 residues (Figs. 4 C vs. 1 A) and shares only 18% identity with the CBM of Hs-laforin (Fig. 4 D). Instead, a sequence search using the CBM of SEX4 shows that it is most similar to another class of CBM, the AMP-activated protein kinase β–glycogen-binding domain (AMPKβ−GBD) family (Polekhina et al., 2003), and not to CBM20 (Fig. 4, C and D). Despite their structural differences, both CBM20 and the AMPKβ-GBD domains interact with individual glycan chains of carbohydrates (Boraston et al., 2004; Polekhina et al., 2005), suggesting that SEX4 could bind starch via its AMPKβ-GBD. Thus, SEX4 contains similar domains to laforin, but the domains are arranged in the opposite orientation (Figs. 1 A vs. 4 A). We next performed BLASTp searches of various databases (Table S1) and found that SEX4 is conserved in all land plants and in Chlamydomonas reinhardtii, a single-cell green alga closely related to the progenitor of land plants (Fig. 4, C and D). Thus, SEX4 likely evolved before or during the establishment of green algae and performs a kingdom-wide function in Plantae.
To ascertain whether SEX4 possesses biochemical properties similar to laforin, we cloned A. thaliana SEX4 and assayed purified recombinant SEX4 protein (At-SEX4; Fig. S1 B). Because the cTP of SEX4 is highly hydrophobic and renders the protein insoluble, we deleted the first 52 amino acids and used purified recombinant HIS-tagged Δ52-SEX4 for our assays (Fig. S1 B). We found that Δ52-SEX4 has a similar specific activity and possesses similar kinetics as Hs-laforin against p-NPP (Fig. 4 E and Table I) and efficiently liberates phosphate from amylopectin (Fig. 4 F). Conversely, mutation of the active site cysteine to serine abolished these activities (Fig. 4, E and F). Additionally, wild-type (Δ52-SEX4) and catalytically inactive SEX4 (Δ52-SEX4-C198S) bind amylopectin similar to Hs-laforin (Fig. 4 G). Importantly, mutations in key conserved AMPKβ-GBD residues that form essential hydrogen bonds with the sugar (Polekhina et al., 2003, 2005) abolish this interaction (Fig. 4 G) while minimally affecting the phosphatase activity of SEX4 (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). These mutations considerably reduced the ability of SEX4 to release phosphate from amylopectin (Fig. S4 B). Thus, like Cm-laforin, SEX4 must also be positioned correctly via the CBM in order for the DSP domain to dephosphorylate amylopectin.
Clearly, SEX4 and the laforins contain both a functional CBM and a DSP domain highly specific for dephosphorylating amylopectin. Additionally, we speculate that they are involved in insoluble carbohydrate metabolism. Because carbohydrate metabolism evolved independently in the kingdom Plantae and kingdom Animalia, the use of similar protein modules to regulate a key feature of carbohydrate metabolism in these lineages is a striking example of convergent evolution and strongly suggests that laforin and SEX4 might be functional equivalents.
SEX4 is a functional equivalent of laforin
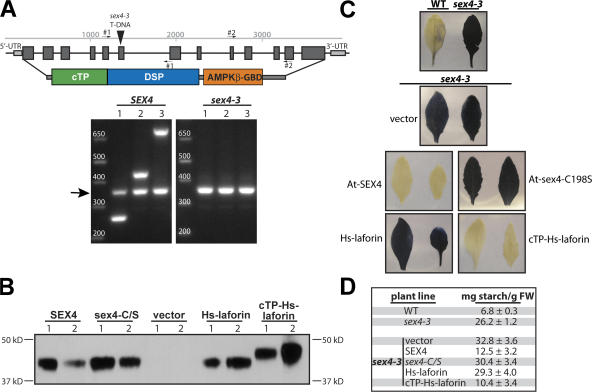
The SEX4 locus was recently mapped in A. thaliana to At3g52180, and multiple mutations in this gene display a starch excess phenotype (Niittyla et al., 2006; Sokolov et al., 2006). One characterized mutation is the sex4-3 allele that contains an Agrobacterium transferred DNA (T-DNA) insertion in the sixth exon (Niittyla et al., 2006) and leads to the disruption of SEX4 expression (Fig. 5 A). Because laforin and SEX4 are the only reported proteins in any kingdom that contain both functional CBM and DSP domains and because mutations in the gene expressing either protein results in aberrant carbohydrate accumulation, we postulated that SEX4 and laforin could be functional equivalents.
Figure 5.
Hs-laforin is a functional equivalent of sex4. (A) sex4-3 disruption and gene expression. A schematic of the At3g51280 locus with exons represented by dark gray boxes, the T-DNA insertion depicted as a black arrowhead, and a schematic of the SEX4 protein. Small arrows denote the primer sets used to confirm the absence of a SEX4 transcript in sex4-3 plants. RT-PCR results from wild-type and sex4-3 isolated mRNA. Lane 1, primer set 1 from the schematic; lane 2, primer set 2; lane 3, the 5′ primer from 1 and the 3′ primer from 2. The arrow indicates the size of the positive control UBC5. (B) Protein expression of transgenes. Tissue samples for Western analysis were taken from two independent T2 plants transformed with empty vector or an HA epitope–tagged transgene as indicated. 30 mg of whole tissue lysate was loaded in each lane. (C) Complementation of the starch excess phenotype. One leaf from the same two independent T2 plants as in Fig. 4 B were decolorized with hot ethanol and stained with iodine. Wild-type (WT) leaves contain little starch and do not stain; conversely, sex4-3 leaves contain high amounts of starch and are darkly stained. Similarly, transgenes that complement the phenotype do not stain, whereas those that do not complement stain dark. (D) Quantitation of the complementation. Starch was quantified from leaves of wild-type, sex4-3, and sex4-3 transgenic plants. Each sample is the mean of replicate samples ± standard error. For sex4-3 transgenic plants, six independent T2 plants isogenic to those used in Fig. 4 (B and C) were used to quantitate the amount of starch in leaves. FW, fresh weight.
To test this hypothesis, we transformed sex4-3 plants to generate stable lines expressing SEX4, sex4-C/S, Hs-laforin, and Hs-laforin fused behind a cTP (cTP–Hs-laforin) to target Hs-laforin to the chloroplast (like SEX4) and monitored protein expression of the transgenes (Fig. 5 B). We then assayed starch accumulation in wild-type, sex4-3, and sex4-3 transgenic plants. As per our prediction, transformants expressing SEX4 and cTP–Hs-laforin no longer displayed the starch excess phenotype, whereas the catalytically inactive sex4-C/S mutant and Hs-laforin transformants still accumulated excess starch (Fig. 5, C and D; and Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). Thus, the cTP–Hs-laforin fusion rescued the starch excess phenotype both qualitatively and quantitatively. Conversely, Hs-laforin lacking the cTP did not rescue any portion of the phenotype. Therefore, Hs-laforin is a functional equivalent of SEX4 that must be targeted to the chloroplast, just like SEX4, to perform the equivalent function.
Discussion
Our studies probe the molecular mechanism of LD. We identified laforin orthologues in specific protists and further showed that Hs-laforin and plant SEX4 are functional equivalents. Our results provide compelling evidence that a laforin-like activity is required to regulate the metabolism of amylopectin-like material across multiple kingdoms. Additionally, they demonstrate the nature of this activity; that is, the dephosphorylation of the carbohydrate itself, thus providing a molecular explanation for LD. Although there are examples of DSPs that dephosphorylate nonproteinacious substrates (such as phosphate and tensin homologue, the myotubularin family, and Sac domain phosphatases that dephosphorylate the inositol head group of phospholipids; Chung et al., 1997; Maehama and Dixon, 1998; Guo et al., 1999; Hughes et al., 2000; Taylor et al., 2000; Robinson and Dixon, 2006), ours is the first example of a family of phosphatases that dephosphorylate complex carbohydrates.
We demonstrate that laforin is not merely restricted to the genomes of vertebrates but is well conserved in the protists T. gondii, E. tenella, T. thermophila, P. tetraurelia, and C. merolae. Laforin's evolutionary lineage shows that it originated in a primitive red alga during early eukaryotic evolution. Despite its early origin, laforin was only maintained by organisms that synthesize floridean starch (such as the aforementioned five protists) and organisms that inhibit the production of insoluble carbohydrates (i.e., all vertebrates). Organisms that no longer performed either of these processes lost laforin. Conversely to laforin, we show that although SEX4 contains similar domains as laforin, its lineage differs in that SEX4 is conserved in all land plants as well as in C. reinhardtii, a close descendent of primitive green algae. Despite their different lineages, Hs-laforin performs the same function as the plant protein SEX4; thus, we propose that laforin and SEX4 are functional equivalents.
It must be noted that although laforin and SEX4 share a common function and similar domains, they are not orthologous proteins. They are not orthologues because (1) although they share similar CBMs, the CBMs belong to different classes and differ considerably with respect to the primary amino acids that are important for binding carbohydrates, and (2) the DSP and CBM of laforin and SEX4 are arranged in opposite orientations. Thus, it is likely that red and green algae independently evolved a phosphatase via convergent evolution that utilizes a similar mechanism to regulate insoluble carbohydrate metabolism.
Despite the independent means by which laforin and SEX4 evolved, they both dephosphorylate the same carbohydrate substrate and constitute a unique family of phosphatases. In addition, we demonstrate that endogenous Cm-laforin localizes around the floridean starch granules. Although most studies thus far suggest a carbohydrate substrate for laforin and SEX4, it is possible that they bind their respective amylopectin-like material (insoluble glycogen and starch, respectively) and dephosphorylate a proteinacious substrate. This proteinacious substrate would likely be involved in regulating carbohydrate metabolism, a process controlled by multiple levels of phosphorylation (Roach, 2002). Although the overall carbohydrate machinery differs substantially between mammals and plants, both systems contain common phosphoproteins that share conserved functions (Preiss et al., 1983; Vikso-Nielsen et al., 2002; Coppin et al., 2005). These proteins would be likely substrate candidates for laforin and SEX4. To address this hypothesis, we tested the majority of the mammalian candidates, but none of them served as a substrate for laforin (Worby et al., 2006; our unpublished data).
It is interesting that laforin and SEX4 are functional equivalents that dephosphorylate a complex carbohydrate and that the mutation of either gene results in the accumulation of insoluble carbohydrates in vertebrates and plants, respectively. Our understanding of the metabolism of insoluble carbohydrates in vertebrate systems is still in its infancy. In contrast, the plant community has made substantial progress in understanding the metabolism of starch (Smith et al., 2005; Zeeman et al., 2007). In plants, it is clear that the phosphorylation of glucose residues within starch is required for its proper accumulation and degradation (Blennow et al., 2002; Smith et al., 2005; Zeeman et al., 2007). In A. thaliana, glucan water dikinase (Ritte et al., 2002) and phosphoglucan water dikinase (Baunsgaard et al., 2005; Kotting et al., 2005) phosphorylate glucose monomers within amylopectin at the C6 and C3 position (Ritte et al., 2006), respectively. As with SEX4, mutations in the genes encoding glucan water dikinase and phosphoglucan water dikinase also yield a starch excess phenotype (Yu et al., 2001; Baunsgaard et al., 2005; Kotting et al., 2005). Phosphorylation is necessary for both starch accumulation and degradation; however, the timing of these phosphorylation and dephosphorylation events is unknown (Smith et al., 2005; Zeeman et al., 2007). Intriguingly, although glycogen, the soluble storage carbohydrate in vertebrates, contains little to no phosphate, detrimental insoluble carbohydrates like LBs are highly phosphorylated, just like amylopectin in plant starch (Schnabel and Seitelberger, 1968; Sakai et al., 1970). Therefore, it appears logical that laforin and SEX4 evolved to perform the critical role of dephosphorylating insoluble carbohydrates to allow their proper degradation.
This basic function of insoluble carbohydrate metabolism provides an intriguing explanation for both the existence of a laforin-like activity in protists and plants and the role of laforin in preventing LD. In protists and plants, carbohydrate dephosphorylation would be necessary for the utilization of insoluble carbohydrates as an energy source. When this activity is absent, these organisms accumulate unusable starch as in the sex4 mutants. In vertebrates, laforin would dephosphorylate nascent insoluble carbohydrates to inhibit the formation of detrimental LBs. In the absence of laforin, these nascent molecules increase in size and number and eventually cause LD.
Our work clearly demonstrates that a laforin-like activity is necessary for the proper metabolism of insoluble carbohydrates. This activity is required throughout multiple kingdoms and regulates an overlooked aspect of carbohydrate metabolism. It is striking that protists and plants have provided new insights into a human neurodegenerative disease involving aberrant carbohydrate metabolism that was described almost 100 yr ago by Lafora and Gluck (Lafora, 1911; Lafora and Gluck, 1911).
Materials and methods
Cloning, vectors, and purification of recombinant proteins
The complete open reading frame of Cm-laforin was cloned from cDNA provided by T. Kuroiwa (Rikkyo University, Tokyo, Japan) and SEX4 from SSP Consortium clone U14967 (Yamada et al., 2003). Cm-laforin and SEX4 were cloned into pET21a (Stratagene) according to standard protocols. A second pET21a SEX4 construct was generated because the full-length protein is largely insoluble. We truncated the first 52 amino acids of SEX4 to generate pET21a Δ52-SEX4. pET21a VHR (Denu et al., 1995) and pET21a Hs-laforin (Wang et al., 2002) have been described previously. Hs-laforin, SEX4, and sex4-C198S were cloned in frame of a triple HA tag into pCHF1 (Neff et al., 1999), which is a modified version of pPZP221 (Hajdukiewicz et al., 1994). pCHF1 contains the 35S cauliflower mosaic virus promoter, the Rubisco terminator from pea, and confers gentamicin resistance for selection in plants. Because Kerk et al. (2006) and Niittyla et al. (2006) demonstrated that the cTP of SEX4 targets SEX4 to the chloroplast, we fused the cTP of SEX4 (nucleotides 1–213) in frame with Hs-laforin and the triple HA tag in pCHF to generate pCHF cTP–Hs-laforin. All point mutations were generated with the QuikChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. All constructs were verified by DNA sequencing. Recombinant proteins were expressed with a carboxy-terminal six-histidine tag in Escherichia coli BL21 (DE3) CodonPlus cells (Stratagene). Fusion proteins were expressed and purified from soluble bacterial extracts using Ni2+-agarose affinity chromatography as described previously (Gentry et al., 2005).
Phosphatase assays
Hydrolysis of p-NPP was performed in 50-μl reactions containing 1× phosphate buffer (0.1 M sodium acetate, 0.05 M bis-Tris, 0.05 M Tris-HCl, and 2 mM DTT at the appropriate pH), 50 mM pNPP, and 100–500 ng of enzyme at 37°C for 1–5 min. The reaction was terminated by the addition of 200 μl of 0.25 M NaOH, and absorbance was measured at 410 nm. We tested the specific activity of each enzyme at pH 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, and 8.0. The optimal pH for each enzyme was as follows: Hs-laforin, pH 5.0; Cm-laforin, pH 5.5; SEX4, pH 6.0; and VHR, pH 6.0. Malachite green assays were performed as described previously (Harder et al., 1994) with the following modifications: 1× phosphate buffer, 100–500 ng of enzyme, and ∼45 μg of amylopectin in a final volume of 20 μl. The reaction was stopped by the addition of 20 μl of 0.1 M N-ethylmaleimide and 80 μl of malachite green reagent. Absorbance was measured at 620 nm. We tested the specific activity of each enzyme at the same pH units as above. The optimal pH for each enzyme was as follows: Hs-laforin, pH 7.0; Cm-laforin, pH 6.0; and SEX4, pH 8.0.
Phylogenetic analyses and sequence alignment
The sequences of laforin and SEX4 orthologues were obtained by performing tBLASTn searches using the GenBank dbEST database or BLASTp and PSI-BLAST (Altschul et al., 1997) searches using GenBank eukaryote genome and nr databases, the C. merolae genome project, Department of Energy Joint Genome Institute Resource, The Institute for Genomic Research, ToxoDB, GeneDB, Genoscope, and Tetrahymena Genome Database. Accession numbers are listed in Tables S2 and S5 (available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1). The web address for each database is listed in Table S1. A list of each genome that we investigated and a reason why an organism's genome lacks laforin is listed in Table S4. Amino acid sequences of laforin orthologues were aligned by ClustalW (Thompson et al., 1994) and refined manually using MacVector. Small subunit ribosomal RNA sequences were obtained by performing BLASTn using GenBank from all organisms and nr databases, and accession numbers are listed in Table S3. The phylogenetic tree was generated from a ClustalW (Thompson et al., 1994) multiple sequence alignment using PROTDIST and FITCH from the PHYLIP 3.65 software package and was displayed using HYPERTREE 1.0.0 (Pfizer; Bingham and Sudarsanam, 2000).
Plant material
Homozygous sex4-3 plants (T-DNA insertion line SALK_102567; Alonso et al., 2003) were isolated by PCR. Stable transgenic plant lines were generated by Agrobacterium-mediated floral dipping (Clough and Bent, 1998), and seeds were sterilized, plated on standard growth medium (Valvekens et al., 1988), and selected for using 100 μg/ml gentamycin per standard protocols (Valvekens et al., 1988; Clough and Bent, 1998). Plants were grown in Promix-HP soil at 22°C with supplemental lighting conditions of 16-h days. To stain starch in leaves, leaves were decolorized in 80% (vol/vol) ethanol, stained with an iodine solution, and destained in water. Starch content was quantified as previously described (Kotting et al., 2005). mRNA was obtained using an RNeasy Plant Mini kit (QIAGEN), and first-strand synthesis was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) according to the manufacture's recommendations. Four primer sets were used to test for the presence of transcripts in wild-type (Columbia) and sex4-3 plants. Three primer sets to the SEX4 transcript and a positive control to UBC5, the UBC5 primer set, was included in each PCR tube. Plant whole leaf lysate was obtained as described previously (Nimchuk et al., 2000).
Antibodies and Western analysis
The α–Hs-laforin and α–Cm-laforin antibodies were generated by immunizing rabbits with recombinant Hs-laforin or α–Cm-laforin, and antibodies were affinity purified from the serum with a HiTrap NHS-activated HP affinity column (GE Healthcare) of Hs-laforin or Cm-laforin protein, respectively. The α–Cm-laforin antibody was generated in a similar manner. Recombinant Hs-laforin and Cm-laforin were detected with their respective primary antibodies followed by goat α–rabbit HRP (GE Healthcare). Recombinant VHR and SEX4 were detected with α-HIS-HRP (Santa Cruz Biotechnology, Inc.). Protein expression of A. thaliana transgenes was monitored by Western analysis using rat anti-HA (clone 3F10; Roche) and goat α–rat HRP (Chemicon).
C. merolae cell culture, immunofluorescence, and immunogold electron microscopy
C. merolae 10D-14 (Toda et al., 1998) was provided by T. Kuroiwa and grown asynchronously at pH 2.5 in 2× Allens's medium at 42°C under continuous illumination as described previously (Minoda et al., 2004). For immunofluorescence, cells were fixed, washed, and blocked as previously described (Nishida et al., 2004). Cells were then probed with 1:100 preimmune serum or 1:1,000 α–Cm-laforin antibody followed by 1:1,000 AlexaFluor488 goat α–rabbit antibody (Invitrogen). Chloroplasts were visualized by their autofluorescence. Immunofluorescence was performed using a light microscope (DMR; Leica) with a PL APO 63× 1.32 NA oil objective (Leica) at room temperature, and images were captured with a CCD camera (C4742-95; Hamamatsu) using OpenLab 4.0.1 software (Improvision). For immunogold EM, cells were fixed, washed, sectioned, and blocked as previously described (Nishida et al., 2003). Sections were immunostained with 1:50 preimmune serum or 1:250 α–Cm-laforin antibody and with 10 nm of gold particle–conjugated goat α–rabbit antibody. Grids were viewed using a transmission electron microscope (1200EX II; JEOL), and images were collected using a digital camera (ORIUS SC600; Gatan) and Digital Micrograph software (Gatan). Photoshop (Adobe) and Illustrator (Adobe) were used to generate figures of all images.
Online supplemental material
Fig. S1 shows purified recombinant Cm-laforin and SEX4. Fig. S2 shows the phosphatase activity of Cm-laforin mutants using p-NPP and amylopectin as substrates. Fig. S3 shows immuno-EM of a C. merolae cell probed with the secondary antibody alone. Fig. S4 shows the phosphatase activity of SEX4 mutants using p-NPP and amylopectin as substrates. Fig. S5 shows the quantitation of starch in wild-type, sex4-3, and transgenic plants. Table S1 provides data about non-NCBI databases, Table S2 provides accession numbers for laforin orthologues, and Table S3 provides small subunit ribosomal RNA accession numbers. Table S4 provides data about the genomes investigated for the presence of laforin, and Table S5 provides accession numbers for AMPKβ-GBD proteins and SEX4 orthologues. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704094/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Tsuneyoshi Kuroiwa, Gregory S. Taylor, Andreas P. Weber, Fred L. Robinson, Neal M. Alto, Michael J. Begley, David J. Pagliarini, Jorrit M. Enserink, Nathan S. Blow, and Matthew J. Rardin for reagents and/or insightful discussions. Immunogold electron microscopy was carried out with the assistance of Dr. Marilyn Farquhar and Timo Meerloo (Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA).
This work was supported by National Institutes of Health (NIH) grants DK18024-31 and DK18849-31 to J.E. Dixon, grants T32 CA09523 and T32 HD 007203-23 to M.S. Gentry, grant T32 DK 07494 to S. Mattoo, and grant T32 GM07752 to R.H. Dowen as well as by the Walther Cancer Institute.
Abbreviations used in this paper: AMPKβ-GBD, AMP-activated protein kinase β–glycogen-binding domain; CBM, carbohydrate-binding module; Cm-laforin, C. merolae laforin; cTP, chloroplast-targeting peptide; DSP, dual specificity phosphatase; Hs-laforin, Homo sapiens laforin; LB, Lafora body; LD, Lafora disease; p-NPP, para-nitrophenylphosphate; SEX4, starch excess 4; Tg-laforin, T. gondii laforin; VHR, VH1 related.
References
- Alonso, A., J. Sasin, N. Bottini, I. Friedberg, I. Friedberg, A. Osterman, A. Godzik, T. Hunter, J. Dixon, and T. Mustelin. 2004. Protein tyrosine phosphatases in the human genome. Cell. 117:699–711. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., A.N. Stepanova, T.J. Leisse, C.J. Kim, H. Chen, P. Shinn, D.K. Stevenson, J. Zimmerman, P. Barajas, R. Cheuk, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 301:653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D.J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., L. Coin, R. Durbin, R.D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E.L.L. Sonnhammer, et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baunsgaard, L., H. Lutken, R. Mikkelsen, M.A. Glaring, T.T. Pham, and A. Blennow. 2005. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 41:595–605. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, D., and L. Medlin. 1998. Algal phylogeny and the origin of land plants. Plant Physiol. 116:9–15. [Google Scholar]
- Bingham, J., and S. Sudarsanam. 2000. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 16:660–661. [DOI] [PubMed] [Google Scholar]
- Blennow, A., T.H. Nielsen, L. Baunsgaard, R. Mikkelsen, and S.B. Engelsen. 2002. Starch phosphorylation: a new front line in starch research. Trends Plant Sci. 7:445–450. [DOI] [PubMed] [Google Scholar]
- Boraston, A.B., D.N. Bolam, H.J. Gilbert, and G.J. Daview. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, S., and G. Karpati. 1981. Sweat gland duct cells in Lafora disease: diagnosis by skin biopsy. Neurology. 31:1564–1568. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 1982. The origin of plastids. Biol. J. Linn. Soc. 17:289–306. [Google Scholar]
- Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellatae, and sporozoan plastid origins and the eukaryote family tree. J. Euk. Microbiol. 46:347–366. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 2004. Only six kingdoms of life. Proc. R. Soc. Lond. B. 271:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E.M., E.J. Young, L. Ianzano, I. Munteanu, X. Zhao, C.C. Christopoulos, G. Avanzini, M. Elia, C.A. Ackerley, N.J. Jovic, et al. 2003. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 35:125–127. [DOI] [PubMed] [Google Scholar]
- Chan, E.M., C.A. Ackerley, H. Lohi, L. Ianzano, M.A. Cortez, P. Shannon, S.W. Scherer, and B.A. Minassian. 2004. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum. Mol. Genet. 13:1117–1129. [DOI] [PubMed] [Google Scholar]
- Chung, J.K., F. Sekiya, H.S. Kang, C. Lee, J.S. Han, S.R. Kin, Y.S. Bae, A.J. Morris, and S.G. Rhee. 1997. Synaptojanin inhibition of phospholipase D activity by hydrolysis of phospatidylinositol 4,5-bisphosphate. J. Biol. Chem. 272:15980–15985. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and A.F. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. [DOI] [PubMed] [Google Scholar]
- Collins, G.H., R.R. Cowden, and A.H. Nevis. 1968. Myoclonus epilepsy with Lafora bodies. An ultrastructural and cytochemical study. Arch. Pathol. 86:239–254. [PubMed] [Google Scholar]
- Coppin, A., F. Dzierszinski, S. Legrand, M. Mortuaire, D. Ferguson, and S. Tomavo. 2003. Developmentally regulated biosynthesis of carbohydrate and storage polysaccharide during differentiation and tissue cyst formation in Toxoplasma gondii. Biochimie. 85:353–361. [DOI] [PubMed] [Google Scholar]
- Coppin, A., J. Varré, L. Lienard, D. Dauvillée, Y. Guérardel, M. Soyer-Gobillard, A. Buléon, S. Ball, and Stanislas Tomavo. 2005. Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J. Mol. Evol. 60:257–267. [DOI] [PubMed] [Google Scholar]
- Coutinho, P.M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach. In Recent Advances in Carbohydrate Bioengineering. H.J. Gilbert, G.J. Davies, B. Henrissat, and B. Svensson, editors. Royal Society of Chemistry, Cambridge. 3–12.
- Denu, J.M., G. Zhou, L. Wu, R. Zhao, J. Yuvaniyama, M.A. Saper, and J.E. Dixon. 1995. The purification and characterization of a human dual- specific protein tyrosine phosphatase. J. Biol. Chem. 270:3796–3803. [DOI] [PubMed] [Google Scholar]
- Denu, J.M., J.A. Stuckey, M.A. Saper, and J.E. Dixon. 1996. Form and function in protein dephosphorylation. Cell. 87:361–364. [DOI] [PubMed] [Google Scholar]
- Dubey, J.P., D.S. Lindsay, and C.A. Speer. 1998. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 11:267–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley, T.M., and W. Martin. 2006. Eukaryotic evolution, changes and challenges. Nature. 440:623–630. [DOI] [PubMed] [Google Scholar]
- Fordham-Skelton, A.P., P. Chilley, V. Lumbreras, S. Reignoux, T.R. Fenton, C.C. Dahm, M. Pages, and J.A. Gatehouse. 2002. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 29:705–715. [DOI] [PubMed] [Google Scholar]
- Ganesh, S., K.L. Agarwala, K. Ueda, T. Akagi, K. Shoda, T. Usui, T. Hashikawa, H. Osada, A.V. Delgado-Escueta, and K. Yamakawa. 2000. Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual-specificity phosphatase associated with polyribosomes. Hum. Mol. Genet. 9:2251–2261. [DOI] [PubMed] [Google Scholar]
- Ganesh, S., K.L. Agarwala, K. Amano, T. Suzuki, A.V. Delgado-Escueta, and K. Yamakawa. 2001. Regional and developmental expression of Epm2a gene and its evolutionary conservation. Biochem. Biophys. Res. Commun. 283:1046–1053. [DOI] [PubMed] [Google Scholar]
- Ganesh, S., A.V. Delgado-Escueta, T. Sakamoto, M.R. Avila, J. Machado-Salas, Y. Hoshii, T. Akagi, H. Gomi, T. Suzuki, K. Amano, et al. 2002. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum. Mol. Genet. 11:1251–1262. [DOI] [PubMed] [Google Scholar]
- Ganesh, S., N. Tsurutani, T. Suzuki, Y. Hoshii, T. Ishihara, A.V. Delgado-Escueta, and K. Yamakawa. 2004. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem. Biophys. Res. Commun. 313:1101–1109. [DOI] [PubMed] [Google Scholar]
- Gentry, M.S., C.A. Worby, and J.E. Dixon. 2005. Insights into Lafora disease: Malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl. Acad. Sci. USA. 102:8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillott, M.A., and S.P. Gibbs. 1980. The cryptomonad nucleomorth: its ultrastructure and evolutionary significance. J. Phycol. 16:558–568. [Google Scholar]
- Guérardel, Y., D. Leleu, A. Coppin, L. Liénard, C. Slomianny, G. Strecker, S. Ball, and S. Tomavo. 2005. Amylopectin biogenesis and characterization in the protozoan parasite Toxoplasma gondii, the intracellular development of which is restricted in the HepG2 cell line. Microbes Infect. 7:41–48. [DOI] [PubMed] [Google Scholar]
- Guo, S., L.E. Stolz, S.M. Lemrow, and J.D. York. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274:12990–12995. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Z. Svab, and P. Maliga. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25:989–994. [DOI] [PubMed] [Google Scholar]
- Harder, K.W., P. Owen, L.K. Wong, R. Aebersold, I. Clark-Lewis, and F.R. Jirik. 1994. Characterization and kinetic analysis of the intracellular domain of human protein tyrosine phosphatase beta (HPTP beta) using synthetic phosphopeptides. Biochem. J. 298:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriman, D.G., J.H. Millar, and A.C. Stevenson. 1955. Progressive familial myoclonic epilepsy in three families: its clinical features and pathological basis. Brain. 78:325–349. [DOI] [PubMed] [Google Scholar]
- Hughes, W.E., F.T. Cooke, and P.J. Parker. 2000. Sac phosphatase domain proteins. Biochem. J. 350:337–352. [PMC free article] [PubMed] [Google Scholar]
- Ianzano, L., J. Zhang, E.M. Chan, X. Zhao, H. Lohi, S.W. Scherer, and B.A. Minassian. 2005. Lafora progressive Myoclonus Epilepsy mutation database-EPM2A and NHLRC1 (EMP2B) genes. Hum. Mutat. 26:397. [DOI] [PubMed] [Google Scholar]
- Kerk, D., T.R. Conley, F.A. Rodriguez, H.T. Tran, M. Nimick, D.G. Muench, and G.B. Moorhead. 2006. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 46:400–413. [DOI] [PubMed] [Google Scholar]
- Kissinger, J.C., B. Gajria, L. Li, I.T. Paulsen, and D.S. Roos. 2003. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 31:234–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotting, O., K. Pusch, A. Tiessen, P. Geigenberger, M. Steup, and G. Ritte. 2005. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 137:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafora, G.R. 1911. Uber des Vorkommen amyloider KJrperchen im innern der Ganglienzellen. Virchows Arch. f. Path. Anat. 205:295. [Google Scholar]
- Lafora, G.R., and B. Gluck. 1911. Beitrag zur histopathologie der myoklonischen epilepsie. Z Ges Neurol Psychiatr. 6:1–14. [Google Scholar]
- Maehama, T., and J.E. Dixon. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273:13375–13378. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., J.B. Anderson, P.F. Cherukuri, C. DeWeese-Scott, L.Y. Geer, M. Gwadz, S. He, D.I. Hurwitz, J.D. Jackson, Z. Ke, et al. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian, B.A., J.R. Lee, J.A. Herbrick, J. Huizenga, S. Soder, A.J. Mungall, I. Dunham, R. Gardner, C.Y. Fong, S. Carpenter, et al. 1998. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 20:171–174. [DOI] [PubMed] [Google Scholar]
- Minoda, A., R. Sakagami, F. Yagisawa, T. Kuroiwa, and K. Tanaka. 2004. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 45:667–671. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., S.M. Nguyen, E.J. Malancharuvil, S. Fujioka, T. Noguchi, H. Seto, M. Tsubuki, T. Honda, S. Takatsuto, S. Yoshida, and J. Chory. 1999. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA. 96:15316–15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittyla, T., S. Comparot-Moss, W.-L. Lue, G. Messerli, M. Trevisan, M.D.J. Seymour, J.A. Gatehouse, D. Villadsen, S.M. Smith, J. Chen, et al. 2006. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J. Biol. Chem. 281:11815–11818. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., E. Marois, S. Kjemtrup, R.T. Leister, F. Katagiri, and J.L. Dangl. 2000. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 101:353–363. [DOI] [PubMed] [Google Scholar]
- Nishida, K., M. Takahara, S.-y. Miyagishima, H. Kuroiwa, M. Matsuzaki, and T. Kuroiwa. 2003. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc. Natl. Acad. Sci. USA. 100:2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, K., O. Misumi, F. Yagisawa, H. Kuroiwa, T. Nagata, and T. Kuroiwa. 2004. Triple immunofluorescent labeling of FtsZ, dynamin, and EF-Tu reveals a loose association between the inner and outer membrane mitochondrial division machinery in the red alga Cyanidioschyzon merolae. J. Histochem. Cytochem. 52:843–849. [DOI] [PubMed] [Google Scholar]
- Peat, S., J.R. Turvey, and J.M. Evans. 1959. The structure of floridean starch. Part II. Enzymic hydrolysis and other studies. J. Chem. Soc. 3341–3344.
- Polekhina, G., A. Gupta, B.J. Michell, B. van Denderen, S. Murthy, S.C. Feil, I.G. Jennings, D.J. Campbell, L.A. Witters, M.W. Parker, et al. 2003. AMPK beta subunit targets metabolic stress sensing to glycogen. Curr. Biol. 13:867–871. [DOI] [PubMed] [Google Scholar]
- Polekhina, G., A. Gupta, B.J. van Denderen, S.C. Feil, B.E. Kemp, D. Stapleton, and M.W. Parker. 2005. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 13:1453–1462. [DOI] [PubMed] [Google Scholar]
- Preiss, J., S.G. Yung, and P.A. Baecker. 1983. Regulation of bacterial glycogen synthesis. Mol. Cell. Biochem. 57:61–80. [DOI] [PubMed] [Google Scholar]
- Ritte, G., J.R. Lloyd, N. Eckermann, A. Rottmann, J. Kossmann, and M. Steup. 2002. The starch-related R1 protein is an alpha-glucan, water dikinase. Proc. Natl. Acad. Sci. USA. 99:7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte, G., M. Heydenreich, S. Mahlow, S. Haebel, O. Kotting, and M. Steup. 2006. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 580:4872–4876. [DOI] [PubMed] [Google Scholar]
- Roach, P.J. 2002. Glycogen and its metabolism. Curr. Mol. Med. 2:101–120. [DOI] [PubMed] [Google Scholar]
- Robinson, F.L., and J.E. Dixon. 2006. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 16:403–412. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Sanoja, R., N. Oviedo, and S. Sanchez. 2005. Microbial starch-binding domain. Curr. Opin. Microbiol. 8:260–267. [DOI] [PubMed] [Google Scholar]
- Sakai, M., J. Austin, F. Witmer, and L. Trueb. 1970. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 20:160–176. [DOI] [PubMed] [Google Scholar]
- Schnabel, R., and F. Seitelberger. 1968. Histophysical and histochemical investigations of myoclonus bodies. Pathol. Eur. 3:218–226. [PubMed] [Google Scholar]
- Schwarz, G.A., and M. Yanoff. 1965. Lafora's disease. Distinct clinico-pathologic form of Unverricht's Syndrome. Arch. Neurol. 12:172–188. [DOI] [PubMed] [Google Scholar]
- Serratosa, J.M., P. Gomez-Garre, M.E. Gallardo, B. Anta, D.B. de Bernabe, D. Lindhout, P.B. Augustijn, C.A. Tassinari, R.M. Malafosse, M. Topcu, et al. 1999. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum. Mol. Genet. 8:345–352. [DOI] [PubMed] [Google Scholar]
- Smith, A.M., S.C. Zeeman, and S.M. Smith. 2005. Starch degradation. Annu. Rev. Plant Biol. 56:73–98. [DOI] [PubMed] [Google Scholar]
- Sokolov, L.N., J.R. Dominguez-Solis, A.L. Allary, B.B. Buchanan, and S. Luan. 2006. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. USA. 103:9732–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.S., T. Maehama, and J.E. Dixon. 2000. Inaugural article: mytotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA. 97:8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., D.G. Higgins, and T.J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, K., H. Takano, S. Miyagishima, H. Kuroiwa, and T. Kuroiwa. 1998. Characterization of a chloroplast isoform of serine acetyltransferase from the thermo-acidiphilic red alga Cyanidioschyzon merolae. Biochim. Biophys. Acta. 1403:72–84. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., M.V. Montagu, and M.V. Lijsebettens. 1988. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA. 85:5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heycop Ten Ham, M.W. 1975. Lafora disease, a form of progressive myoclonus epilepsy. In Handbook of Clinical Neurology. Vol. 15. P.J Vinken and G.W. Bruyn, editors. Elsevier Science Publishing Co., Inc., New York. 382–422.
- Van Hoof, F., and M. Hageman-Bal. 1967. Progressive familial myoclonic epilepsy with lafora bodies. Electron microscopic and histochemical study of a cerebral biopsy. Acta Neuropathol. (Berl.). 7:315–326. [DOI] [PubMed] [Google Scholar]
- Vikso-Nielsen, A., P. Hao-Jie Chen, H. Larsson, A. Blennow, and B.L. Moller. 2002. Production of highly phosphorylated glycopolymers by expression of R1 in Escherichia coli. Carbohydr. Res. 337:327–333. [DOI] [PubMed] [Google Scholar]
- Viola, R., P. Nyvall, and M. Pedersén. 2001. The unique features of starch metabolism in red algae. Proc. Biol. Sci. 268:1417–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., J.A. Stuckey, M.J. Wishart, and J.E. Dixon. 2002. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J. Biol. Chem. 277:2377–2380. [DOI] [PubMed] [Google Scholar]
- Weber, A.P., M. Linka, and D. Bhattacharya. 2006. Single, ancient origin of a plastid metabolite translocator family in Plantae from an endomembrane-derived ancestor. Eukaryot. Cell. 5:609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby, C.A., M.S. Gentry, and J.E. Dixon. 2006. Laforin: a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 281:30412–30418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., J. Lim, J.M. Dale, H. Chen, P. Shinn, C.J. Palm, A.M. Southwick, H.C. Wu, C. Kim, M. Nguyen, et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 302:842–846. [DOI] [PubMed] [Google Scholar]
- Yokoi, S., J. Austin, and F. Witmer. 1967. Isolation and characterization of Lafora bodies in two cases of myoclonus epilepsy. J. Neuropathol. Exp. Neurol. 26:125–127. [PubMed] [Google Scholar]
- Yokoi, S., J. Austin, F. Witmer, and M. Sakai. 1968. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch. Neurol. 19:15–33. [DOI] [PubMed] [Google Scholar]
- Yu, T.S., H. Kofler, R.E. Hausler, D. Hille, U.I. Flugge, S.C. Zeeman, A.M. Smith, J. Kossmann, J. Lloyd, G. Ritte, et al. 2001. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 13:1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama, J., J.M. Denu, J.E. Dixon, and M.A. Saper. 1996. Crystal structure of the dual specificity protein phosphatase VHR. Science. 272:1328–1331. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., S.M. Smith, and A.M. Smith. 2007. The diurnal metabolism of leaf starch. Biochem. J. 401:13–28. [DOI] [PubMed] [Google Scholar]
- Zolnierowicz, S. 2000. Type 2A protein phosphatase, the complex regulator of numerous signaling pathways. Biochem. Pharmacol. 60:1225–1235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.