Abstract
Neuronal differentiation involves the formation and extension of neuronal processes. We have identified a novel regulator of neurite formation and extension, the neurite outgrowth multiadaptor, NOMA-GAP, which belongs to a new family of multiadaptor proteins with RhoGAP activity. We show that NOMA-GAP is essential for NGF-stimulated neuronal differentiation and for the regulation of the ERK5 MAP kinase and the Cdc42 signaling pathways downstream of NGF. NOMA-GAP binds directly to the NGF receptor, TrkA, and becomes tyrosine phosphorylated upon receptor activation, thus enabling recruitment and activation of the tyrosine phosphatase SHP2. Recruitment of SHP2 is required for the stimulation of neuronal process extension and for sustained activation of ERK5 downstream of NOMA-GAP. In addition, we show that NOMA-GAP promotes neurite outgrowth by tempering activation of the Cdc42/PAK signaling pathway in response to NGF. NOMA-GAP, through its dual function as a multiadaptor and RhoGAP protein, thus plays an essential role downstream of NGF in promoting neurite outgrowth and extension.
Introduction
Signaling by ligands of receptor tyrosine kinases (RTKs) is critical for the stimulation and regulation of neuronal differentiation. Neurotrophins, for instance, act through specific members of the Trk RTK family to activate multiple signaling pathways that alter protein activities and gene expression and thereby promote neuronal differentiation and function (for review see Reichardt, 2006). Targeted deletion of the neurotrophin nerve growth factor (NGF), or of its receptor TrkA, results in decreased innervation of the spinal cord, hippocampus, and cerebral cortex as well as of target organs such as the skin. In addition, NGF/TrkA signaling is essential for the survival of particular subgroups of neurons in trigeminal, sympathetic, and dorsal root ganglia (DRG) (Crowley et al., 1994; Smeyne et al., 1994).
Binding of neurotrophins to Trk receptors results in receptor dimerization, autophosphorylation, and in the subsequent activation of multiple signaling pathways such as the ERK/MAP kinase, phosphatidylinositol 3-kinase (PI-3K), and phospholipase C-γ (PLC-γ) pathways. Activation of the ERK1/2 MAP kinase pathway is directed through the recruitment of adaptor proteins such as Shc, Grb2, FRS2, SH2-B, and Gab1 to the receptor complex, through multiple redundant mechanisms (for review see Reichardt, 2006). In addition, the SH2 domain containing tyrosine phosphatase, SHP2, is necessary for the sustained, but not for the transient, activation of the ERK1/2 MAP kinase pathway and for the stimulation of neuronal differentiation downstream of NGF and other growth factors (Goldsmith and Koizumi, 1997; Wright et al., 1997; Hadari et al., 1998; for review see Neel et al., 2003; Rosário and Birchmeier, 2003). SHP2 becomes activated through a conformational change induced by binding of the SHP2 SH2 domains to phosphorylated tyrosine residues on multiadaptor proteins (Lechleider et al., 1993; Pluskey et al., 1995; Ong et al., 1997; Hof et al., 1998).
Another ERK MAP kinase, ERK5 (also known as Big MAP kinase), is also activated downstream of neurotrophins, although the exact mechanism involved is not known (Cavanaugh et al., 2001; Watson et al., 2001). ERK1/2 and ERK5 have similar kinase domains and have been shown to share many downstream substrates, but there are also important differences (for review see Wang and Tournier, 2006). Although both kinases are required for neuronal differentiation in Xenopus, ERK1/2 appears to regulate neuronal induction while ERK5 regulates subsequent neuronal differentiation (Pera et al., 2003; Kuroda et al., 2005; Nishimoto et al., 2005). Moreover, ERK5 but not ERK1/2 promotes transcription and cellular survival after NGF-stimulation of distal axons in mature neurons (Watson et al., 2001).
The Rho family GTPases Rac and Cdc42 become activated downstream of a variety of stimuli, including NGF, and regulate various aspects of neuronal differentiation, from neurite formation and axon outgrowth to growth cone function and dendritic maturation (Lamoureux et al., 1997; Yasui et al., 2001; Aoki et al., 2004). Cycling of these GTPases between GTP- and GDP-bound states is essential for their function and is regulated by binding to guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs). Loss-of-function mutations in certain of these regulators are associated with human diseases that affect the nervous system, including mental retardation and the degenerative motor neuron disease, amyotrophic lateral sclerosis (for review see Govek et al., 2005).
We have here identified a novel regulator of neuronal process extension, the neurite outgrowth multiadaptor GTPase-activating protein, NOMA-GAP, which belongs to a new family of multiadaptor proteins with RhoGAP activity. We show that NOMA-GAP is required for NGF-stimulated neurite outgrowth and extension and for the regulation of the ERK5 and Cdc42 pathways downstream of NGF. NOMA-GAP is directly associated with the NGF receptor, TrkA, and functions both as a GAP for Cdc42 and, upon tyrosine phosphorylation, as a multiadaptor protein, to recruit SHP2, Shc, and Grb2 proteins. We show that recruitment of SHP2 by NOMA-GAP is required for the extension of neuronal processes and for the activation of the ERK5 MAP kinase. Furthermore, we demonstrate that NOMA-GAP negatively regulates the activation of Cdc42 and PAK after stimulation by NGF and that this activity is also critical for the formation of neuronal processes.
Results
Novel family of neuronal RhoGAP domain–containing adaptor proteins
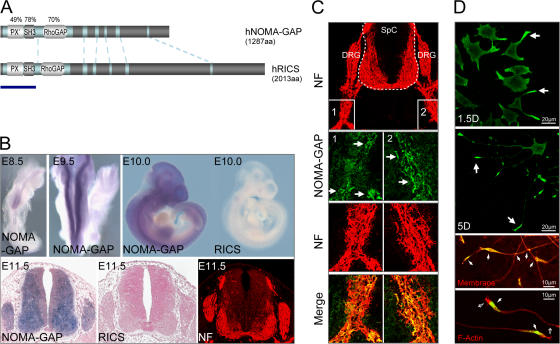
We have searched for novel regulators of Rho family GTPases, which are involved in the control of neuronal differentiation, by expression profiling of E12-E14 mouse spinal cords and DRG (unpublished data). We identified a gene highly expressed in differentiating neurons that codes for a protein containing a RhoGAP domain, an incomplete Phox (PX) domain, and an SH3 domain. We have named this gene neurite outgrowth multiadaptor RhoGAP protein, NOMA-GAP (Fig. 1 A). NOMA-GAP has previously been entered in the human genome database (GenBank) under the name sorting nexin 26 (Gene ID 115703) due to the presence of the PX-like domain, which represents the only structural consensus between true sorting nexins and NOMA-GAP. An alternatively spliced form of the murine NOMA-GAP homologue, which is not present in humans, has been described as a regulator of insulin-stimulated glucose uptake in adipocytes (Chiang et al., 2003). As we have identified an alternative function, we will use the name NOMA-GAP (Fig. 1 A).
Figure 1.
Structure and expression pattern of NOMA-GAP and RICS. (A) Schematic representation of the structure of human NOMA-GAP and RICS proteins. Phox (PX), Phox-like (‘PX’), SH3, and RhoGAP domains are indicated. Additional homology regions are shaded light blue. The degree of amino acid identity of particular domains between the proteins is given in percentages. Novel RICS N-terminal sequences are indicated by a thick blue line. (B) Expression of NOMA-GAP mRNA (blue-purple) is shown in whole mount and in mid-thoracic transverse sections of murine embryos (E8.5 to E11.5). Multiple probes to distinct portions of the NOMA-GAP and RICS gene were tested, with similar results. Sections from E11.5 embryos have been counterstained with Nuclear Fast Red (pink). Immunostaining for neurofilament (NF) of an adjacent section is shown in red. (C) Localization of endogenous NOMA-GAP protein (green) in transverse sections of mouse embryos (E11). NF staining is shown in red. (D) Localization of endogenous NOMA-GAP in NGF-stimulated PC12 cells. NOMA-GAP staining is shown in green. The two bottom panels show magnifications of the growth cones of PC12 cells stimulated with NGF for 5 d and co-stained for endogenous NOMA-GAP (green) and either total membrane or polymerized actin as indicated (red). Filled arrows indicate selected points of NOMA-GAP accumulation; empty arrows indicate microspikes emanating from neurite tips.
Sequence comparison of NOMA-GAP with human GenBank sequences revealed another gene sharing 47% overall sequence identity as well as structural similarity with huNOMA-GAP (Fig. 1 A). Partial sequences of the rat and mouse homologues, which lack the N-terminal region containing the PX and SH3 domains, had been previously named RICS, GC-GAP, or GRIT (Nakamura et al., 2002; Okabe et al., 2003; Zhao et al., 2003). Given the high degree of structural and sequence identity between these two genes, we propose that they form a novel family of signaling molecules (Fig. 1 A; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200609146/DC1).
NOMA-GAP is first expressed in the neural tube of mouse embryos as early as E8.5, and during development of the central nervous system, expression increases along the anterior-posterior axis (Fig. 1 B, top left panels). Analysis of transverse sections shows that NOMA-GAP expression precedes terminal neuronal differentiation, as determined by staining for the post-mitotic neuronal marker neurofilament (NF; Fig. 1 B, bottom panels). Strong expression is also seen in the peripheral nervous system in the DRGs (Fig. 1 B). NOMA-GAP is also expressed in other neuronal tissues such as in the brain and in the eye (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200609146/DC1; and unpublished data). RICS is not expressed in the spinal cord during stages of neuronal differentiation, E8.5 to E14 (Fig. 1 B). We have also analyzed the location of NOMA-GAP protein in transverse sections of mouse embryos, using specific antibodies raised against the C terminus of NOMA-GAP (Fig. 1 C; see Materials and methods). NOMA-GAP protein is found in distinct punctate staining along, and at the ends, of axons projecting from the spinal cord (Fig. 1 C). To localize NOMA-GAP at the cellular level, we have used PC12 neuronal precursor cells, which differentiate upon the addition of NGF (Greene and Tischler, 1976). NOMA-GAP is evenly distributed in a fine punctate pattern around the cell periphery in unstimulated cells. Stimulation with NGF results in the extension of neuronal processes and the preferential accumulation of NOMA-GAP punctate staining at these early protrusions. NOMA-GAP is also present at the growth cones of differentiated cells (Fig. 1 D, indicated by arrows). We have controlled for the higher level of membrane at the growth cones by co-staining samples with a lipophilic styryl dye (in red; Fig. 1 D, third panel). In addition, co-staining of polymerized actin, shows that NOMA-GAP is concentrated in the body of the growth cone but absent in the microspikes emanating from the neurite tips (in red; Fig. 1 D, indicated by arrows in last panel).
NOMA-GAP is required for NGF-induced neurite extension
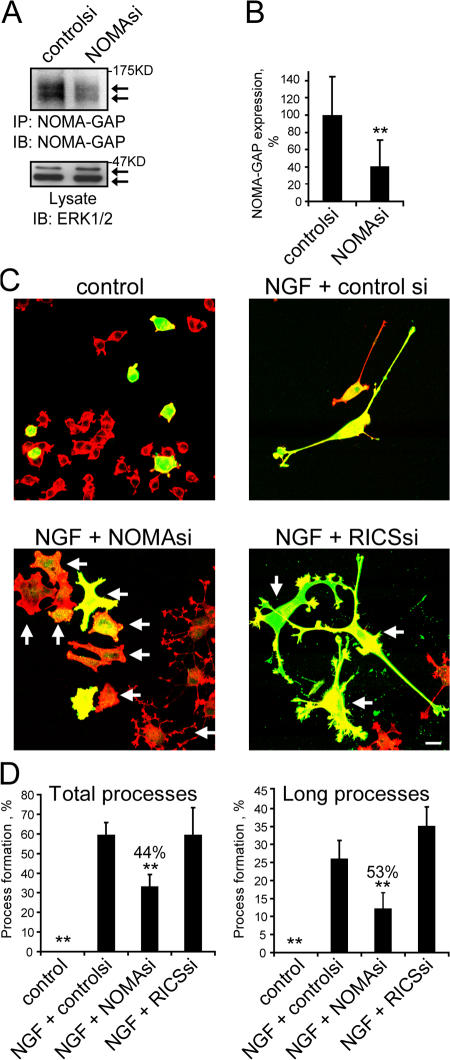
We have analyzed the role of NOMA-GAP during NGF-stimulated neuronal differentiation using siRNA directed against rat NOMA-GAP in PC12 cells, which express both NOMA-GAP and RICS (Fig. S1 C). Transfection of siRNA to NOMA-GAP significantly reduced NOMA-GAP expression as visualized both in immunoprecipitates (Fig. 2 A) and in fixed cells immunostained for endogenous NOMA-GAP (Fig. 2 B). NGF stimulates the transient cell flattening of PC12 cells followed by neurite outgrowth and extension over several days (Greene and Tischler, 1976). We determined the proportion of transfected cells forming neurites (>30 μm long; total processes) or containing long axonal-like processes (>100 μm long; long processes). Remarkably, down-regulation of NOMA-GAP expression by siRNA inhibited the extension of neuronal processes and resulted in cell flattening and spreading (Fig. 2 C, indicated by arrows). The formation of long neuronal processes was inhibited by over 50% in response to down-regulation of NOMA-GAP (Fig. 2 D). Down-regulation of RICS, on the other hand, did not inhibit neurite outgrowth and elongation (Fig. 2 D; Nasu-Nishimura et al., 2006). These data suggest that NOMA-GAP is required for NGF-stimulated neurite outgrowth.
Figure 2.
NOMA-GAP is required for NGF-stimulated process extension. (A) Down-regulation of endogenous NOMA-GAP protein in NGF-stimulated PC12 cells transfected with control siRNA (controlsi) or siRNA directed against rat NOMA-GAP (NOMAsi). NOMA-GAP immunoprecipitates (IP) were immunoblotted (IB) for NOMA-GAP. Lysates were also immunoblotted for ERK1/2. (B) Down-regulation of immunofluorescent staining for endogenous NOMA-GAP in NGF-stimulated siRNA-transfected PC12 cells. Cells were transfected with control siRNA or NOMA-GAP siRNA in the presence of limiting amounts of the transfection marker, GFP, and immunostained for endogenous NOMA-GAP. Mean pixel intensity of NOMA-GAP staining per GFP-positive cell is shown. The mean staining of controlsi (n = 40) and NOMAsi (n = 56) samples were compared with a t test for unequal variances and found to be significantly different (P < 0.0005; see Materials and methods). (C) Effect of down-regulation of NOMA-GAP expression on NGF-stimulated differentiation in PC12 cells. Cells transfected with control siRNA, NOMA-GAP siRNA, or RICS siRNA were stained for polymerized actin (red) and the transfection marker GFP (green) 72 h after transfection. Arrows indicate selected transfected cells. Bar, 20 μm. (D) Quantification of neuronal process extension in siRNA-transfected PC12 cells (five independent experiments). Total processes, cells bearing processes >30 μm long; long processes, cells bearing processes >100 μm long. The means of the four samples were compared with the multiple test of ANOVA and found to be significantly different (P < 0.0005). The error probabilities of pairwise tests were corrected by Bonferroni. The means of the control and NGF+NOMAsi samples were found to be significantly different from the mean of the NGF+controlsi sample (P < 0.0005 in both cases for Total processes and P < 0.0005 and P < 0.001, respectively for Long processes; summarized by asterisks; see Materials and methods). Percentages denote level of inhibition.
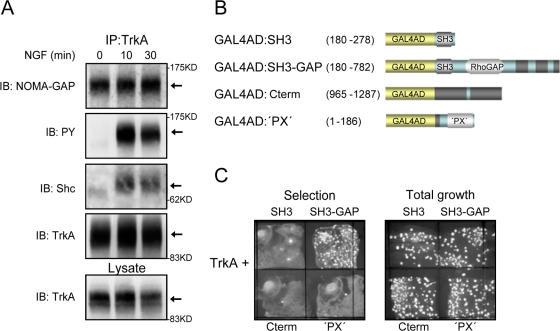
We then examined whether NOMA-GAP is part of the signaling complex that becomes recruited to the NGF receptor, TrkA. Immunoprecipitation of endogenous TrkA from PC12 cells resulted in the coimmunoprecipitation of endogenous NOMA-GAP and Shc proteins (Fig. 3 A). Unlike Shc, NOMA-GAP formed a constitutive, NGF-independent complex with the receptor. NGF-stimulation resulted in the strong tyrosine phosphorylation of the associated NOMA-GAP (Fig. 3 A). We confirmed that NOMA-GAP interacts directly with TrkA by expressing N- and C-terminal deletion mutants of huNOMA-GAP with the cytoplasmic domain of huTrkA in the yeast two-hybrid system (Fig. 3 B). Expression of a deletion mutant of NOMA-GAP containing the SH3 and RhoGAP domains (SH3-GAP) interacted with the TrkA cytoplasmic domain and enabled growth of the transfected yeast on selection plates (Fig. 3 C). No interaction was seen with the N-terminal 186 amino acids containing the PX domain (‘PX’), the C-terminal region of NOMA-GAP (Cterm), or with the isolated SH3 domain of NOMA-GAP (Fig. 3, B and C). Together, these data show that NOMA-GAP interacts directly with the cytoplasmic domain of TrkA, through an N-terminal region that includes residues 279–782.
Figure 3.
NOMA-GAP interacts with TrkA. (A) Coimmunoprecipitation of endogenous NOMA-GAP and TrkA from NGF-stimulated PC12 cells. TrkA immunoprecipitates were immunoblotted (IB) as indicated. (B) Diagrammatic representation of the GAL4 activation domain: NOMA-GAP fusion constructs used for yeast two-hybrid analysis. (C) Interaction of huNOMA-GAP with the cytoplasmic domain of huTrkA in the yeast two-hybrid system. The growth of transformed yeast on plates selecting for protein interaction (Selection plate) or for the presence of both plasmids (Total growth plate) is shown.
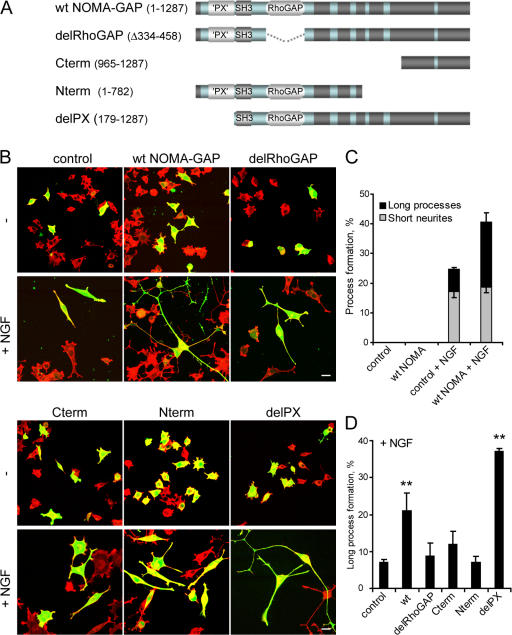
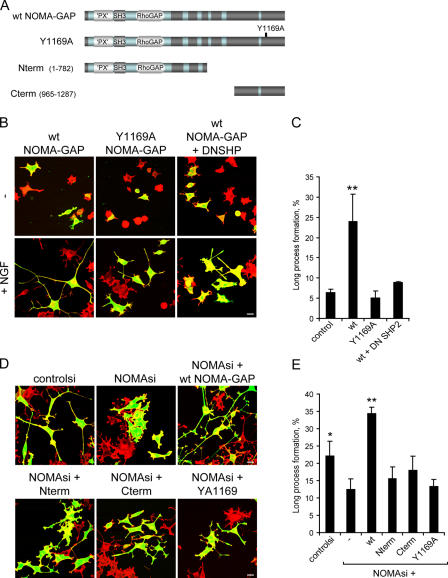
We then dissected the function of the different domains of NOMA-GAP in PC12 cells (Fig. 4; Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200609146/DC1). Transfected cells were stimulated for a short period (48 h) with NGF, during which time control cells start to differentiate, but have not yet formed long neuronal processes (Fig. 4 B, top left column). Samples were then scored for the proportion of transfected cells bearing short neurites (30–100 μm), long processes (>100 μm), or no neurites. Expression of NOMA-GAP alone did not affect cellular morphology during this time period. However, NOMA-GAP strongly synergized with NGF in the formation and extension of neuronal processes (Fig. 4 B, top middle column; quantified in Fig. 4 C). NOMA-GAP mutants, where the RhoGAP domain (delRhoGAP), the whole N-terminal region (containing the PX, SH3, and RhoGAP domains; Cterm), or the C-terminal region (Nterm) had been deleted, failed to induce the extension of long neuronal processes. The formation of shorter neurites was not affected (Fig. 4 B; quantified in Fig. 4 D). This suggests that both the RhoGAP domain, and sequences in the C-terminal region of NOMA-GAP are required for the stimulation of neurite extension. The PX domain, on the other hand, negatively regulates NOMA-GAP function, as deletion of this domain (delPX) stimulates NOMA-GAP–induced process outgrowth (Fig. 4 B, bottom right column; quantified in Fig. 4 D). PX domains have been reported to negatively regulate protein activity in other proteins with tandem PX-SH3 domains (Yuzawa et al., 2004).
Figure 4.
NOMA-GAP stimulates the extension of long neuronal processes. (A) Schematic representation of wild-type (wt) and deletion mutant NOMA-GAP constructs. (B) NOMA-GAP stimulates the extension of long neuronal processes in PC12 cells stimulated with low levels of NGF for 48 h. Samples were stained as described in Fig. 2 C. (C) Summary of the average percentage of GFP-positive cells bearing short or long neuronal processes in three independent experiments. (D) Quantification of the effect of different NOMA-GAP deletion mutants on the extension of long neuronal processes in three independent experiments. The means of the different samples were compared as described in Fig. 2 D. The means of the wt and delPX samples were found to be significantly different from the mean of the control sample (P < 0.01 and P < 0.0005, respectively).
NOMA-GAP recruits the tyrosine phosphatase SHP2
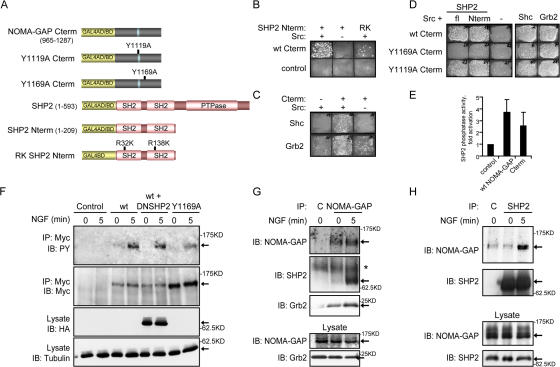
Next we examined the signaling function of the C-terminal region of NOMA-GAP (residues 965–1287), which is rich in tyrosine residues that lie within consensus sites for the binding of SH2 domains. We performed a modified yeast two-hybrid screen using this portion of NOMA-GAP and a human brain cDNA library (HY4004AH). An activated mutant of the tyrosine kinase Src (Y416F Y527F) was coexpressed from the same vector to enable tyrosine phosphorylation of NOMA-GAP (see also Materials and methods). We identified several clones encoding the N-terminal region of the tyrosine phosphatase SHP2 as well as full-length clones of the adaptor proteins Grb2 and Shc (Fig. 5, B and C). The C terminus of NOMA-GAP contains a consensus binding site for the Grb2 SH3 domain (PXXPXR; Simon and Schreiber, 1995), which may explain weak binding of Grb2 to NOMA-GAP in the absence of tyrosine phosphorylation (Fig. 5 C). Both full-length and the N terminus of SHP2 interacted with the C terminus of NOMA-GAP in a Src-dependent fashion (Fig. 5, B and D). The N terminus of SHP2 contains two SH2 domains, and binding to NOMA-GAP is abrogated by mutation of the conserved arginine residues in the phosphotyrosine binding pockets of these domains (RK mutant, Fig. 5 B; O'Reilly and Neel, 1998). This indicates that interaction with SHP2 is mediated through phosphorylated tyrosine residues on NOMA-GAP. To identify the SHP2-binding site on NOMA-GAP, we mutated all tyrosine residues in the C-terminal region of NOMA-GAP found in the consensus YXXI/V/L sequence for SHP2 binding (for review see Neel et al., 2003; Rosário and Birchmeier, 2003). Mutation of a single tyrosine in the C terminus of NOMA-GAP, residue 1169, abolished the direct interaction of NOMA-GAP with SHP2 but had no effect on the binding of Grb2 or Shc (Fig. 5, A and D). Mutation of other tyrosine residues, for example tyrosine 1119, had no effect on SHP2 binding to NOMA-GAP.
Figure 5.
The C terminus of NOMA-GAP acts as a docking site for multiple signaling effectors. (A) Schematic representation of the NOMA-GAP and SHP2 constructs used for yeast two-hybrid analysis (where C-term refers to C terminus of NOMA-GAP; fl to full-length SHP2; Nterm to the N terminus of SHP2). (B–D) Yeast growth under selection for the reconstitution of GAL4 transcriptional activity. (B) Interaction of NOMA-GAP Cterm with wild-type and RK mutant SHP2 Nterm. (C) Interaction of NOMA-GAP Cterm with Shc and Grb2. (D) Interaction of point mutants of NOMA-GAP Cterm with SHP2, Shc, and Grb2. (E) NOMA-GAP activates SHP2 phosphatase activity. Recombinant wt GST-SHP2 incubated with lysates derived from serum-stimulated NIH3T3 cells expressing full-length NOMA-GAP or NOMA-GAP Cterm, was recovered by precipitation with glutathione-Sepharose beads and phosphatase activity measured on pNPP. The average of two independent experiments is shown. (F) Tyrosine phosphorylation of wild-type and Y1169A NOMA-GAP after NGF stimulation. Myc-tagged wt and Y1169A NOMA-GAP were immunoprecipitated from NGF-stimulated and unstimulated transiently transfected PC12 cells. Precipitates and lysate were immunoblotted as indicated. (G) Coimmunoprecipitation of endogenous SHP2 and Grb2 with endogenous NOMA-GAP from unstimulated and NGF-stimulated PC12 cells. NOMA-GAP and beads-alone control (C) precipitates and lysate were immunoblotted as indicated. Asterisk marks a nonspecific band. (H) Coimmunoprecipitation of endogenous NOMA-GAP with endogenous SHP2 from unstimulated and NGF-stimulated PC12 cells. SHP2 and beads-alone control (C) precipitates were immunoblotted as indicated.
SHP2 is activated through a conformational change induced by binding of tyrosine-phosphorylated proteins to its SH2 domains (Hof et al., 1998). Because NOMA-GAP binds to SHP2 through the SHP2 SH2 domains, it should lead to the activation of this phosphatase. We confirmed this by incubating recombinant wild-type GST-tagged SHP2 with lysates from serum-stimulated cells expressing full-length or the C terminus of NOMA-GAP. Recombinant SHP2 was recovered by incubation with glutathione-Sepharose beads, and phosphatase activity assayed on the artificial substrate pNPP. Indeed, expression of both full-length and the C terminus of NOMA-GAP stimulated SHP2 phosphatase activity (Fig. 5 E).
To test whether the SHP2 binding site on NOMA-GAP, residue 1169, is a major phosphorylation site downstream of NGF, we analyzed tyrosine phosphorylation of Myc-tagged wild-type and Y1169A NOMA-GAP immunoprecipitated from NGF-stimulated PC12 cells. Wild-type NOMA-GAP is weakly phosphorylated on tyrosine residues in cells grown in reduced serum and becomes strongly phosphorylated upon stimulation with NGF (Fig. 5 F). Mutation of residue 1169 significantly reduced tyrosine phosphorylation of NOMA-GAP, indicating that this is a major tyrosine phosphorylation site downstream of NGF. Expression of dominant-negative SHP2, which contains a 31-amino acid deletion in the phosphatase domain (DNSHP2; O'Reilly and Neel, 1998), did not significantly alter tyrosine phosphorylation on NOMA-GAP (Fig. 5 F), suggesting that NOMA-GAP is not a major substrate for SHP2.
We confirmed that NOMA-GAP associates with SHP2 downstream of NGF signaling in two-way immunoprecipitations of endogenous proteins from NGF-stimulated PC12 cells. Immunoprecipitation of NOMA-GAP resulted in the coimmunoprecipitation of SHP2 only upon stimulation with NGF (Fig. 5 G). NOMA-GAP also associated with Grb2, and this was enhanced by stimulation with NGF. Immunoprecipitation of endogenous SHP2 confirmed that NGF stimulates the association of NOMA-GAP with SHP2 in PC12 cells (Fig. 5 H).
SHP2 is required for NOMA-GAP–induced signaling and neurite extension
We interfered with SHP2 function downstream of NOMA-GAP by either mutating the SHP2 binding site on full-length NOMA-GAP (Y1169A, Fig. 6 A) or by coexpressing DNSHP2. Interfering with SHP2 function inhibited NOMA-GAP-induced extension of neuronal processes in NGF-stimulated PC12 cells (Fig. 6 B; quantified in Fig. 6 C). We then analyzed whether wild-type and mutant human NOMA-GAP could rescue the inhibition of NGF-induced neurite outgrowth caused by down-regulation of endogenous NOMA-GAP (Fig. 6 D). Expression of wild-type human NOMA-GAP, but not of Y1169A or N- and C-terminal deletion mutants, rescued NGF-stimulated neurite outgrowth in PC12 cells treated with NOMA-GAP siRNA (Fig. 6 D; quantified in Fig. 6 E).
Figure 6.
SHP2 is required for NOMA-GAP–stimulated neurite extension. (A) Schematic representation of full-length wild-type, mutant, and deletion NOMA-GAP constructs. (B and C) SHP2 is required for NOMA-GAP–stimulated process extension. PC12 cells were transfected with wild-type and mutant NOMA-GAP and, where indicated, dominant-negative SHP2 (DNSHP), in the presence or absence of low levels of NGF. Samples were stained for polymerized actin (red) and for GFP cotransfected at limiting amounts (green) 48 h after transfection. Bar, 20 μm. (C) Quantification of four independent experiments. The means of the different samples were compared as described in Fig. 2 D. Only the mean of the wt sample was found to be significantly different from the control mean (P < 0.008). (D and E) Expression of full-length human NOMA-GAP, but not the full-length Y1169A point mutant nor C- and N-terminal deletion mutants, rescues NGF-stimulated process formation in NOMA-GAP siRNA (NOMAsi)-treated PC12 cells. Samples were stained as before. Bars, 20 μm. (E) Quantification of the proportion of GFP-positive cells bearing long neuronal processes (three independent experiments). The means of the different samples were compared as described in Fig. 2 D. Only the means of the controlsi (P < 0.037) and NOMAsi+wt NOMA-GAP (P < 0.0005) samples were found to be significantly different from NOMAsi.
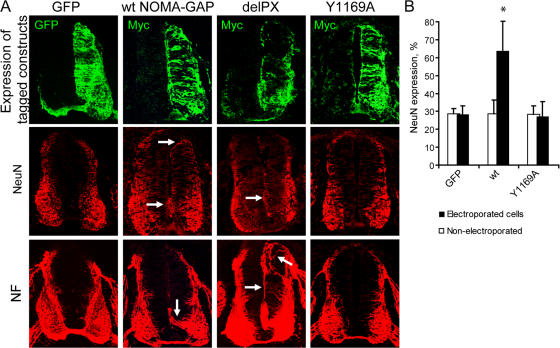
We also analyzed the function of NOMA-GAP in the spinal cord by using in ovo electroporation of chick embryos. Using this technique, cells in the proliferative ependymal layer on one side (Fig. 7 A; right-hand side, top row) of the spinal cord are electroporated at stages preceding endogenous neuronal differentiation. These cells then populate that entire half of the spinal cord. Expression of wild-type or delPX human NOMA-GAP resulted in the premature differentiation of cells in the proliferative ependymal layer (as shown by staining for the post-mitotic neuronal marker NeuN; quantified in Fig. 7 B) and the projection of neuronal processes from these cells (neurofilament staining; indicated by arrows, second column, Fig. 7 A). This was abrogated by mutation of the SHP2 binding site in NOMA-GAP (Y1169A, last column, Fig. 7 A; quantified in Fig. 7 B). Together, the above data indicate that recruitment of SHP2 through the C-terminal domain is required for the function of NOMA-GAP in neuronal process extension and differentiation.
Figure 7.
NOMA-GAP stimulates neuronal differentiation in the developing chick spinal cord. (A) Mid-thoracic transverse sections of the spinal cords of chick embryos electroporated 48 h earlier, at HH stages 15–17, with GFP or Myc-tagged wild-type and mutant NOMA-GAP constructs. The electroporated side is shown on the right-hand side of the spinal cord. Adjacent sections were stained for expression of the tagged-electroporated protein (green) and the neuronal markers NeuN and NF (red). Arrows indicate up-regulation of NeuN staining in the proliferative ependymal layer and the extension of neuronal processes in NOMA-GAP–expressing cells. (B) Quantification of NeuN expression. Cell nuclei were counterstained with TOTO-3 for identification of individual cells for counting (see Materials and methods). The percentage of GFP- or NOMA-GAP–expressing cells expressing NeuN is shown as black bars, while the proportion of NeuN expressing cells in the nonelectroporated side of the same embryos is shown as white bars. Five embryos were each analyzed for wt and Y1169A NOMA-GAP and two embryos for GFP electroporations. The means of the different samples were compared as described in Fig. 2 D. Only the mean of the wt electroporated cells was found to be significantly different from the respective GFP mean (P < 0.022).
NOMA-GAP regulates the activation of the ERK5 MAP kinase downstream of NGF
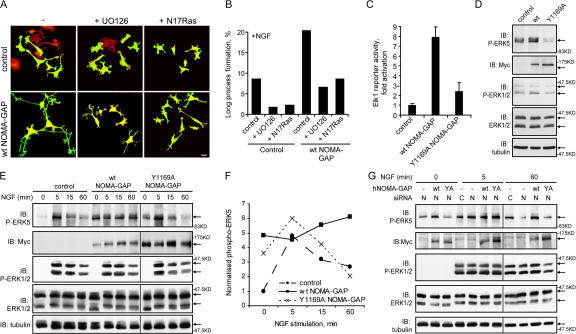
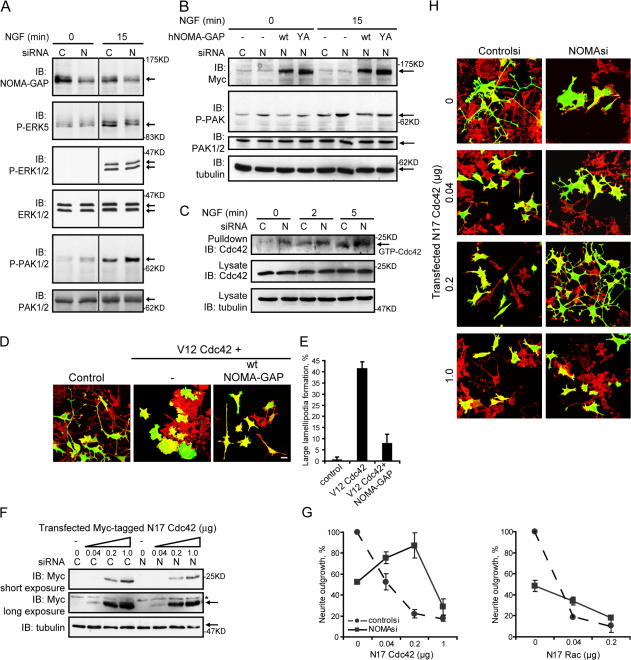
SHP2 has been shown to regulate the activation of the Ras/ERK1/2 MAP kinase pathway (for review see Neel et al., 2003; Rosário and Birchmeier, 2003). Indeed, NOMA-GAP–stimulated neurite extension in PC12 cells was inhibited by the MAP kinase pathway inhibitor U0126 or by a dominant-negative mutant of Ras (N17Ras; Fig. 8 A, quantified in Fig. 8 B). Moreover, overexpression of wild-type NOMA-GAP resulted in the strong activation of the Elk1 reporter in epithelial cells (Fig. 8 C). Activity was dependent on interaction with SHP2, as mutation of the SHP2 binding site Y1169 strongly inhibited transcriptional activation of Elk1. We also analyzed whether NOMA-GAP could stimulate MAP kinase activation in PC12 cells. PC12 cells expressing wild-type or Y1169A NOMA-GAP were stimulated with NGF either for 24 h (Fig. 8 D) or for shorter time periods (Fig. 8 E). To our surprise, NOMA-GAP did not significantly alter the phosphorylation (and thus activation) of the classical MAP kinases, ERK1 and 2, but resulted in the sustained elevated activation of ERK5. Cells expressing Y1169A NOMA-GAP, on the other hand, showed an elevation in the transient activation of ERK5 but then a rapid loss in ERK5 phosphorylation (Fig. 8 E, quantified in Fig. 8 F). A similar result was observed upon coexpression of DNSHP2 with NOMA-GAP (unpublished data). Down-regulation of endogenous NOMA-GAP in PC12 cells, on the other hand, resulted in a decrease in the sustained (1 h), but not in transient (5 min), activation of ERK5 and had no effect on the activation of ERK1/2 (Fig. 8 G). Furthermore, ERK5 activation in NOMA-GAP down-regulated PC12 cells could be rescued by coexpression of wild-type but not of Y1169A NOMA-GAP (Fig. 8 G). NOMA-GAP thus regulates sustained activation of ERK5 downstream of NGF, through recruitment of SHP2.
Figure 8.
NOMA-GAP regulates the ERK5 MAP kinase pathway. (A and B) Ras/MAP kinase activity is required for NOMA-GAP induced neurite extension. PC12 cells were transiently transfected with control or wt NOMA-GAP expression constructs in the presence or absence of an expression construct for N17Ras and stimulated 24 h later with NGF in the presence or absence of 20 μM U0126. (A) Samples were fixed and stained 72 h later for the GFP transfection marker (green) and for polymerized actin (red). Bars, 20 μm. (B) Quantification of the proportion of GFP-expressing cells bearing long neuronal processes (>100 μm). (C) NOMA-GAP activates ERK-dependent Elk1 transcriptional activation of a firefly luciferase reporter gene in RK13 epithelial cells. Samples are normalized on renilla luciferase expression. The average of duplicate experiments is shown. (D–F) NOMA-GAP activates ERK5 MAP kinase in PC12 cells. PC12 cells were transiently transfected with the indicated Myc-tagged NOMA-GAP constructs or with control vector and stimulated with NGF for 24 h (D) or for the indicated times (E). Lysates were analyzed by Western blot as indicated. (F) Quantification of the levels of phospho-ERK5 normalized on the α-tubulin loading control for the experiment shown in E. (G) NOMA-GAP is required for the sustained activation of ERK5 downstream of NGF. PC12 cells were transfected with control (C) or rat NOMA-GAP (N) siRNA in the presence or absence of expression constructs for wt and Y1169A huNOMA-GAP and stimulated 24 h later with NGF for the indicated times. Lysates were immunoblotted as indicated. Vertical lines denote nonconsecutive lanes from the same gel and Western blot.
NOMA-GAP negatively regulates the Cdc42/PAK signaling pathway downstream of NGF
Binding assays of the RhoGAP domain of NOMA-GAP with dominant-active and -negative forms of Cdc42 and other RhoGTPases showed that NOMA-GAP preferentially binds to active Cdc42 (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200609146/DC1). We studied whether NOMA-GAP regulated the Cdc42 signaling pathway downstream of NGF in PC12 cells. We used antibodies to the autophosphorylation sites on the S/T kinase PAK 1/2, which lies downstream of Cdc42 in NGF-stimulated PC12 cells (Obermeier et al., 1998). Down-regulation of NOMA- GAP resulted in the inhibition of ERK5 as observed before, but also in a strong elevation in the autophosphorylation, and thus activation, of PAK (Fig. 9 A). Furthermore, we could rescue this elevation by coexpression of hNOMA-GAP (Fig. 9 B). A partial rescue was seen upon coexpression of the SHP2 binding mutant, Y1169A NOMA-GAP (Fig. 9 B), suggesting that SHP2 may play a small role in the regulation of PAK downstream of NOMA-GAP. We also analyzed the levels of GTP-Cdc42 in NOMA-GAP siRNA-treated PC12 cells, using a pull-down assay with the Cdc42/Rac-interactive binding (CRIB) domain of PAK (Fig. 9 C). Down-regulation of NOMA-GAP resulted in increased Cdc42-GTP in NGF-stimulated and unstimulated PC12 cells. Thus, NOMA-GAP is an important negative regulator of Cdc42/PAK signaling downstream of NGF.
Figure 9.
NOMA-GAP negatively regulates Cdc42 and PAK downstream of NGF. (A and B) PC12 cells were transfected with control (C) or rat NOMA-GAP (N) siRNA in the absence (A) or presence (B) of expression constructs for wt and Y1169A huNOMA-GAP as described in Fig. 8 G. Lysates were immunoblotted as indicated. Vertical lines denote nonconsecutive lanes from the same gel and Western blot. (C) GST-PAK CRIB pull-downs were performed on lysates of PC12 cells transfected 24 h earlier with control or rat NOMA-GAP siRNA and stimulated with NGF. Pull-downs and lysates were immunoblotted as indicated. (D and E) PC12 cells were transiently transfected with dominant-active (V12) Cdc42, wt NOMA-GAP, and a GFP transfection marker as indicated and stimulated with NGF 24 h after transfection. (D) Samples were stained 72 h after transfection for polymerized actin (red) and GFP (green). Bar, 20 μm. (E) Quantification of the proportion of cells forming large lamellipodia (>30 μm diameter) in duplicate samples. (F–H) PC12 cells were transfected with control siRNA or NOMA-GAP siRNA in the presence of increasing levels of an expression construct for Myc-tagged dominant-negative (N17) Cdc42 and a GFP transfection marker. Cells were stimulated with NGF 4 h after transfection and were then either lysed 48 h after transfection and analyzed for expression of Myc-tagged N17 Cdc42 (F) or stained 72 h after transfection for polymerized actin (red) and GFP (green) (H). Bar, 20 μm. Asterisk marks a nonspecific band. (G) Quantification of the proportion of NGF-stimulated control or NOMA-GAP siRNA-transfected cells bearing neurites (>30 μm) in the presence of increasing levels of N17Cdc42 or N17Rac. The normalized average of two independent experiments is shown.
Expression of dominant-active Cdc42 (V12 Cdc42) in PC12 cells is inhibitory to neurite outgrowth, and induces instead cell flattening and spreading as well as filopodia formation (Nobes and Hall, 1995; Daniels et al., 1998; Aoki et al., 2004). This could be reversed by coexpression of NOMA-GAP (Fig. 9, D and E). PC12 cells treated with NOMA-GAP siRNA exhibit a similar, although less extensive, phenotype (Fig. 2 D and Fig. 9 H). We asked whether the inhibition of neurite outgrowth caused by down-regulation of NOMA-GAP is due to the elevated activation of Cdc42 observed in these cells. We therefore titrated dominant-negative Cdc42 (N17Cdc42) into NGF-stimulated NOMA-GAP or control siRNA-treated PC12 cells (Fig. 9 F). As has been previously observed (Daniels et al., 1998), expression of N17Cdc42 in controlsi-treated NGF-stimulated cells resulted in inhibition of neurite outgrowth and the cells showed a round undifferentiated phenotype (Fig. 9 H, first column; quantified in Fig. 9 G). Down-regulation of NOMA-GAP leads to cell flattening. Expression of low levels of N17Cdc42 (but not N17Rac) in these cells, however, resulted in a decrease in cell spreading and stimulated the extension of neurites (Fig. 9, G and H). Thus, NOMA-GAP enables neurite outgrowth downstream of NGF signaling by tempering Cdc42 activation. At higher levels of N17Cdc42, neurite outgrowth is also inhibited in these cells, indicating that this GTPase has a function, albeit at controlled levels, in neurite outgrowth.
Discussion
We have here identified a novel regulator of neuronal process extension, the neurite outgrowth multiadaptor RhoGAP protein, NOMA-GAP, which has both multiadaptor and enzymatic functions that are essential for the transduction of signals and biological activity of NGF/TrkA. In particular, we demonstrate that NOMA-GAP plays two distinct and crucial roles downstream of NGF signaling: promotion of SHP2/ERK5 signaling and tempering the activation of the Cdc42/PAK signaling pathway.
Regulation of SHP2 and ERK5 by NOMA-GAP
NOMA-GAP synergizes with NGF in promoting the extension of neuronal processes in PC12 cells and induces neuronal differentiation in the developing chick spinal cord. We show that NOMA-GAP is constitutively associated with the NGF receptor, TrkA, through a direct phosphotyrosine-independent interaction involving sequences in the N terminus of NOMA-GAP. Activation of TrkA results in tyrosine phosphorylation of NOMA-GAP, although it is not clear whether this is direct. NGF-stimulated tyrosine phosphorylation of NOMA-GAP, however, enables the recruitment of the tyrosine phosphatase SHP2.
The tyrosine phosphatase SHP2 has been shown to control the sustained activation of the ERK1/2 MAP kinases and to be required for the extension of neurites downstream of NGF (Wright et al., 1997, for review see Neel et al., 2003). ERK5 is another ERK kinase that is activated by neurotrophins and that in cardiomyocytes is regulated by SHP2 (Nakaoka et al., 2003; for review see Wang and Tournier, 2006). We have analyzed the interaction of NOMA-GAP with SHP2, and demonstrate that it involves the N-terminal SHP2 SH2 domains and phosphorylation of tyrosine 1169 in the C terminus of NOMA-GAP. This type of interaction has been shown to result in an activating conformational change in SHP2 (Lechleider et al., 1993; Ong et al., 1997; Hof et al., 1998; Cunnick et al., 2001), and indeed, interaction with NOMA-GAP results in activation of SHP2. Furthermore, we demonstrate that association of SHP2 with NOMA-GAP is required for the extension of neuronal processes.
NOMA-GAP–stimulated neurite extension is inhibited by the ERK MAP kinase pathway inhibitor U0126 and by dominant-negative Ras. However, both ERK1/2 and ERK5 are sensitive to U0126 and can be activated downstream of Ras in PC12 cells (English et al., 1998; Kamakura et al., 1999). We demonstrate that NOMA-GAP is specifically required for the sustained activation of ERK5, but not of ERK1/2, downstream of NGF. Furthermore, SHP2 plays a critical role downstream of NOMA-GAP in the activation of ERK5.
Several other multiadaptor proteins have been shown to interact with TrkA and to activate ERK1/2 MAP kinases (for review see Reichardt, 2006). However, their relative importance is poorly understood. Nevertheless, Shc has been shown to mediate transient activation of ERK1/2 kinases, while both FRS-2 and the membrane-spanning protein ARMS/Kidins220 have been proposed to regulate sustained activation of ERK1/2 (Kao et al., 2001; Arevalo et al., 2004, 2006). The molecular mechanisms linking the TrkA to ERK5 activation were unknown. This study, thus, identifies the first multiadaptor protein, NOMA-GAP, that specifically regulates ERK5 downstream of NGF.
Negative regulation of Cdc42 by NOMA-GAP downstream of NGF
Unlike other multiadaptor proteins associated with TrkA, NOMA-GAP also contains an enzymatic domain, the RhoGAP domain, which specifically binds to active forms of the RhoGTPase, Cdc42. Previous in vitro GTPase assays using the isolated RhoGAP domain of NOMA-GAP, report a similar preference for Cdc42 (Chiang et al., 2003). Cdc42 regulates various aspects of neuronal differentiation including neurite outgrowth and extension (for review see Govek et al., 2005). However, paradoxically, both active and inactive mutants of Cdc42 have been reported to inhibit neurite outgrowth and neuronal differentiation (Luo et al., 1994; Allen et al., 2000; Aoki et al., 2004). NGF-driven differentiation of PC12 cells normally proceeds through a two-step process where cells initially spread and produce lamellipodia and unstable filopodia and then extend stable neurites (Greene and Tischler, 1976; Aoki et al., 2004). Expression of a dominant-active mutant of Cdc42 results not in neurite outgrowth but in filopodia formation and cell spreading (Nobes and Hall, 1995; Daniels et al., 1998; Aoki et al., 2004), which can be counteracted by coexpression of NOMA-GAP. Indeed, the RhoGAP domain of NOMA-GAP is required for the extension of neuronal processes. Furthermore, we show that down-regulation of NOMA-GAP in PC12 cells results in elevated levels of active Cdc42 and the Cdc42 effector, PAK, which can be rescued by reexpression of NOMA-GAP. Thus, NOMA-GAP is an important negative regulator of Cdc42 signaling downstream of NGF. This signaling function is essential to permit neurite outgrowth. PC12 cells with down-regulated NOMA-GAP undergo cell spreading and fail to extend neurites. Moreover, neurite outgrowth can be rescued by coexpression of low levels of dominant-negative Cdc42. Higher levels of dominant-negative Cdc42 are also inhibitory to neurite outgrowth in PC12 cells with down-regulated NOMA-GAP. This suggests that although excessive activation of Cdc42, as induced by NGF in the absence of NOMA-GAP, is counterproductive to neurite outgrowth and extension, low levels or spatially restricted activation of Cdc42 are still required. Interestingly, GTP-Cdc42 has been shown by FRET analysis to be restricted to the microspikes projecting from the neurite tips of NGF-stimulated PC12 cells and to be absent in the tips themselves (Aoki et al., 2004). We show that the opposite is true for NOMA-GAP. Negative roles for Cdc42 and Cdc42 effectors have also been described by others. For example, the Cdc42 effectors and actin polymerization regulators N-WASP and Toca-1 have been shown to negatively regulate neurite extension in NGF-stimulated PC12 cells and in hippocampal neurons (Strasser et al., 2004; Kakimoto et al., 2006). Additionally, in Drosophila neurons Cdc42 has been shown to inhibit axon growth and guidance through a PAK-LIMK-cofilin pathway (Ng and Luo, 2004). Work with PAK and LIMK null mice also suggests that these Cdc42 effectors are required for the later maturation of dendritic spines but not for axon outgrowth (for review see Boda et al., 2006).
Finally, the role of NOMA-GAP in the promotion of neuronal process outgrowth may have implications for human pathology. We have demonstrated that NOMA-GAP is a necessary player in NGF-stimulated signaling. In humans, TrkA mutations are associated with congenital insensitivity to pain with anhydrosis (Indo et al., 1996), while reduced NGF signaling may be involved in the pathology of Alzheimer's disease and Downs syndrome (Capsoni et al., 2000; Salehi et al., 2006). Moreover, NOMA-GAP regulates at least two signaling pathways which are key to the regulation of neurite outgrowth and neuronal function: SHP2/ERK5 and Cdc42/PAK. Mutational activation of SHP2 is linked to the multi-symptomatic Noonan syndrome and to various human malignancies, in particular pediatric leukemias and neuroblastoma (Tartaglia et al., 2003; Bentires-Alj et al., 2004).On the other hand, loss-of-function of several regulators of Cdc42, including huPak3, is associated with human mental retardation (for review see Govek et al., 2005). Analysis of NOMA-GAP signaling and function has provided novel insights into the regulation of neurite outgrowth and extension by NGF and may therefore lead to a better understanding into the development of human neurodegenerative conditions.
Materials and methods
Plasmid construction, RT-PCR, and reagents
Full-length human NOMA-GAP was subcloned from ESTs AW166303, AL137579, and BG772651 (RZPD Deutshces Ressourcenzentrum für Genomforschung GmbH, Berlin, Germany) into the mammalian expression vector pEFmyc.2. The N and C termini of NOMA-GAP and full-length human SHP2 were subcloned into the yeast expression vectors pGBT9, pGADT7 (CLONTECH Laboratories, Inc.), and pSrcBridge. pSrcBridge was generated by subcloning nonmyristoylated (G2E) active (Y416F Y527F) chicken Src into the second multiple cloning site of pBridge (CLONTECH Laboratories, Inc.). NOMA-GAP deletion and point mutants were subcloned into pEFmyc.2. NOMA-GAP C terminus was subcloned into pcDNA3Flag (Invitrogen). Wild-type and dominant-negative (delP mutation; provided by Dr. A.M. Bennett, Yale University School of Medicine, New Haven, CT; Tang et al., 1995) SHP2 were subcloned into pEF HA.6. Point mutations were inserted by PCR using oligonucleotide primers containing respective base substitutions. Y527F Src pEF and all pEF vectors have been described previously (Rosário et al., 1999). Control siRNA and NOMA-GAP siRNA were purchased from Dharmacon (siCONTROL Non-Targeting siRNA pool D-001206-13-20 and siGENOME SMARTpool D-050940, respectively). A pool of the following NOMA-GAP siRNAs was used: GUACAGAAAUGGAGGACAUUU, GGACAGACCAGAAGUUACUUU, and GAGGUCCUGUUCAGCGAUAUU. The siRNAs were also tested singly with similar effects. RICS siRNA has been previously described (Zhao et al., 2003). The following antibodies were used: Anti-Neurofilament 68, anti-α-tubulin, and anti-phospho-ERK1/2 (from Sigma-Aldrich); anti-HA tag (Roche); anti-NeuN antibody (Chemicon International); anti-Myc (A14), anti-SHP2 (C18), and anti-Trk (C14) antibodies from Santa Cruz Biotechnology, Inc.; Fast-Track anti-NOMA-GAP antibody (AbCam); anti-GFP antibody (Invitrogen); anti-Shc antibody (Upstate Biotechnology); anti- phospho-ERK5 (Biosource International); anti-ERK1/2, anti-phospho-tyrosine, anti-PAK1/2, and anti-phospho PAK1/2 from Cell Signaling Technology; and anti-Grb2, anti-Cdc42, and anti-phospho tyrosine PY20 from BD Transduction Laboratories. Polymerized actin was stained with Phalloidin-Texas red (Sigma-Aldrich), the lipophilic styryl dye FM 1-43FX (Invitrogen) was used to stain cell membranes and TOTO-3 (Invitrogen) was used to stain nuclei for counting NeuN staining in chick embryos. All fluorescent secondary antibodies (Cy2, Cy3, and Cy5) were from Jackson Laboratories. Phospho-specific and total protein immunoblotting for a given protein was performed simultaneously on duplicate Western blots. U0126 was from Calbiochem.
Cell culture, transfection, and in ovo electroporation
PC12 cells were grown at 37°C and 5% CO in DME (Invitrogen) supplemented with 10% horse serum and 5% fetal calf serum. Stimulations were carried after overnight culture in media with reduced serum (2.5% horse and 1.25% fetal calf serum) with 100 μg/μl (or 25 μg/μl for low levels) of NGF (Promega). DNA transfections were performed according to the manufacturer's instructions using LipofectAMINE 2000 (Invitrogen). 40 ng of pEGFP-C1 (CLONTECH Laboratories, Inc.) were used as a transfection marker. For transfections, PC12 cells were seeded onto poly-d-lysine/laminin-coated coverslips (BD Biosciences) or on poly-dl-ornithine/laminin (Sigma-Aldrich) coated tissue culture plates and transfected 24 h later. Fertilized chick eggs (Charles River Laboratories) were incubated at 38°C in a humidified incubator and unilateral in ovo electroporations were performed at room temperature using a T820 electro-squareporator (BTX, Inc.).
Immunofluorescence and in situ hybridization
For immunofluorescence, PC12 cells were prepared as described previously (Rosário et al., 2001). siRNA-transfected PC12 cells were stimulated with NGF 4 h after transfection and analyzed 72 h after transfection. PC12 cells overexpressing NOMA-GAP constructs were stimulated with low levels of NGF 24 h after transfection and analyzed 72 h after transfection. In PC12 differentiation assays, at least 150 GFP-positive cells were scored from random fields per condition and per experiment for the presence of short (30–100 μm) or long (>100 μm) processes. Mean pixel intensity of immunofluorescent staining for endogenous NOMA-GAP was determined for GFP-positive cells from jointly stained samples using LSM 5 Pascal version 3.2 software. Immunofluorescent staining of chick embryos was performed on 12-μm cryosections. Samples were analyzed at room temperature using a confocal microscope (LSM 510 META) equipped with Plan-Neofluar 40× 0.75 objective (all from Carl Zeiss MicroImaging, Inc.) and the software package LSM 5 Pascal version 3.2. In situ hybridizations were performed at room temperature using digoxygenin-labeled (Roche) RNA probes as previously described (Huelsken et al., 2000). Photos were taken at room temperature with an AxioCam HRC camera mounted on an Axioskop microscope with 10× and 20× Plan-Neofluar objectives (all from Carl Zeiss MicroImaging, Inc.). Images were adjusted for contrast and image size using Adobe Photoshop 7.0.
Statistical analysis of results
Results were analyzed using the statistical package SPSS. The multiple test of ANOVA was used to test the null hypotheses of equality of means of different groups in experiments with multiple conditions. The error probabilities of particular pairwise tests were corrected by Bonferroni. A t test was used to compare the means of experiments with only two conditions. Probabilities are summarized as asterisks above the graphs as follows: *, P < 0.05; **, P < 0.01.
Cell lysis, immunoprecipitations, and Rho family GTPase pull-downs
For endogenous coimmunoprecipitations and for whole-cell lysates, PC12 cells were lysed on ice in IP buffer (50 mM Tris, 100 mM NaCl, 1 mM EDTA, and 1% vol/vol Triton X-100, pH 7.4) containing protease and phosphatase inhibitors (10 μg/ml Leupeptin, 10 μg/ml Aprotinin, 5 μg/ml Pepstatin A, 1 mM Benzamidine, 0.5 μg/ml Microcystin LR, and 1 mM Na3VO4; all from Sigma-Aldrich). For immunoprecipitations of Myc-tagged NOMA-GAP, lysates were prepared on ice in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% vol/vol NP-40, 0.1% wt/vol SDS, and 0.5% wt/vol deoxycholate, pH 8.0) containing protease and phosphatase inhibitors. Immunoprecipitations were performed at 4°C for 1–2 h using equal amounts of protein (determined by Bradford assay) and using antibodies precoupled to protein G–Sepharose 4 Fast Flow (GE Healthcare) or, for Myc immunoprecipitations with anti-Myc agarose beads (Sigma-Aldrich), and subsequently washed thoroughly with lysis buffer. Cdc42 pull-downs were performed as described previously (Ogita and Takai, 2006) using recombinant GST-PAK CRIB (see Fig. S4). PC12 lysates for pull-downs were prepared on ice in pull-down buffer (50 mM Tris, 100 mM NaCl, 1% vol/vol Triton X-100, 0.1% wt/vol SDS, 0.5% wt/vol deoxycholate, and 1 mM DTT, pH 7.4) containing protease and phosphatase inhibitors, incubated for 30 min at 4°C with GST-PAK CRIB precoupled to Glutathione-Sepharose 4B beads (GE Healthcare), and then washed thoroughly in IP buffer containing 1 mM DTT and protease inhibitors. Images were processed and quantified using Adobe Photoshop 7.0.
SHP2 phosphatase and Elk1 transactivation assays
For SHP2 phosphatase assays, NIH3T3 cells were lysed on ice 24 h after transfection in Hepes buffer (25 mM Hepes, 100 mM NaCl, 1% vol/vol Triton X-100, 40% Glycerol, and 1 mM EDTA, pH 7.0) containing protease inhibitors, clarified by centrifugation, and incubated with recombinant GST-tagged wild-type SHP2 at 4°C (see Fig. S4). GST-SHP2 was recovered by incubation with Glutathione-Sepharose beads. SHP2 activity was assayed at room temperature on para-Nitrophenylphosphate (pNPP) as described previously (Niu et al., 1999). Elk1 transactivation assays were performed using the PathDetect Elk1 trans-Reporting System (Stratagene). Firefly and renilla (for normalization) luciferase activities were determined using the Dual-Luciferase Reporter Assay System kit (Promega) 24 h after transfection of RK13 epithelial cells.
Yeast two-hybrid analysis
Plasmids were cotransfected into yeast (S. cerevisiae Y190, AH109, and L40 strains; CLONTECH Laboratories, Inc.) as previously described (Rosário et al., 1999).), and plated on selection (−His-Leu-Trp+/− Met) or on total growth plates (−Leu-Trp) and grown at 32°C in a humidified incubator. A human brain cDNA library (HY4004AH; CLONTECH Laboratories, Inc.) was used for screening.
Online supplemental material
Fig. S1 shows NOMA-GAP family of multiadaptors. Fig. S2 shows expression of NOMA-GAP in the murine brain and eye. Fig. S3 shows expression of huNOMA-GAP deletion and mutant proteins in PC12 cells. Fig. S4 shows recombinant GST-SHP2 and GST-PAK CRIB proteins. Fig. S5 shows binding of RhoGTPase proteins to the NOMA-GAP RhoGAP domain. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200609146/DC1.
Supplementary Material
Acknowledgments
We would like to thank Stefan Britsch (Berlin) for the microarray work on mouse spinal cords. We would also like to thank Christina Eichhorn (Berlin) for carrying out statistical analysis of our data, Alexandra Klaus (Berlin) for help with in situ hybridizations, and Hendrik Wildner (Berlin) for advice on in ovo electroporations. In addition, we would like to acknowledge Anton Bennett for providing dominant-negative SHP2. We also thank Carmen Birchmeier, Alistair Garrett, and Dietmar Zechner (Berlin) for critical reading of the manuscript and for helpful discussions.
This work was partly funded through a Marie Curie fellowship (M. Rosário) and through a grant from the German National Genome Research Network (NGFN).
Abbreviations used in this paper: DRG, dorsal root ganglia; GAP, GTPase activating protein; NOMA-GAP, neurite outgrowth multiadaptor RhoGAP.
References
- Allen, M.J., X. Shan, and R.K. Murphey. 2000. A role for Drosophila Drac1 in neurite outgrowth and synaptogenesis in the giant fiber system. Mol. Cell. Neurosci. 16:754–765. [DOI] [PubMed] [Google Scholar]
- Aoki, K., T. Nakamura, and M. Matsuda. 2004. Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J. Biol. Chem. 279:713–719. [DOI] [PubMed] [Google Scholar]
- Arevalo, J.C., H. Yano, K.K. Teng, and M.V. Chao. 2004. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. EMBO J. 23:2358–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo, J.C., D.B. Pereira, H. Yano, K.K. Teng, and M.V. Chao. 2006. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J. Biol. Chem. 281:1001–1007. [DOI] [PubMed] [Google Scholar]
- Bentires-Alj, M., J.G. Paez, F.S. David, H. Keilhack, B. Halmos, K. Naoki, J.M. Maris, A. Richardson, A. Bardelli, D.J. Sugarbaker, et al. 2004. Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 64:8816–8820. [DOI] [PubMed] [Google Scholar]
- Boda, B., I. Nikonenko, S. Alberi, and D. Muller. 2006. Central nervous system functions of PAK protein family: from spine morphogenesis to mental retardation. Mol. Neurobiol. 34:67–80. [DOI] [PubMed] [Google Scholar]
- Capsoni, S., G. Ugolini, A. Comparini, F. Ruberti, N. Berardi, and A. Cattaneo. 2000. Alzheimer-like neurodegeneration in aged antinerve growth factor transgenic mice. Proc. Natl. Acad. Sci. USA. 97:6826–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh, J.E., J. Ham, M. Hetman, S. Poser, C. Yan, and Z. Xia. 2001. Differential regulation of mitogen-activated protein kinases ERK1/2 and ERK5 by neurotrophins, neuronal activity, and cAMP in neurons. J. Neurosci. 21:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, S.H., J. Hwang, M. Legendre, M. Zhang, A. Kimura, and A.R. Saltiel. 2003. TCGAP, a multidomain Rho GTPase-activating protein involved in insulin-stimulated glucose transport. EMBO J. 22:2679–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley, C., S.D. Spencer, M.C. Nishimura, K.S. Chen, S. Pitts-Meek, M.P. Armanini, L.H. Ling, S.B. McMahon, D.L. Shelton, A.D. Levinson, et al. 1994. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 76:1001–1011. [DOI] [PubMed] [Google Scholar]
- Cunnick, J.M., L. Mei, C.A. Doupnik, and J. Wu. 2001. Phosphotyrosines 627 and 659 of Gab1 constitute a bisphosphoryl tyrosine-based activation motif (BTAM) conferring binding and activation of SHP2. J. Biol. Chem. 276:24380–24387. [DOI] [PubMed] [Google Scholar]
- Daniels, R.H., P.S. Hall, and G.M. Bokoch. 1998. Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 17:754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J.M., G. Pearson, R. Baer, and M.H. Cobb. 1998. Identification of substrates and regulators of the mitogen-activated protein kinase ERK5 using chimeric protein kinases. J. Biol. Chem. 273:3854–3860. [DOI] [PubMed] [Google Scholar]
- Goldsmith, B.A., and S. Koizumi. 1997. Transient association of the phosphotyrosine phosphatase SHP-2 with TrkA is induced by nerve growth factor. J. Neurochem. 69:1014–1019. [DOI] [PubMed] [Google Scholar]
- Govek, E.E., S.E. Newey, and L. Van Aelst. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49. [DOI] [PubMed] [Google Scholar]
- Greene, L.A., and A.S. Tischler. 1976. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA. 73:2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadari, Y.R., H. Kouhara, I. Lax, and J. Schlessinger. 1998. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell. Biol. 18:3966–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof, P., S. Pluskey, S. Dhe-Paganon, M.J. Eck, and S.E. Shoelson. 1998. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 92:441–450. [DOI] [PubMed] [Google Scholar]
- Huelsken, J., R. Vogel, V. Brinkmann, B. Erdmann, C. Birchmeier, and W. Birchmeier. 2000. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148:567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indo, Y., M. Tsuruta, Y. Hayashida, M.A. Karim, K. Ohta, T. Kawano, H. Mitsubuchi, H. Tonoki, Y. Awaya, and I. Matsuda. 1996. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat. Genet. 13:485–488. [DOI] [PubMed] [Google Scholar]
- Kakimoto, T., H. Katoh, and M. Negishi. 2006. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J. Biol. Chem. 281:29042–29053. [DOI] [PubMed] [Google Scholar]
- Kamakura, S., T. Moriguchi, and E. Nishida. 1999. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 274:26563–26571. [DOI] [PubMed] [Google Scholar]
- Kao, S., R.K. Jaiswal, W. Kolch, and G.E. Landreth. 2001. Identification of the mechanisms regulating the differential activation of the mapk cascade by epidermal growth factor and nerve growth factor in PC12 cells. J. Biol. Chem. 276:18169–18177. [DOI] [PubMed] [Google Scholar]
- Kuroda, H., L. Fuentealba, A. Ikeda, B. Reversade, and E.M. De Robertis. 2005. Default neural induction: neuralization of dissociated Xenopus cells is mediated by Ras/MAPK activation. Genes Dev. 19:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux, P., Z.F. Altun-Gultekin, C. Lin, J.A. Wagner, and S.R. Heidemann. 1997. Rac is required for growth cone function but not neurite assembly. J. Cell Sci. 110:635–641. [DOI] [PubMed] [Google Scholar]
- Lechleider, R.J., R.M. Freeman Jr., and B.G. Neel. 1993. Tyrosyl phosphorylation and growth factor receptor association of the human corkscrew homologue, SH-PTP2. J. Biol. Chem. 268:13434–13438. [PubMed] [Google Scholar]
- Luo, L., Y.J. Liao, L.Y. Jan, and Y.N. Jan. 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8:1787–1802. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., M. Komiya, K. Sone, E. Hirose, N. Gotoh, H. Morii, Y. Ohta, and N. Mori. 2002. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 22:8721–8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka, Y., K. Nishida, Y. Fujio, M. Izumi, K. Terai, Y. Oshima, S. Sugiyama, S. Matsuda, S. Koyasu, K. Yamauchi-Takihara, et al. 2003. Activation of gp130 transduces hypertrophic signal through interaction of scaffolding/docking protein Gab1 with tyrosine phosphatase SHP2 in cardiomyocytes. Circ. Res. 93:221–229. [DOI] [PubMed] [Google Scholar]
- Nasu-Nishimura, Y., T. Hayashi, T. Ohishi, T. Okabe, S. Ohwada, Y. Hasegawa, T. Senda, C. Toyoshima, T. Nakamura, and T. Akiyama. 2006. Role of the Rho GTPase-activating protein RICS in neurite outgrowth. Genes Cells. 11:607–614. [DOI] [PubMed] [Google Scholar]
- Neel, B.G., H. Gu, and L. Pao. 2003. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28:284–293. [DOI] [PubMed] [Google Scholar]
- Ng, J., and L. Luo. 2004. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 44:779–793. [DOI] [PubMed] [Google Scholar]
- Nishimoto, S., M. Kusakabe, and E. Nishida. 2005. Requirement of the MEK5-ERK5 pathway for neural differentiation in Xenopus embryonic development. EMBO Rep. 6:1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, T., X. Liang, J. Yang, Z. Zhao, and G.W. Zhou. 1999. Kinetic comparison of the catalytic domains of SHP-1 and SHP-2. J. Cell. Biochem. 72:145–150. [DOI] [PubMed] [Google Scholar]
- Nobes, C.D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62. [DOI] [PubMed] [Google Scholar]
- Obermeier, A., S. Ahmed, E. Manser, S.C. Yen, C. Hall, and L. Lim. 1998. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 17:4328–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita, H., and Y. Takai. 2006. Activation of Rap1, Cdc42, and rac by nectin adhesion system. Methods Enzymol. 406:415–424. [DOI] [PubMed] [Google Scholar]
- Okabe, T., T. Nakamura, Y.N. Nishimura, K. Kohu, S. Ohwada, Y. Morishita, and T. Akiyama. 2003. RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-D-aspartate receptor signaling. J. Biol. Chem. 278:9920–9927. [DOI] [PubMed] [Google Scholar]
- Ong, S.H., Y.P. Lim, B.C. Low, and G.R. Guy. 1997. SHP2 associates directly with tyrosine phosphorylated p90 (SNT) protein in FGF-stimulated cells. Biochem. Biophys. Res. Commun. 238:261–266. [DOI] [PubMed] [Google Scholar]
- O'Reilly, A.M., and B.G. Neel. 1998. Structural determinants of SHP-2 function and specificity in Xenopus mesoderm induction. Mol. Cell. Biol. 18:161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera, E.M., A. Ikeda, E. Eivers, and E.M. De Robertis. 2003. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 17:3023–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskey, S., T.J. Wandless, C.T. Walsh, and S.E. Shoelson. 1995. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J. Biol. Chem. 270:2897–2900. [DOI] [PubMed] [Google Scholar]
- Reichardt, L.F. 2006. Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1545–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosário, M., and W. Birchmeier. 2003. How to make tubes: signaling by the Met receptor tyrosine kinase. Trends Cell Biol. 13:328–335. [DOI] [PubMed] [Google Scholar]
- Rosário, M., H.F. Paterson, and C.J. Marshall. 1999. Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J. 18:1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosário, M., H.F. Paterson, and C.J. Marshall. 2001. Activation of the Ral and phosphatidylinositol 3′ kinase signaling pathways by the ras-related protein TC21. Mol. Cell. Biol. 21:3750–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi, A., J.D. Delcroix, P.V. Belichenko, K. Zhan, C. Wu, J.S. Valletta, R. Takimoto-Kimura, A.M. Kleschevnikov, K. Sambamurti, P.P. Chung, et al. 2006. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 51:29–42. [DOI] [PubMed] [Google Scholar]
- Simon, J.A., and S.L. Schreiber. 1995. Grb2 SH3 binding to peptides from Sos: evaluation of a general model for SH3-ligand interactions. Chem. Biol. 2:53–60. [DOI] [PubMed] [Google Scholar]
- Smeyne, R.J., R. Klein, A. Schnapp, L.K. Long, S. Bryant, A. Lewin, S.A. Lira, and M. Barbacid. 1994. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 368:246–249. [DOI] [PubMed] [Google Scholar]
- Strasser, G.A., N.A. Rahim, K.E. VanderWaal, F.B. Gertler, and L.M. Lanier. 2004. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 43:81–94. [DOI] [PubMed] [Google Scholar]
- Tang, T.L., R.M. Freeman Jr., A.M. O'Reilly, B.G. Neel, and S.Y. Sokol. 1995. The SH2-containing protein-tyrosine phosphatase SH-PTP2 is required upstream of MAP kinase for early Xenopus development. Cell. 80:473–483. [DOI] [PubMed] [Google Scholar]
- Tartaglia, M., C.M. Niemeyer, A. Fragale, X. Song, J. Buechner, A. Jung, K. Hahlen, H. Hasle, J.D. Licht, and B.D. Gelb. 2003. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34:148–150. [DOI] [PubMed] [Google Scholar]
- Wang, X., and C. Tournier. 2006. Regulation of cellular functions by the ERK5 signalling pathway. Cell. Signal. 18:753–760. [DOI] [PubMed] [Google Scholar]
- Watson, F.L., H.M. Heerssen, A. Bhattacharyya, L. Klesse, M.Z. Lin, and R.A. Segal. 2001. Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat. Neurosci. 4:981–988. [DOI] [PubMed] [Google Scholar]
- Wright, J.H., P. Drueckes, J. Bartoe, Z. Zhao, S.H. Shen, and E.G. Krebs. 1997. A role for the SHP-2 tyrosine phosphatase in nerve growth-induced PC12 cell differentiation. Mol. Biol. Cell. 8:1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui, H., H. Katoh, Y. Yamaguchi, J. Aoki, H. Fujita, K. Mori, and M. Negishi. 2001. Differential responses to nerve growth factor and epidermal growth factor in neurite outgrowth of PC12 cells are determined by Rac1 activation systems. J. Biol. Chem. 276:15298–15305. [DOI] [PubMed] [Google Scholar]
- Yuzawa, S., K. Ogura, M. Horiuchi, N.N. Suzuki, Y. Fujioka, M. Kataoka, H. Sumimoto, and F. Inagaki. 2004. Solution structure of the tandem Src homology 3 domains of p47phox in an autoinhibited form. J. Biol. Chem. 279:29752–29760. [DOI] [PubMed] [Google Scholar]
- Zhao, C., H. Ma, E. Bossy-Wetzel, S.A. Lipton, Z. Zhang, and G.S. Feng. 2003. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J. Biol. Chem. 278:34641–34653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.