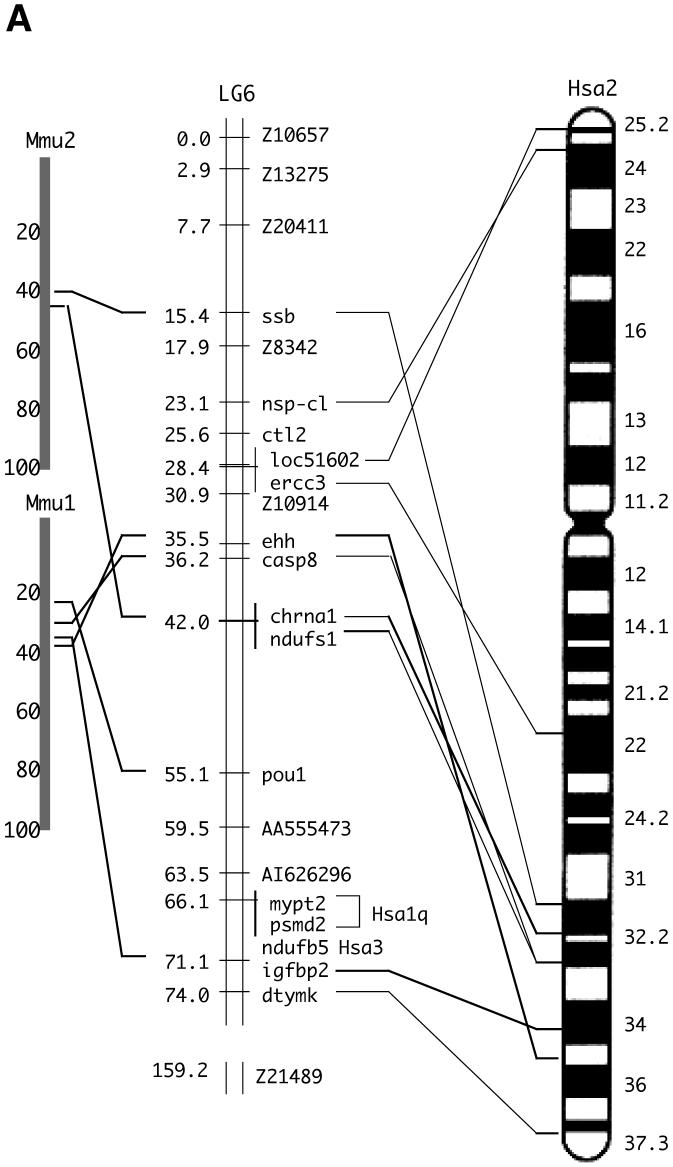
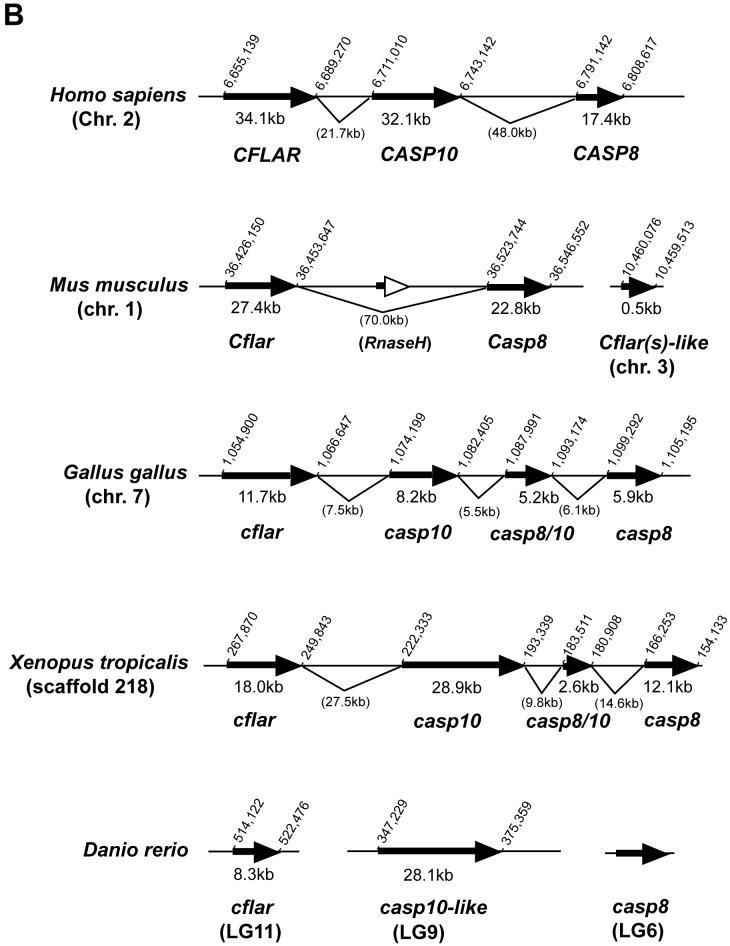
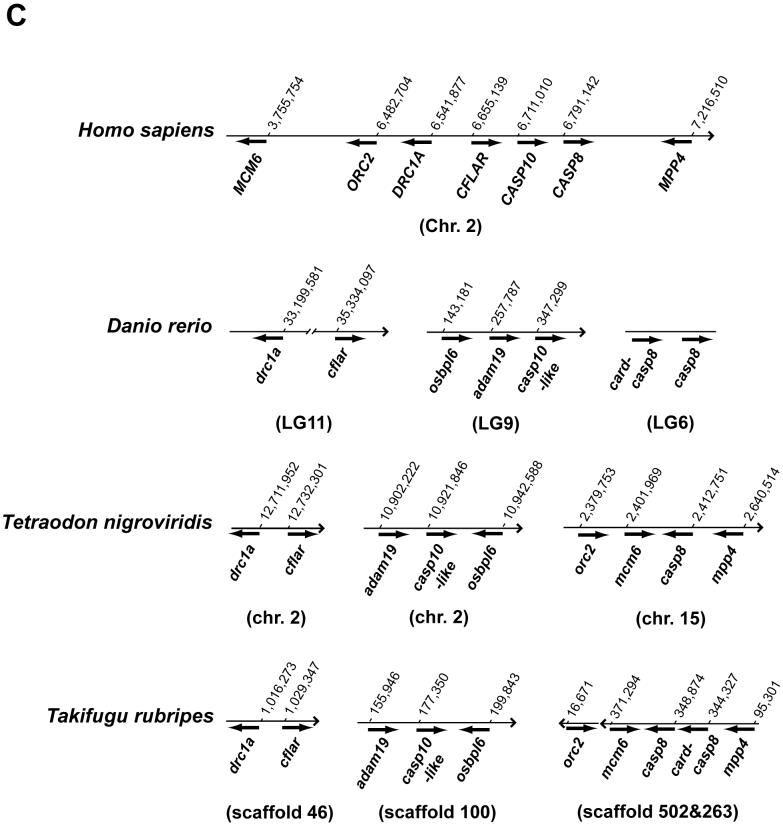
Fig. 2.
Chromosomal analyses of the caspase-8 gene and its orthologs.
(A) Chromosomal mapping of the zebrafish casp8 gene. Center, zebrafish linkage group 6 (LG6), numbers indicate distance from the top of the chromosome in centiMorgans, with casp8 at 36.2 centiMorgans. Left and right, locations of mouse (left) and human (right) orthologs of human Chr. 2 genes mapped on LG6. (B) Physical map of the region including the casp8 gene and its related genes in vertebrates. The bold arrows indicate the coding region and orientation of the gene. In human, the CASP8, CASP10 and CFLAR genes form a cluster on Chr. 2. In zebrafish, the casp8, casp10 and cflar genes localize on different chromosomes. Rodents have lost the Casp10 gene following the insertion of RnaseH. The chicken and frog have an additional caspase-8/10-homologus gene, casp8/10 between the casp8 and casp10 genes. (C) Comparative mapping of the CASP8, CASP10 and CFLAR genes in human and fish. The bold arrows indicate the coding region and orientation of the gene. The casp8-like gene resides near the casp8 gene on both zebrafish and fugu chromosomes. Abbreviated genes: adam19; a disintegrin and metalloproteinase domain 19; DRC1A, down-regulated by CTNNB1 protein A, MCM6, minichromosome maintenance protein 6; MPP4; MAGUK p55 subfamily member 4; osbpl6, oxysterol-binding protein-like protein 6; ORC2, origin recognition complex, subunit 2.