Abstract
In humans, UDP glucuronosyltransferase (UGT) operates in opposition to glucuronidase (GUS) to control activity of diverse metabolites such as hormones, by reversible conjugation with glucuronic acid. Previous data revealed that, as in mammals, these enzymes are required for plant life in that a UGT from Pisum sativum (PsUGT1) controls plant development by opposing endogenous glucuronidase (GUS) activity thereby modulating the duration of the cell cycle. Here we report that a small family of genes (AtUGT85A1, 2, 3, 4, 5, and 7) homologous to pea PsUGT1 exists in the Arabidopsis genome. The AtUGT85A encoded-proteins are predicted to be membrane-associated enzymes. Three genes (AtGUS1, AtGUS2, and AtGUS3) that are homologous to a GUS encoding gene from Scutellaria baicalensis (sGUS) were identified. The AtGUS encoded proteins are predicted to be secretory (AtGUS1) as well as membrane-associated (AtGUS2 and AtGUS3) enzymes. Both AtUGT85A and AtGUS genes, like PsUGT1, exhibit localized, tissue-specific expression, mainly in areas of active cell division with possible involvement in cell cycle regulation.
Keywords: UDP-glycosyltransferase, glucuronidase, Arabidopsis AtUGT85A family, Lethality
Glycosylation is a critical metabolic pathway with diverse roles in cellular processes [1]. In plants, glycosylation of phytochemicals by the addition of glucose or other sugars generally results in enhanced water solubility and lower chemical reactivity, allowing long-term storage in vacuoles or cell walls [2]. Reversible conjugation of hormones such as auxin and cytokinin may be important in ‘homeostasis’ for the regulation of physiologically active hormone levels [3]. In other cases, conjugation of plant hormones might accompany or introduce irreversible deactivation.
Based on amino acid sequence similarities, glycosyltransferases (GTs) from diverse species have been classified into 87 families (http://afmb.cnrs-mrs.fr/CAZY/), with hundreds of GTs in the Arabidopsis thaliana genome. GTs are the families of carbohydrate-active enzymes that add sugars to diverse substrates [37, 38]. Family 1 GTs (‘UGTs’) are predicted to transfer nucleotide-diphosphate-activated sugars to a diverse array of low-molecular weight ‘secondary’ metabolites [4-6, 34]. Family 1 GTs are classified based on the presence of a 44-50 amino acid C-terminal consensus sequence. This sequence, thought to represent the nucleotide-sugar binding site, is termed the plant secondary product GT (PSPG) consensus [39], and has been identified in 120 putative PSGTs in Arabidopsis [6]. In family 1 GTs, 120 GTs in Arabidopsis are classified into 14 groups (i.e., groups A-N) based on their substrate specificities [12, 36].
Family 1 GTs has been extensively studied both in Arabidopsis and human. In mammals, UGTs coordinate the cellular activity of endogenous signal molecules such as steroid hormones and detoxify xenobiotic molecules from the environment [7, 8]. Polymorphisms among human UGT genes are associated with increased susceptibility to diseases including cancer, and loss-of-function mutations can be lethal [9-11]. In mammals, UGTs transfer sugar from UDP-glucuronic acid to endogenous acceptors. In contrast, plant UGTs transfer glucose from UDP-glucose to acceptors including hormones and small molecules [35, 36].
Despite their long-established role in animal metabolism, UGT activities in plants historically have received relatively little attention [12, 13]. Predicted substrates of plant UGTs include diverse chemicals such as flavonoids, terpenes, auxin, cytokinin, salicylic acid, and sterols, which are postulated to play key roles in plant development, metabolism, and defense [14, 15, 34]. As such they comprise important targets for agronomic and medicinal applications [16]. To date, however, information about the expression, function, substrate, substrate specificity, and biological effects in plants is limited to a few individual UGTs [17-21]. One obstacle to defining function is the chemical complexity and diversity of the metabolites that may be modified by UGT activities. Plants produce >9000 species of the flavonoid class alone, and a single flavonoid, quercetin, can exist as 350 distinctive glycosides by the action of UGTs [17, 22].
In previous studies, we isolated a Pisum sativum UDP-glucuronosyltransferase gene (PsUGT1) and established that its expression is localized to meristems and is tightly correlated with the induction of mitosis [20, 21]. The PsUGT1 enzyme, when expressed in Neurospora crassa, was shown to glycosylate a single metabolite from pea root extracts [21]. Inhibiting PsUGT1 expression by antisense mRNA expression under the control of the PsUGT1 promoter resulted in lethality in pea and alfalfa.
As a part of the characterization of PsUGT1 expression, efforts were made to develop a reporter gene using the PsUGT1 promoter fused to the standard reporter gene E. coli uidA encoding glucuronidase (GUSA) [23]. Surprisingly, when expressed in coordination with meristem-localized UGT expression, GUSA expression was lethal. Two explanations could account for this result. First, the lethal effect of the PsUGT1 promoter-controlled expression of GUSA, which removes glucuronic acid from diverse substrates, could verify the importance of PsUGT1 by the artifactual neutralization of its biological activity when the two genes are expressed in the same time and place. Alternatively, PsUGT1 might operate normally in conjunction with an endogenous GUS activity, which operates in tandem with PsUGT1 to modulate activity of specific signals controlling the cell cycle [23]. Predictions of this hypothesis are that (1) a gene or genes encoding GUS are present in the pea genome; (2) GUS activity is expressed at the same time and place as PsUGT1; and (3) GUS activity exerts physiological effects in opposition to those of PsUGT1 by reversing the enzyme’s glucuronidation of biologically active metabolites.
In microorganisms and animals, GUS operates in tandem with UGTs in the reversible conjugation of a range of biologically active molecules [40, 41, 42]. In vertebrates, GUS is expressed in most if not all tissues. Though its normal substrates are not well characterized, it has been implicated in functions ranging from sperm cytoskeleton structure [43] to regulation of thyroid hormone levels in cardiac fibroblasts [44]. Its key role in metabolism has made it a prime target for genetic therapies in humans [45, 46].
One plant GUS-encoding gene has been isolated from Scutellaria baicaliensis (sGUS) [24, 25]. In preliminary studies to examine the hypothesis that PsUGT1 operates in conjunction with a GUS enzyme in legumes, efforts to identify GUS genes in pea using probes from sGUS were unsuccessful (unpublished). In this study, we exploit the Arabidopsis genome as an alternative model to test predictions of the hypothesis that co-localized UGT and GUS activities in meristems control key aspects of plant development. We report that six genes (AtUGT85A1, 2, 3, 4, 5, and 7) exhibit homology to pea PsUGT1, and that three genes (AtGUS1, AtGUS2, and AtGUS3), which are homologous to sGUS, are also present in the Arabidopsis genome. We used these genes as tools to examine the hypothesis that members of the AtUGT85A gene family are expressed in coordination with AtGUS genes, with opposing effects on physiology and development.
Results
To initiate studies for using the Arabidopsis model to characterize the relations between UGT and GUS activities in plant development, the Arabidopsis database was searched. In Arabidopsis, six AtUGT85A genes, named AtUGT85A4, 1, 3, 5, 2, and 7, homologous to PsUGT1, and three AtGUS genes, named AtGUS1, AtGUS2, and AtGU3, homologous to Scutellaria sGUS were identified (Fig. 1). Phylogenetic analysis showed that six AtUGT85A genes together with PsUGT1 belong to group G of family 1 glycosyltransferases (Fig. 2) [12, 34, 35, 36].
Fig. 1.
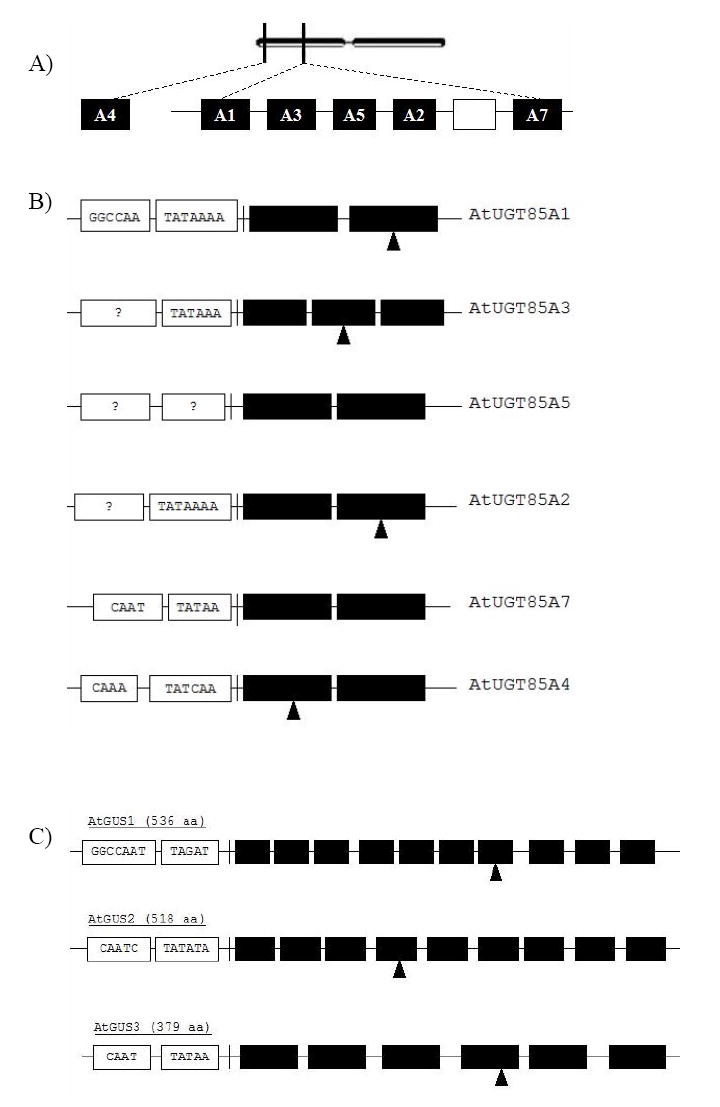
Structure of six AtUGT85A genes and three AtGUS genes in Arabidopsis.
(A) Structure of AtUGT85A1, 2, 3, 4, 6, and 7 on chromosome 1. Five AtUGT85A genes are clustered together in an 18 kb region on the left arm of chromosome 1. AtUGT85A4 is located at the end of left arm of chromosome 1. Open rectangle between A2 and A7 indicates a pseudo gene. (B) Each AtUGT85A gene has its own transcription unit including a TATAA box and a CAAT box. In AtUGT85A5, TATAA and CAAT boxes are not found. All the AtUGT85A genes have introns with G/T and A/G conserved splicing sequences. The second intron in AtUGT85A3 and the intron in AtUGT85A5 do not contain conserved splicing sequences. AtUGT85A1 and a partial 5’-region of AtUGT85A3 were found in BAC clone F12K8 (AC006551). A partial sequence of the 3’-region of AtUGT85A3, and complete sequences of AtUGT85A3, 2, and 7 were found in BAC clone T16E15 (AC068562). A complete sequence of AtUGT85A4 was found in BAC clone F12K8 (AC006551). The closed triangles under AtUGT85A1, 2, 3, and 4, indicate the T-DNA insertion site. (C) Structure of three AtGUS genes on chromosome 5. Each gene has its own transcription unit including a TATAA box and a CAAT box. Nine introns are found in the AtGUS1 gene, 8 introns are found in the AtGUS2 gene, and 5 introns are found in the AtGUS3 gene; i indicates intron. G/T and A/G indicate conserved splicing sequences for intron excision. The closed triangles under AtGUS1, AtGUS2, and AtGUS3 indicate the T-DNA insertion site. The AtGUS1 gene was found in the P1 clone MFB13 (AB010073). The AtGUS2 and AtGUS3 genes were found in the P1 clone MXM12 (AB005249).
Fig. 2.

A phylogenetic tree showing the relationship of the AtUGT85A-encoded proteins with each other. Two subgroups are found; AtUGT85A1 and AtUGT85A3 belong to the same subgroup. AtUGT85A2 and AtUGT85A7 belong to another subgroup. The tree was generated using CLUSTAL W with a gap-opening penalty of 10 and a gap extension penalty of 0.1.
A family of Arabidopsis AtUGT85A genes related to pea PsUGT1
Six genes are clustered together in an 18 kb region on the left arm of chromosome 1 (Fig. 1A). AtUGT85A4 is located at the end of left arm of chromosome 1. The AtUGT85A sequences have 63-66% DNA sequence identity and 57-64% amino acid sequence identity with the coding region in PsUGT1 (Table 2).
Table 2.
Comparisons of mRNA and protein identities between the predicted full-length sequences of the AtUGT85A1, AtUGT85A3, AtUGT85A5, AtUGT85A2, AtUGT85A7, and AtUGT85A4. Pair-wise analysis was done using San Diego Supercomputer Center Biology workbench (http://workbench.sdsc.edu/), Optimal Global Sequence Alignment (ALIGN) with Gap open penalty: -16.00 and Gap extension penalty: -4.
| mRNA | AtUGT85A1 | AtUGT85A3 | AtUGT85A5 | AtUGT85A2 | AtUGT85A7 | AtUGT85A4 |
|---|---|---|---|---|---|---|
| PsUGT1 | 66% | 65% | 65% | 65% | 65% | 63% |
| AtUGT85A1 | 81% | 78% | 80% | 78% | 68% | |
| AtUGT85A3 | 77% | 78% | 77% | 67% | ||
| AtUGT85A5 | 80% | 76% | 67% | |||
| AtUGT85A2 | 79% | 68% | ||||
| AtUGT85A7 | 67% | |||||
| Protein | AtUGT85A1 | AtUGT85A3 | AtUGT85A5 | AtUGT85A2 | AtUGT85A7 | AtUGT85A4 |
| PsUGT1 | 61% | 61% | 60% | 64% | 61% | 57% |
| AtUGT85A1 | 78% | 72% | 79% | 74% | 62% | |
| AtUGT85A3 | 72% | 78% | 75% | 60% | ||
| AtUGT85A5 | 77% | 73% | 59% | |||
| AtUGT85A2 | 79% | 61% | ||||
| AtUGT85A7 | 58% | |||||
| mRNA | AtGUS1 | AtGUS2 | AtGUS3 | |||
| SGUS | 54% | 53% | 54% | |||
| AtGUS1 | 75% | 55% | ||||
| AtGUS2 | 53% | |||||
| HPSE | 47% | 46% | 47% | |||
| Protein | AtGUS1 | AtGUS2 | AtGUS3 | |||
| sGUS | 38% | 39% | 30% | |||
| AtGUS1 | 73% | 34% | ||||
| AtGUS2 | 33% | |||||
| HPSE | 26% | 28% | 18% |
Five AtUGT85A genes have a TATAA box located upstream of the transcription initiation site (Fig. 1B). AtUGT85A1, 7, and 4 genes contain a CAAT box upstream of the TATAA box, whereas AtUGT85A2, 3, and 5 genes lack a CAAT box. AtUGT85A5, in contrast, lacks a TATAA box. The sequences of the six AtUGT85A genes are conserved within the family. With the exception of AtUGT85A3, which has 2 introns, each gene contains a single intron. Furthermore, except for AtUGT85A5 and the second intron of AtUGT85A3, each intron has G-T/A-G conserved splicing sequences at the 5’ and 3’ ends.
A phylogenetic tree analysis in mRNA and encoded protein sequences shows that AtUGT85A3 and AtUGT85A1 belong to the same subfamily, and AtUGT85A2 and AtUGT85A7 belong to another subfamily (Fig. 2). Pair-wise comparisons of the DNA sequences among family members show that each AtUGT85A mRNAs shares 67% to 81% identity (Table 2). Pair-wise comparisons of the amino acid sequences among the family members show that each of the AtUGT85A-encoded proteins share 58% to 79% identity (Table 2). AtUGT85A genes have less similarity (45-50% identity in DNA sequences, and 25-30% identity in protein sequences, not shown) with any other known glycosyltransferases, including UDP-glucosyltransferases.
The greatest amino acid identity is found in the predicted UDP-sugar binding domain of 50 amino acids at the C-terminal of protein (Fig. 3). This PSPG (plant secondary product glycosyltransferase) motif, which is found in most UGTs, also contains highly conserved ‘H(C/S)GWNS’ residues for direct interaction with uracil moiety of UDP-sugar (Fig. 3) [26]. Together with PsUGT1, the Arabidopsis AtUGT85A genes belong to the GT1 family of plant glycosyltransferases (Family GT1; http://afmb.cnrs-mrs.fr/CAZY/). In the Arabidopsis genome, 14 groups of ~120 secondary metabolism UGT genes in family GT1 were identified [12, 4]. According to this classification, AtUGT85A genes belong to group G.
Fig. 3.

Comparison of amino acid sequences with a region of the deduced PsUGT1 (AF034743) together with the conserved UDP-glucuronic acid binding site and corresponding regions in Arabidopsis AtUGT85As (lower six sequences). Shaded sequences represent absolute matches between the conserved UDP-glucuronic acid-binding site in PsUGT1 and AtUGT85As. Box of dotted line indicates highly conserved ‘H(C/S)GWNS’ residues for direct interaction with uracil moiety of UDP-sugar.
The PsUGT1-encoded protein was predicted to be a membrane-associated enzyme [21]. The WoLF PSORT program (http://psort.nibb.ac.jp/) predicts that all AtUGT85A-encoded proteins are membrane-associated enzymes, targeting chloroplasts and ER. All the AtUGT85A proteins contain the xKQxxEF motif for microsome retention (Table 3). The AtUGT85A1 protein contains the ER membrane retention signal QKSQ at the C-terminus, and the AtUGT85A3 protein has the ER membrane retention signal GSRF at the N-terminus and KIPN at the C-terminus (Table 3). All six AtUGT85A-encoded proteins have several post-translational modification sites, including signature motifs for O-β-GlcNAc attachment sites, and serine-, threonine-, tyrosine-phosphorylation sites (Table 4). This suggests that all the AtUGT proteins are extensively modified post-translationally.
Table 3.
Prediction of membrane-association of AtUGT85A encoded-proteins and AtGUS encoded-proteins. The presence of N-terminal signal peptide was determined by SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/). The certainty to be membrane-associated was determined by using WoLF PSORT Prediction (http://psort.nibb.ac.jp/). The PsUGT1-encoded protein is membrane-associated [21]. The AtGUS1-encoded protein has an 85% chance of being extracellularly secreted.
| xKQxxEF (Lysosomal/Golgi Loc. Sig.) | ER membrane retention signal | |
|---|---|---|
| AtUGT85A1 | present | QKSQ at C-terminus |
| AtUGT85A3 | present | GSRF at N-terminus/KIPN at C-terminus |
| AtUGT85A5 | present | absent |
| AtUGT85A2 | present | absent |
| AtUGT85A4 | present | absent |
| AtUGT85A7 | present | absent |
| PsUGT1 | present | absent |
| N-terminal hydrophobic signal peptide | ||
| AtGUS1 | present | |
| AtGUS2 | present | |
| AtGUS3 | present |
Table 4.
Post-translational modification of AtUGT85A and AtGUS-encoded proteins. Prediction of O-β-GlcNAc attachment sites was determined by YinOYang 1.2 program (http://www.cbs.dtu.dk/services/YinOYang/). Prediction of phosphorylation sites was determined by NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/). O-β-GlcNAc = N-acetyl glucosamine.
| Conserved UDP-GA binding site | No. of O-b-GlcNAc attachment sites | No. of Serine phosphorylation | No. of Threonine phosphorylation | No. of Tyrosine phosphorylation | |
|---|---|---|---|---|---|
| AtUGT85A1 | present | 4 | 5 | 5 | 2 |
| AtUGT85A3 | present | 6 | 10 | 6 | 1 |
| AtUGT85A5 | present | 3 | 12 | 5 | 2 |
| AtUGT85A2 | present | 3 | 12 | 5 | 3 |
| AtUGT85A7 | present | 4 | 14 | 4 | 3 |
| AtUGT85A4 | present | 7 | 8 | 6 | 3 |
| PsUGT1 | present | 2 | 4 | 5 | 3 |
| Leucine zipper motif | No. of O-β-GlcNAc attachment sites | No. of Serine phosphorylation | No. of Threonine phosphorylation | No. of Tyrosine phosphorylation | |
| AtGUS1 | present | 5 | 8 | 4 | 8 |
| AtGUS2 | present | 7 | 14 | 4 | 8 |
| AtGUS3 | absent | 5 | 11 | 5 | 6 |
A family of Arabidopsis genes related to Scutellaria sGUS
Three genes with significant DNA and amino acid similarities to Scutellaria sGUS [24] have been identified from the Arabidopsis database. Except these three AtGUS genes, no other similar genes were identified in the Arabidopsis genome. These genes, AtGUS1, AtGUS2, and AtGUS3 have 53-54% nucleotide sequence identity with the sGUS coding region, and 30-39% amino acid sequence identity with the sGUS-encoded protein (Table 2). AtGUS1, AtGUS2, and AtGUS3 also have 46-47% nucleotide sequence identity with the human heparanase (HPSE) coding region, and 18-28% amino acid sequence identity with the HPSE-encoded protein (Table 2) [27]. Pair-wise comparisons between AtGUS1, AtGUS2, and AtGUS3 mRNAs show that AtGUS mRNAs share 53-75% identity. Pair-wise comparison of the amino acid sequences between AtGUS1 and AtGUS2-encoded proteins show a strong identity (73%), and a weak identity (33%) between AtGUS1/2-encoded proteins and AtGUS3-encoded protein (Table 2). A phylogenetic tree analysis in AtGUS-encoded protein sequences shows that AtGUS proteins are closely related to the Scutellaria sGUS protein and distantly related to the human heparanase enzymes (not shown).
AtGUS1 is located in the right arm of chromosome 5, AtGUS2 is located in the left arm of chromosome 5, and AtGUS3 is located in the middle of chromosome 5. All three AtGUS genes have a TATAA box and a CAAT box located upstream of the transcription initiation site. The AtGUS1 gene contains 9 introns, the AtGUS2 gene contains 8 introns, and the AtGUS3 gene contains 5 introns (Fig. 1C). In AtGUS1, the second and fourth introns have G-T/A-G conserved splicing sequences, whereas the third and seventh introns have G-T/A-G conserved splicing sequences in AtGUS2. In AtGUS3, the first and third introns have G-T/A-G conserved splicing sequences. The remaining introns in the AtGUS1, AtGUS2, and AtGUS3 genes do not contain conserved splicing sequences.
The SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) predicts the presence of an N-terminal hydrophobic signal peptide in three AtGUS-encoded proteins (Table 3). The WoLF PSORT program (http://psort.nibb.ac.jp/) predicts that the AtGUS1-encoded protein is secreted and that AtGUS2 and AtGUS3-encoded proteins are membrane associated enzymes. Both AtGUS1- and AtGUS2-encoded proteins contain a leucine zipper motif. For the AtGUS1 encoded-protein, this motif (LNREEYHLSPKDGDLRSKIMLL) is located at 470-491 and for the AtGUS2 encoded-protein the leucine zipper motif (LNREEYHLTPENGVLRSKTMVL) is at 452-473. No leucine zipper motif was found in the predicted AtGUS3-encoded protein. In both AtGUS1 and AtGUS2 proteins, two motifs, an ATP/GTP binding site and leucine zipper, are present. Together with sGUS, the Arabidopsis AtGUS1 and AtGUS2 genes belong to the plant glycoside hydrolase family 79 (GH79; http://afmb.cnrs-mrs.fr/CAZY/). AtGUS encoded-proteins have several post-translational modification sites including O-β-GlcNAc attachment sites, and serine-, threonine-, tyrosine-phosphorylation sites (Table 4), suggesting that AtGUS proteins are extensively modified post-translationally.
Expression profiles of AtUGT85A genes and AtGUS genes
In previous studies, PsUGT1 expression was localized to regions of active cell division in leaves, roots, and flowers [19-21]. If the AtUGT85A family carries out a similar role in development, then similar expression patterns are predicted to occur in Arabidopsis. To characterize expression patterns of the AtUGT85A gene family members, three complementary approaches were used: (1) reverse transcriptase-polymerase chain reaction (RT-PCR) with gene specific primers (Table 1, Fig. 4); (2) promoter∷uidA expression in transgenic Arabidopsis plants (Fig. 5A-F); and (3) whole mount in situ hybridization (WISH) (Fig. 5G-I). The AtGUS family was analyzed in parallel to examine the hypothesis that these three enzymes operate in tandem to carry out reversible glucuronidation of specific metabolites.
Table 1.
Primers used for promoter∷uidA cloning, WISH probe generation, and RT-PCR.
| AtUGT85A1 Promoter 5’ | AGTCAGAAAGCTTTCTACTTATAATTCAATTTTCTCTCG |
| AtUGT85A1 Promoter 3’ | TGCGACAGGATCCTGGTTTGCCTTTGAAATAAGCCA |
| AtUGT85A3 Promoter 5’ | ACTGATGGTCGACTTCTATAATGATTACTAAGTAAGTG |
| AtUGT85A3 Promoter 3’ | AGTACTCGGATCCTCTATCTCTCTTTTGCCAAAAGAC |
| AtUGT85A5 Promoter 5’ | CAGTCGAAAGCTTAAGATTAATTGAAAACGTAAGATTGTC |
| AtUGT85A5 Promoter 3’ | AGCTGACGGATCCTGTTGATCCGCCGGAGAAGCTC |
| AtUGT85A2 Promoter 5’ | CAGTCGAAAGCTTGTGGAGTTTGCTTGGGGTTTAGC |
| AtUGT85A2 Promoter 3’ | TGCAGTCGGATCCGATTCCTTTACGCTTATAGGACTC |
| AtUGT85A7 Promoter 5’ | ACTGAATTCTAGAAGGTCGAGACAGTGGTTAGAGAG |
| AtUGT85A7 Promoter 3’ | AGTCAGAGGATCCGGCTTTTGTGCGTTATGAACAAC |
| AtUGT85A4 Promoter 5’ | CCTCGATTTAATTAAATTTTCTTGAGTAACTCCTTCTCTG |
| AtUGT85A4 Promoter 3’ | CTCGTGAGGCGCGCCAGTGTATCGTCCCATAATTGATAGT |
| AtGUS1 Promoter 5’ | CTAGGCCTGCAGATATTTCCTCGTTGATCATCAATGACA |
| AtGUS1 Promoter 3’ | GCTTGCGGTACCGTTCTTGGAGTTGACAACACTTCATC |
| AtGUS2 Promoter 5’ | TCAGTAGAAGCTTCAACATACTTCCAAACTGACTATG |
| AtGUS2 Promoter 3’ | AGTCAGAGGATCCTGTTTTCTCAGGGACTAGAAGAAG |
| AtGUS3 Promoter 5’ | CTAGGCCTGCAGATAACTAACCGGAGCATTGACTAC |
| AtGUS3 Promoter 3’ | GCTTGCGGTACCGAAAAATGAGATCTCATTTGGTG |
| AtUGT85A1-WISH 5’ | ATGTCTGAAGCTTGGATCCAAAACTCACATTTGTTCATTACAA |
| AtUGT85A1-WISH 3’ | ATGTCTGTCTAGAGGTACCTGAGTTATGAATG ATCTGAGATC |
| AtUGT85A2-WISH 5’ | ATGTCTGGGTACCAAGCTTCCCAAATTAA TTACTCATTTACTC |
| AtUGT85A2-WISH 3’ | ATGTCTGTCTAGACTCGAGGAACAATTTTTTATCAAAACAAAGTC |
| AtUGT85A7-WISH 5’ | ATGTCTGGGTACCAAGCTTACTGTCTCGCTCCATTCAAAGAG |
| AtUGT85A7-WISH 3’ | ATGTCTGTCTAGACTCGAGCTTTAAAAGGAGATAGACCCTTCTC |
| AtGUS1-WISH 5’ | ATGTCTGAAGCTTGAATTCTCTGCTCTGTTTTGAGATCCAAC |
| AtGUS1-WISH 3’ | ATGTCTGTCTAGAGGTACCTCGTTCCATGTTGCTACCAAAAG |
| AtGUS2-WISH 5’ | ATGTCTGAAGCTTGGATCCACAAGAAACTTGACAGCTTCTC |
| AtGUS2-WISH 3’ | ATGTCTGTCTAGAGGTACCGAGATGACAGGGGGTGGACTC |
| AtGUS3-WISH 5’ | ATGTCTGAAGCTTATGGCTTATCGTCAAATTTTGGC |
| AtGUS3-WISH 3’ | ATGTCTGTCTAGAGCTCTTTTATAAGACTTCATGTGC |
| AtUBC8-5’ | ATGGCTTCGAAACGGATCTTGAAGG |
| AtUBC8-3’ | AGCCCATGGCATACTTCTGAGTCC |
| AtUGT85A1 RT-PCR 5’ | TATCTAGGATCCATGGGATCTCAGATCATTCATAAC |
| AtUGT85A1 RT-PCR 3’ | GTCTAGAAGCTTTTAATCCTGTGATTTTTGTCCCAAAAG |
| AtUGT85A3 RT-PCR 5’ | GTCAGTGCATGCATGGGATCCCGTTTTGTTTCTAAC |
| AtUGT85A3 RT-PCR 3’ | GACTCTCTGCAGTTACGTGTTAGGGATCTTTCCCAAG |
| AtUGT85A5 RT-PCR 5’ | TACTGAGGATCCATGGCGTCTCATGCTGTTACAAGCG |
| AtUGT85A5 RT-PCR 3’ | AGTCTAGTCGACCTACTCCCCTAAAAGAACCTTGTCAAC |
| AtUGT85A2 RT-PCR 5’ | GCACGTGGATCCATGGGATCTCATGTCGCACAAAAAC |
| AtUGT85A2 RT-PCR 3’ | GCAGTCAAGCTTCTACTCCCCTAAAAGAACCTTATTA |
| AtUGT85A7 RT-PCR 5’ | GCACTGGGATCCATGGAATCTCATGTTGTTCATAAC |
| AtUGT85A7 RT-PCR 3’ | GCACTGGGATCCATGGAATCTCATGTTGTTCATAAC |
| AtUGT85A4 RT-PCR 5’ | TCAGAGATCTAGAGGATCCATGGAACAACATGGCGGTTCTAGCTC |
| AtUGT85A4 RT-PCR 3’ | TAGCATCGGTACCTTAGGTCGATCTAATCGTGTGACATG |
| AtGUS1 RT-PCR 5’ | TATCATGGATCCAACATGGAACGAACCACCTTGG |
| AtGUS1 RT-PCR 3’ | ATACTAGTCGACTCAAGAACAAGCAGGAGCATCAAAG |
| AtGUS2 RT-PCR 5’ | ATATCGGCATGCATGGCTCAAGAAATGAAACGTGC |
| AtGUS2 RT-PCR 3’ | TATCAGGTACCTCATGAACAAGCAGAAGCATCAAA |
| AtGUS3 RT-PCR 5’ | ATGTCTGAAGCTTATGGCTTATCGTCAAATTTTGGC |
| AtGUS3 RT-PCR 3’ | ATGTCTGTCTAGAGCTCTTTTATAAGACTTCATGTGC |
Fig. 4.
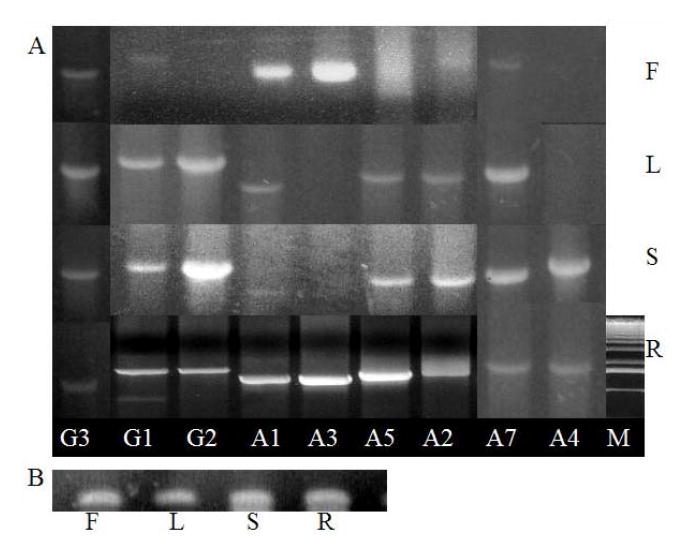
Expression of three AtGUS genes and six AtUGT85A genes (A) and AtUBC8 as an internal loading control for RNA (B) in flowers, leaves, stems, siliques, and roots. All six AtUGT85As and three AtGUS generated products corresponding to the full-length mRNAs visible in ethidium bromide-stained gels. G3=AtGUS3, G1=AtGUS1, G2=AtGUS2, A1=AtUGT85A1, A3=AtUGT85A3, A5=AtUGT85A5, A2=AtUGT85A2, A7=AtUGT85A7, A4=AtUGT85A4, F=Flower, L=Leaf, St/Si=Stem/ Silique, R=Root, M=DNA size marker. MW scale applies directly to only G1-A2 of R, others are from other gels, aligned accordingly manually in the composite image (using the MW markers in the other gels)
Fig. 5.
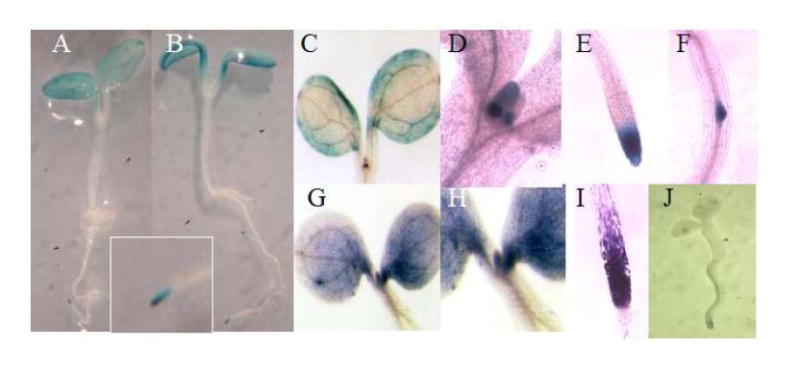
Localization of AtUGT85A and AtGUS expression by expression of (A) AtUGT85A-promoter∷uidA and (B) AtGUS-promoter∷uidA and (G-I) WISH. Because expression pattern is very similar in the six AtUGT85A genes and three AtGUS genes, only representative examples are shown. (A) AtUGT85A1-promoter∷uidA expression is mainly localized in the leaf periphery, lateral root initials, and the root apex (insert). (B) AtGUS1-promoter∷uidA expression is mainly localized in the leaf periphery, lateral root initials, and the root apex. (C-F) AtUGT85A1-promoter∷uidA expression in leaves, leaf primordia, root tips and lateral root initials. (G-I) AtUGT85A1 mRNA detected throughout leaves, leaf primordia, and root tips. (J) Sense control seedlings hybridized by sense RNA probe show no background staining.
Each approach revealed similar results. All six AtUGT85A genes and the three AtGUS genes showed similar expression patterns, with strong expression in root tips and leaf peripheries where cell division is active (Fig. 4, 5). This expression pattern, like that of PsUGT1 [20, 21], is similar to patterns of auxin distribution [47] revealed by auxin-responsive expression of the DR5∷uidA reporter [48]. A previous study revealed altered auxin distribution in plants with altered PsUGT1 expression, in correlation with altered growth, life cycle, and morphology [20].
Silenced expression of AtUGT85A∷uidA reporter gene in the T2 generation
With the exception of the AtUGT85A3 promoter, all AtUGT85A-promoter∷uidA constructs with 1.5-2.0 kb upstream sequences were expressed in the T1 generation of transgenic Arabidopsis. The AtUGT85A5-promoter∷uidA construct was not expressed in transgenic Arabidopsis. The reason for the silencing of AtUGT85A5-promoter∷uidA is not known at this time. Interestingly, with the exception of the AtUGT85A7-promoter, AtUGT85A-promoter∷uidA expression in transgenic plants, became silent in the T2 generation. Homozygous selection of β-glucuronidase (uidA) expression under the control of AtUGT85A promoters may be lethal after the T2 generation. These findings are consistent with previous observations suggesting that uidA expression in the root meristems in correlation with PsUGT1, is not compatible with life [20, 21, 23].
Discussion
In Arabidopsis, the duration of the life cycle can be altered in response to ectopic expression of PsUGT1, which also causes a change in auxin distribution in regions of active cell division [20]. These results suggested that the pea gene interferes with a similar process controlled by endogenous UGT activity in Arabidopsis. If so, then a PsUGT1-related gene, with similar expression patterns and effects on development, is predicted to be present in Arabidopsis. The results of the current study are consistent with this hypothesis. In contrast to pea and alfalfa, each of which have a single UGT1 gene, Arabidopsis has six PsUGT1-homologous sequences, which were named AtUGT85A1, 2, 3, 4, 5, and 7. They are clustered together in a small region of chromosome 1 except AtUGT85A4, which is located at the end of left arm of chromosome 1. This redundancy suggests possible gene duplication during evolution. The six genes have 67-81% DNA sequence identity to each other with 1-2 intron sequences at the middle of the coding region. They have less similarity (45-50% similarity in DNA sequences, and 25-30% similarity in protein sequences) with any other known glycosyltransferases, including UDP-glucosyltransferases. All six AtUGT85A encoded-proteins have a highly conserved UDP-sugar binding site (i.e. PSPG motif) with strong identity to the UDP-glucuronic acid-binding site in PsUGT1. In plants, the conserved UDP-sugar binding site has 44-50 amino acids and binds UDP-glucose [5], UDP-xylose [29], and UDP-glucuronic acid [21, 28].
Previously, we found that meristem-localized, inducible expression of an E. coli uidA gene under the control of PsUGT1 promoter is lethal to plant development [19-21]. In microorganisms and animals, GUS operates in tandem with UGTs in the reversible conjugation of a range of biologically active molecules [7, 13]. In vertebrates, GUS is expressed in most if not all tissues. Although its normal substrates are not well characterized, it has been implicated in functions ranging from sperm cytoskeleton structure [30] to regulation of thyroid hormone levels in cardiac fibroblasts [31]. When endogenous GUS activity in the pea root cap meristem of normal plants is inhibited by an enzyme inhibitor, the cell cycle is predictably altered [23]. This could be explained by the fact that, as in animals, GUS and UGT work together to regulate cellular levels of a product controlling cell division and/or to prevent the accumulation of toxic products to levels that are incompatible with life. To test this model, we searched for genes encoding glucuronidase activities in Arabidopsis. Three AtGUS genes with high similarity with Scutellaria sGUS were identified, and the AtGUS1/2-encoded proteins were predicted to comprise an endo-β-glucuronidase (GH family 79) with similarity to heparanases of human, mouse, and rat.
If the AtUGT85A and AtGUS genes operate in tandem to modulate activity of a common substrate, then their products are predicted to co-localize within the same tissues at the same time in development. The tissue-specific expression pattern of AtGUS genes is similar to the tissue-specific expression pattern of the AtUGT85A genes; e.g., the AtGUS genes are expressed mainly in root tips, leaf primordia, and the leaf periphery. This finding is consistent with the possibility that both the AtGUS genes and AtUGT85A genes are involved in the same cellular processes. The only endogenous plant GUS gene characterized to date, baicaleinase, encodes an endo-β-glucuronidase [24]. This enzyme, isolated from roots of Scutellaria baicalensis, removes glucuronic acid from baicalein (5,6,7-trihydroxyflavone) and scutellarin (5,6,7,4’-tetrahydroxyflavone). Levvy [14] established that the enzyme is similar in specificity and pH optimum (4.5) to mouse liver β-glucuronidase and is inhibited by saccharide 1,4-lactone. He suggested that because baicalein (BA) forms 10-20% of the dry weight of the S. baicalensis root, baicaleinase must play an important role in the metabolism of the plant, and that products such as saccharide-lactone, which inhibit its activity, “might act as a selective plant growth regulator.”
Conclusions
Our results are consistent with the hypothesis that UGT and GUS enzymes in plants are co-localized in plants, and may comprise a system allowing plants to control growth by coordinating internal and external signals through reversible glucuronidation. Localization of AtUGT85A mRNAs and AtGUS mRNAs in areas of intense cell division is consistent with possible involvement in cell cycle regulation. The availability of Arabidopsis mutants with altered expression of individual family members provides necessary tools to begin analysis of the natural substrates of these enzymes in different tissues, and how they operate to modulate growth and development.
Methods
Plant materials and growth conditions
The Arabidopsis ecotype Columbia was grown in greenhouse soil at 22°C and 80% relative humidity with 16 h of light and at 20°C with 8 h of dark.
Arabidopsis gene bank search and phylogenetic analysis of AtUGT85A and AtGUS genes
To identify the PsUGT1 and sGUS homologous sequences, Arabidopsis database was searched. Six AtUGT85A genes and three AtGUS genes were identified. Coding regions of fifty AtUGT genes including six AtUGT85A genes representing 14 groups (groups A–N) of AtUGTs were collected to draw a phylogenetic tree. Rooted phylogenetic tree was drawn by CLUSTAL W. Gap open penalty was 10 and gap extension penalty was 0.1. K-tuple size 1, gap penalty 3, top diagonals 5, and window size 5 were used.
PCR and RT-PCR
For amplifications of genomic or cDNA templates, PCR reactions were performed with 1.25 mM dNTPs, 5 μM each of the primers, 1x Taq buffer, and 0.5 units of of Taq polymerase (Roche, Indianapolis, IN) in a volume of 20 μL. The amplification program consisted of an initial 94°C cycle for 2 min followed by 25 cycles of 94°C, 15 s; (Tm-5)°C, 15 s; and 65-70°C, 60-120 s, and a final extension at 70°C for 6 min. Reverse transcription reactions for RT-PCR were done using the Superscript II reverse transcriptase system following recommendations from the manufacturer (Gibco-BRL, Rockville, MD). Gene-specific primers were generated using PRIMERCHECK and PRIMERTM softwares in SDSC Biology Workbench (http://workbench.sdsc.edu/). Gene-specific primers were used to generate first-strand cDNAs, and that primer was used as the reverse primer in the PCR reaction (Gibco-BRL, Rockville, MD). All six AtUGT85A cDNAs and the three AtGUS cDNAs were cloned by reverse transcriptase-polymerase chain reaction (RT-PCR) into pCRII vectors with gene specific primers (Table 1). The correct identity of the amplified cDNA clones was verified by sequencing the DNA sequence before use in in situ hybridization. For AtUGT85A5, RT-PCR generated a sequence of 1452 bp, which is 202 bp longer than the published AtUGT85A5 mRNA (AY039897). AtUBC8 mRNA was used as an internal loading control because it is expressed in all of the plant organs examined [32].
Preparation of probes for in situ hybridization
The plasmids carrying cDNAs corresponding to the genes for AtUGT85A and AtGUS were linearized for T3 or T7 polymerase-directed RNA synthesis, and sense and antisense strands were synthesized for each by standard procedures. RNA was labeled by incorporating digoxigenin-conjugated UTP (Roche Applied Science, Indianapolis, IN).
For whole mount in situ hybridization (WISH), 7- to 10-day old Arabidopsis seedlings were fixed in 4% paraformaldehyde and stored in methanol at −20°C until needed. WISH was by standard procedures, as described previously [21]. The hybridization buffer is composed of 50% formamide, 5x SSC, 1 mg/ml RNA, 1x Denhardt’s solution, 0.1% Tween 20, 5 mM EDTA, pH 8.0. In situ hybridization was performed in glass vials (4.5 ml) overnight at 45°C. Both sense and antisense probes (100-500 ng probe/ml) were used for each gene analyzed. Seedlings were washed in decreasing concentrations of hybridization buffer diluted with increasing concentrations of 2x SSC. The final rinse was in 0.2x SSC at 45°C. After two rinses in maleate buffer (100 mM sodium maleate, pH 7.5, 150 mM NaCl), the root tips were incubated in northern block (5% blocking reagent in maleate buffer, Roche Applied Science, Indianapolis, IN) at 45°C for 60 min. Northern block (5% blocking reagent in maleate buffer) was replaced with fresh northern block containing a 1:2000 dilution of anti-digoxigenin-alkaline phosphatase (Roche Applied Science, Indianapolis, IN) and the tissues were incubated at room temperature for 1 h. Seedlings were rinsed in two changes of maleate buffer for 30 min each, incubated in buffer no. and 3 (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2) and 5 mM levamisole (Sigma, St. Louis, MO) for 5 min, and then placed in color solution (buffer no. 3,5 mM levamisole, 4.5 μL/ml nitroblue tetrazolium; Roche Applied Science, Indianolis, IN) and 3.5 μL/ml X-phosphate solution (Roche Applied Science, Indianapolis, IN). Seedlings were placed in the dark and color development was monitored from 1 to 24 h. Seedlings were mounted on glass slides in ethanol, and photographed. Chlorophyll was removed with methanol, which causes some loss of label in the leaf periphery due to loss o alkaline phosphatase substrates, but allows observation of labeling in photosynthetic tissues.
Generation of transgenic plants expressing AtUGT85A promoter∷uidA and AtGUS promoter∷uidA
Transgenic plants expressing AtUGT85A promoter∷uidA and AtGUS promoter∷uidA were generated as follows. Upstream promoter sequences of AtUGT85A and AtGUS genes were cloned into the pBI101 vector; i.e. 1.58 kb of AtUGT85A1, 1.48 kb of AtUGT85A3, 1.35 kb of AtUGT85A5, 0.93 kb of AtUGT85A2, 1.1 kb of AtUGT85A7, 1.5 kb of AtUGT85A4, 1.24 kb of AtGUS1, 1.47 kb of AtGUS2, and 1.54 kb of AtGUS3 upstream promoter sequences were PCR-amplified and cloned into the restriction enzyme sites of the pBI101 vector and transformed into Agrobacterium tumefaciens ASE.
Four-week-old plants were transformed by floral dip as described previously [33]. Seeds harvested from transformed plants were grown on Murashige and Skoog (MS) selection plates containing kanamycin. For selection of transgenic plants, kanamycin-resistant primary plants were analyzed for the presence of the uidA gene by PCR. Transgenic plants were maintained in a controlled environmental chamber with 16 h of light (mixed fluorescent bulbs) at 22°C and 8 h of dark at 20°C. Because of the silencing of the E. coli uidA gene in the T2 transgenic plants, GUS staining used plants derived from T1 seeds for AtUGT85A promoter∷uidA and AtGUS promoter∷uidA expression. Three- to 10-d-old seedlings were stained by 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, according to the manufacturer’s instructions. Samples were observed and photographed using a stereomicroscope or a light microscope.
Accession numbers
GenBank accession numbers for AtUGT85A4, 1, 3, 5, 2, and 7 are NM_106476.3, NM_102089, NM_102088, AY765462, NM_102086, and NM_102085, respectively. GenBank accession numbers for AtGUS1, 2, and 3 are NM_125518, NM_120865, and NM_122885. The GenBank accession number for Human heparanse HSPE is AF144325. The GenBank accession number for AtUBC8 is NC_003076.
Acknowledgments
The project was supported in part by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number #2003-35304-13362;
 and Department of Energy, # defg0394er20164, to MCH and HHW; by NIH/NCCAMSPSO AT00151 to the Center for Dietary Supplement Research Botanicals (CDSRB) at UCLA (A.M.H., Director, Agricultural Botany Core and H.-H.W., Junior Investigator), and by a grant from the Shanbrom Family Foundation to A.M.H. B.R.J. was supported in part by Daegu University Research Grant, 2004, and in part by Daegu University RRC Program. All plasmids and transgenic plant seeds will be available at the Ohio State University Arabidopsis stock center.
and Department of Energy, # defg0394er20164, to MCH and HHW; by NIH/NCCAMSPSO AT00151 to the Center for Dietary Supplement Research Botanicals (CDSRB) at UCLA (A.M.H., Director, Agricultural Botany Core and H.-H.W., Junior Investigator), and by a grant from the Shanbrom Family Foundation to A.M.H. B.R.J. was supported in part by Daegu University Research Grant, 2004, and in part by Daegu University RRC Program. All plasmids and transgenic plant seeds will be available at the Ohio State University Arabidopsis stock center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jaeken J, Matthijs G. Congenital disorders of glycosylation. Ann Rev Genomics Hum Genet. 2001;2:129–51. doi: 10.1146/annurev.genom.2.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Dixon RA. Phytoestrogens. Ann Rev Plant Biol. 2004;55:225–261. doi: 10.1146/annurev.arplant.55.031903.141729. [DOI] [PubMed] [Google Scholar]
- 3.Szerszen JB, Szczyglowski K, Bandurski RS. Iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science. 1994;265:1699–1701. doi: 10.1126/science.8085154. [DOI] [PubMed] [Google Scholar]
- 4.Gachon CMM, Langlois-Meurinne M, Saindrenan P. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends in Plant Sci. 2005;10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Bowles D, Lim E-K, Poppenberger B, Vaistij FE. Glycosyltransferases of liphophilic small molecules. Ann Rev Plant Biol. 2006;57:567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- 6.Paquette S, Moller BL, Bak S. On the origin of family 1 plant glycosyltransferases. Phytochem. 2003;62:399–413. doi: 10.1016/s0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- 7.Nebert DW. Proposed role of drug-metabolizing enzymes: regulation of steady state levels of the ligands that effect growth, homeostasis, differentiation, and neuroendocrine functions. Mol Endocrinol. 1991;5:1203–14. doi: 10.1210/mend-5-9-1203. [DOI] [PubMed] [Google Scholar]
- 8.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Ann Rev Pharma Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 9.Bock KW. Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochem Pharmacol. 2003;66:691–6. doi: 10.1016/s0006-2952(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 10.Bosma PJ, et al. Sequence of exons and the flanking regions of human bilirubin- UDP-glucuronosyltransferase gene complex and identification of a genetic mutation in a patient with Crigler-Najjar syndrome, type I. Hepatology. 1992;15:941–947. doi: 10.1002/hep.1840150531. [DOI] [PubMed] [Google Scholar]
- 11.Strassburg CP, Vogel A, Kneip S, Tukey RH, Manns MP. Polymorphisms of the human UDP-glucuronosyltransferase (UGT) 1A7 gene in colorectal cancer. Gut. 2002;50:851–856. doi: 10.1136/gut.50.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross J, Li Y, Lim EK, Bowles DJ. Higher plant glycosyltransferases. Genome Biology. 2001;2:3004–3006. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- 14.Bowles D. A multigene family of glycosyltransferases in a model plant, Arabidopsis thaliana. Biochem Soc Trans. 2002;30:301–6. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 15.Dixon RA, Steele CL. Flavonoids and isoflavonoids - a gold mine for metabolic engineering. Trends Plant Sci. 1999;4:394–400. doi: 10.1016/s1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- 16.Dixon RA, Ferreira D. Genistein. Phytochem. 2002;60:205–211. doi: 10.1016/s0031-9422(02)00116-4. [DOI] [PubMed] [Google Scholar]
- 17.Jones P, Messner B, Nakajima J, Schaffner AR, Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. Biol Chem. 2003;278:43910–843918. doi: 10.1074/jbc.M303523200. [DOI] [PubMed] [Google Scholar]
- 18.Quiel JA, Bender J. Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J Biol Chem. 2003;278:6275–6281. doi: 10.1074/jbc.M211822200. [DOI] [PubMed] [Google Scholar]
- 19.Woo HH, Hirsch AM, Hawes MC. Altered susceptibility to infection by bacteria and fungi in alfalfa roots with altered cell cycle. Plant Cell Rep. 2004;22:967–973. doi: 10.1007/s00299-004-0787-x. [DOI] [PubMed] [Google Scholar]
- 20.Woo HH, Faull KF, Hirsch AM, Hawes MC. Altered life cycle in Arabidopsis thaliana plants expressing PsUGT1, a UDP-glucuronosyltransferase encoding gene from Pisum sativum. Plant Physiol. 2003;133:538–548. doi: 10.1104/pp.103.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo HH, Orbach M, Hirsch AM, Hawes MC. Meristem-localized inducible expression of a UDP-glycosyltransferase gene is essential for growth and development in pea and alfalfa. Plant Cell. 1999;11:2303–2316. doi: 10.1105/tpc.11.12.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams CA, Grayer RJ. Anthocyanins and other flavonoids. Nat Prod Rep. 2004;21:539–73. doi: 10.1039/b311404j. [DOI] [PubMed] [Google Scholar]
- 23.Wen F, Woo HH, Hirsch AM, Hawes MC. Lethality of inducible, meristem-localized ectopic β-glucuronidase expression in plants. Plant Mol Biol Reporter. 2004;22:7–14. [Google Scholar]
- 24.Sasaki K, Taura F, Shoyama Y, Morimoto S. Molecular characterization of a novel GUS from Scutellaria baicalensis Georgi. J Biol Chem. 2000;275:27466–27472. doi: 10.1074/jbc.M004674200. [DOI] [PubMed] [Google Scholar]
- 25.Levvy GA. Baicalinase, a plant beta-glucuronidase. Biochem J. 1954;58:462–469. doi: 10.1042/bj0580462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hans J, Brandt W, Vogt T. Site-directed mutagenesis and protein 3D-homology modelling suggest a catalytic mechanism for UDP-glucose-dependent betanidin 5-O-glucosyltransferase from Dorotheanthus bellidiformis. Plant J. 2004;39:319–333. doi: 10.1111/j.1365-313X.2004.02133.x. [DOI] [PubMed] [Google Scholar]
- 27.Dong J, Kukula AK, Toyoshima M, Nakajima M. Genomic organization and chromosome localization of the newly identified human heparanase gene. Gene. 2000;253:171–178. doi: 10.1016/s0378-1119(00)00251-1. [DOI] [PubMed] [Google Scholar]
- 28.Woo HH, Jeong BR, Hawes MC, Hirsch AM. Expression of closely related UDP-glycosyltransferases from pea and Arabidopsis are involved in gravitropic response and leaf development. Physiol Plant. In press. [Google Scholar]
- 29.Hou B, Lim EK, Higgins GS, Bowles DJ. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem. 2004;279:47822–47832. doi: 10.1074/jbc.M409569200. [DOI] [PubMed] [Google Scholar]
- 30.Lopes CH, LaFalci VS, Silva CE, Brandelli A. GUS is associated with goat sperm cytoskeleton. J Exp Zoo. 2001;289:146–152. [PubMed] [Google Scholar]
- 31.Van der Heide SM, Visser TJ, Everts ME, Klaren PH. Metabolism of thyroid hormones in cultured cardiac fibroblasts of neonatal rats. J Endocrinol. 2002;174:111–119. doi: 10.1677/joe.0.1740111. [DOI] [PubMed] [Google Scholar]
- 32.Woo HH, Brigham LA, Hawes MC. Primary structure of the mRNA encoding a 16.5-kDa ubiquitin-conjugating enzyme of Pisum sativum. Gene. 1994;148:369–370. doi: 10.1016/0378-1119(94)90715-3. [DOI] [PubMed] [Google Scholar]
- 33.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Bowles D, Isayenkova J, Lim E-K, Poppenberger B. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Lim E-K, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis. The EMBO J. 2004;23:2915–2922. doi: 10.1038/sj.emboj.7600295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Baldauf S, Lim E-K, Boweles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. JBC. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- 37.Keeggstra K, Raikhel N. Plant glycosyltransferases. Curr Opin Plant Biol. 2001;4:219–224. doi: 10.1016/s1369-5266(00)00164-3. [DOI] [PubMed] [Google Scholar]
- 38.Henrissat B, Davies GJ. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000;124:1515–1519. doi: 10.1104/pp.124.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gachon CMM, Langlois-Meurinne M, Henry Y, Saindrenan P. Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: functional and evolutionary implications. Plant Mol Biol. 2005;58:229–245. doi: 10.1007/s11103-005-5346-5. [DOI] [PubMed] [Google Scholar]
- 40.Gilissen LJW, Metz PLJ, Stiekema WJ, Nap J-P. Biosafety of E. coli β-glucuronidase (GUS) in plants. Transgenic Res. 1998;7:157–163. doi: 10.1023/a:1008832711805. [DOI] [PubMed] [Google Scholar]
- 41.Nebert DW. Proposed role of drug-metabolizing enzymes: regulation of steady state levels of the ligands that effect growth, homeostasis, differentiation, and neuroendocrine functions. Mol Endocrinol. 1991;5:1203–14. doi: 10.1210/mend-5-9-1203. [DOI] [PubMed] [Google Scholar]
- 42.Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–6. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- 43.Lopes CH, LaFalci VS, Silva CE, Brandelli A. GUS is associated with goat sperm cytoskeleton. J Exper Zool. 2001;289:146–52. [PubMed] [Google Scholar]
- 44.Van der Heide SM, Visser TJ, Everts ME, Klaren PH. Metabolism of thyroid hormones in cultured cardiac fibroblasts of neonatal rats. J Endocrinol. 2002;174:111–119. doi: 10.1677/joe.0.1740111. [DOI] [PubMed] [Google Scholar]
- 45.deGroot FMH, Damen EWP, Scheeren HW. Anticancer prodrugs for application in monotherapy: Targeting hypoxia, tumor-associated enzymes, and receptors. Cur Med Chem. 2001;8:1093–1122. doi: 10.2174/0929867013372634. [DOI] [PubMed] [Google Scholar]
- 46.Unak T. Potential use of radiolabeled glucuronide prodrugs with Auger and/or alpha emitters in combined chemo- and radio-therapy of cancer. Cur Pharma Design. 2000;6:1127–1142. doi: 10.2174/1381612003399798. [DOI] [PubMed] [Google Scholar]
- 47.Aloni R, Schwalm K, Langhans M, Ullrich CI. Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta. 2003;216:841–53. doi: 10.1007/s00425-002-0937-8. [DOI] [PubMed] [Google Scholar]
- 48.Ulmasov T, Murfett J, Hagen G, Guilfoyle T. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]


