Abstract
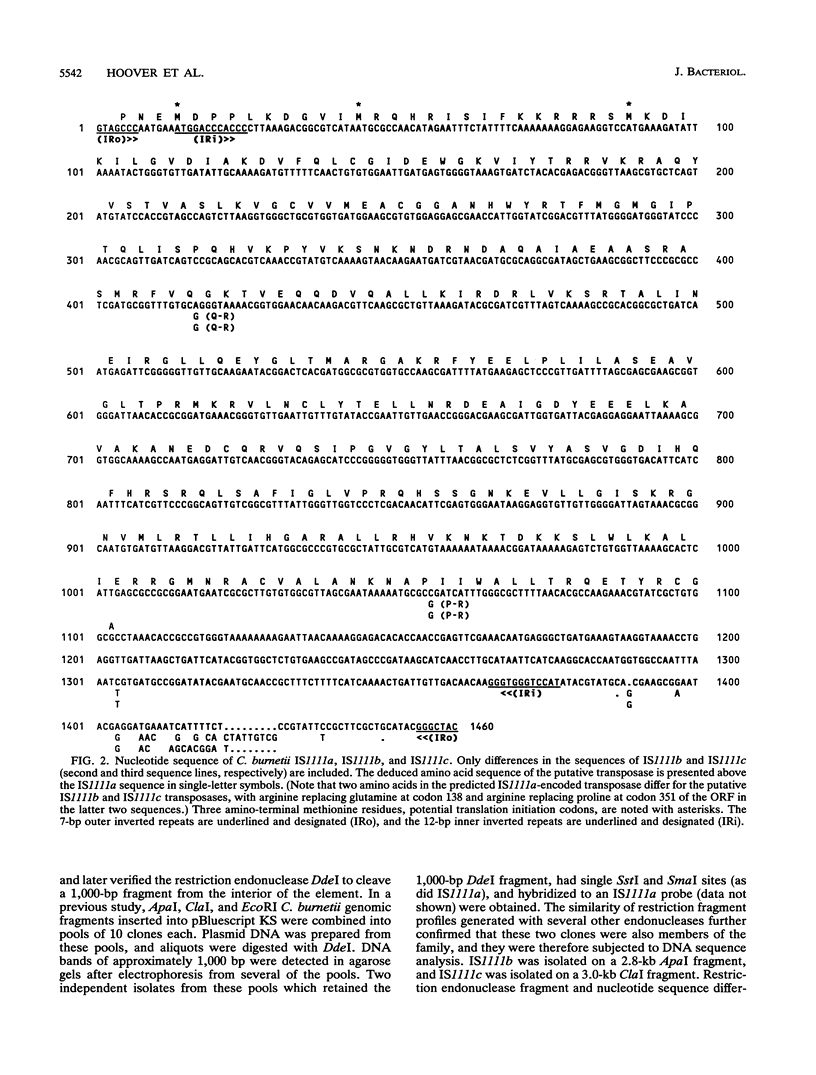
A DNA fragment located on the 3' side of the Coxiella burnetii htpAB operon was determined by Southern blotting to exist in approximately 19 copies in the Nine Mile I genome. The DNA sequences of this htpAB-associated repetitive element and two other independent copies were analyzed to determine the size and nature of the element. The three copies of the element were 1,450, 1,452, and 1,458 bp long, with less than 2% divergence among the three sequences. Several features characteristic of bacterial insertion sequences were discovered. These included a single significant open reading frame that would encode a 367-amino-acid polypeptide which was predicted to be highly basic, to have a DNA-binding helix-turn-helix motif, to have a leucine zipper motif, and to have homology to polypeptides found in several other bacterial insertion sequences. Identical 7-bp inverted repeats were found at the ends of all three copies of the element. However, duplications generated by many bacterial mobile elements in the recipient DNA during insertion events did not flank the inverted repeats of any of the three C. burnetii elements examined. A second pair of inverted repeats that flanked the open reading frame was also found in all three copies of the element. Most of the divergence among the three copies of the element occurred in the region between the two inverted repeat sequences in the 3' end of the element. Despite the sequence changes, all three copies of the element have retained significant dyad symmetry in this region.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akporiaye E. T., Rowatt J. D., Aragon A. A., Baca O. G. Lysosomal response of a murine macrophage-like cell line persistently infected with Coxiella burnetii. Infect Immun. 1983 Jun;40(3):1155–1162. doi: 10.1128/iai.40.3.1155-1162.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby M. K., Bergquist P. L. Cloning and sequence of IS1000, a putative insertion sequence from Thermus thermophilus HB8. Plasmid. 1990 Jul;24(1):1–11. doi: 10.1016/0147-619x(90)90020-d. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. Structural basis of DNA-protein recognition. Trends Biochem Sci. 1989 Jul;14(7):286–290. doi: 10.1016/0968-0004(89)90066-2. [DOI] [PubMed] [Google Scholar]
- Bruton C. J., Chater K. F. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolor A3(2). Nucleic Acids Res. 1987 Sep 11;15(17):7053–7065. doi: 10.1093/nar/15.17.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton P. R., Kordová N., Paretsky D. Electron microscopic studies of the rickettsia Coxiella burneti: entry, lysosomal response, and fate of rickettsial DNA in L-cells. Can J Microbiol. 1971 Feb;17(2):143–150. doi: 10.1139/m71-025. [DOI] [PubMed] [Google Scholar]
- Burton P. R., Stueckemann J., Welsh R. M., Paretsky D. Some ultrastructural effects of persistent infections by the rickettsia Coxiella burnetii in mouse L cells and green monkey kidney (Vero) cells. Infect Immun. 1978 Aug;21(2):556–566. doi: 10.1128/iai.21.2.556-566.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S. J., Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990 Feb;6(2):36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Foster S. G., Tobek I. Physical and genetic analysis of IS110, a transposable element of Streptomyces coelicolor A3(2). Mol Gen Genet. 1985;200(2):235–239. doi: 10.1007/BF00425429. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Clark-Curtiss J. E., Docherty M. A. A species-specific repetitive sequence in Mycobacterium leprae DNA. J Infect Dis. 1989 Jan;159(1):7–15. doi: 10.1093/infdis/159.1.7. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Dasch G. A., Ching W. M., Kim P. Y., Pham H., Stover C. K., Oaks E. V., Dobson M. E., Weiss E. A structural and immunological comparison of rickettsial HSP60 antigens with those of other species. Ann N Y Acad Sci. 1990;590:352–369. doi: 10.1111/j.1749-6632.1990.tb42242.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. P., Tizard M. L., Moss M. T., Thompson J., Winterbourne D. J., McFadden J. J., Hermon-Taylor J. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 1989 Nov 25;17(22):9063–9073. doi: 10.1093/nar/17.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Grosskinsky C. M., Jacobs W. R., Jr, Clark-Curtiss J. E., Bloom B. R. Genetic relationships among Mycobacterium leprae, Mycobacterium tuberculosis, and candidate leprosy vaccine strains determined by DNA hybridization: identification of an M. leprae-specific repetitive sequence. Infect Immun. 1989 May;57(5):1535–1541. doi: 10.1128/iai.57.5.1535-1541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T. Estimation of the cytoplasmic pH of Coxiella burnetii and effect of substrate oxidation on proton motive force. J Bacteriol. 1983 May;154(2):591–597. doi: 10.1128/jb.154.2.591-597.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T. The role of lipopolysaccharides in the virulence of Coxiella burnetii. Ann N Y Acad Sci. 1990;590:27–32. doi: 10.1111/j.1749-6632.1990.tb42203.x. [DOI] [PubMed] [Google Scholar]
- Hackstadt T., Williams J. C. Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc Natl Acad Sci U S A. 1981 May;78(5):3240–3244. doi: 10.1073/pnas.78.5.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T., Williams J. C. Stability of the adenosine 5'-triphosphate pool in Coxiella burnetii: influence of pH and substrate. J Bacteriol. 1981 Nov;148(2):419–425. doi: 10.1128/jb.148.2.419-425.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T., Williams J. C. pH dependence of the Coxiella burnetii glutamate transport system. J Bacteriol. 1983 May;154(2):598–603. doi: 10.1128/jb.154.2.598-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Heinzen R. A., Frazier M. E., Mallavia L. P. Nucleotide sequence of Coxiella burnetii superoxide dismutase. Nucleic Acids Res. 1990 Nov 11;18(21):6437–6437. doi: 10.1093/nar/18.21.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen R. A., Mallavia L. P. Cloning and functional expression of the Coxiella burnetii citrate synthase gene in Escherichia coli. Infect Immun. 1987 Apr;55(4):848–855. doi: 10.1128/iai.55.4.848-855.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D. J., Lydiate D. J., Hopwood D. A. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolor A3(2). Mol Microbiol. 1989 Oct;3(10):1307–1318. doi: 10.1111/j.1365-2958.1989.tb00112.x. [DOI] [PubMed] [Google Scholar]
- Hendrix L., Mallavia L. P. Active transport of proline by Coxiella burnetii. J Gen Microbiol. 1984 Nov;130(11):2857–2863. doi: 10.1099/00221287-130-11-2857. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hoover T. A., Williams J. C. Characterization of Coxiella burnetii pyrB. Ann N Y Acad Sci. 1990;590:485–490. doi: 10.1111/j.1749-6632.1990.tb42258.x. [DOI] [PubMed] [Google Scholar]
- Hu J. C., O'Shea E. K., Kim P. S., Sauer R. T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990 Dec 7;250(4986):1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsrud L. DNA sequence of the transposable element IS1. Mol Gen Genet. 1979 Jan 31;169(2):213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Leskiw B. K., Mevarech M., Barritt L. S., Jensen S. E., Henderson D. J., Hopwood D. A., Bruton C. J., Chater K. F. Discovery of an insertion sequence, IS116, from Streptomyces clavuligerus and its relatedness to other transposable elements from actinomycetes. J Gen Microbiol. 1990 Jul;136(7):1251–1258. doi: 10.1099/00221287-136-7-1251. [DOI] [PubMed] [Google Scholar]
- Lydiate D. J., Ikeda H., Hopwood D. A. A 2.6 kb DNA sequence of Streptomyces coelicolor A3(2) which functions as a transposable element. Mol Gen Genet. 1986 Apr;203(1):79–88. doi: 10.1007/BF00330387. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. Insertion element IS1 encodes two structural genes required for its transposition. J Mol Biol. 1984 Aug 5;177(2):229–245. doi: 10.1016/0022-2836(84)90454-6. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxon M. E., Wigboldus J., Brot N., Weissbach H. Structure-function studies on Escherichia coli MetR protein, a putative prokaryotic leucine zipper protein. Proc Natl Acad Sci U S A. 1990 Sep;87(18):7076–7079. doi: 10.1073/pnas.87.18.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnick M. F., Heinzen R. A., Frazier M. E., Mallavia L. P. Characterization and expression of the cbbE' gene of Coxiella burnetii. J Gen Microbiol. 1990 Jun;136(6):1099–1107. doi: 10.1099/00221287-136-6-1099. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990 Sep 7;62(5):945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Vodkin M. H., Williams J. C. The Mycobacterium tuberculosis 65-kilodalton antigen is a heat shock protein which corresponds to common antigen and to the Escherichia coli GroEL protein. Infect Immun. 1988 Feb;56(2):446–451. doi: 10.1128/iai.56.2.446-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin M. H., Williams J. C. A heat shock operon in Coxiella burnetti produces a major antigen homologous to a protein in both mycobacteria and Escherichia coli. J Bacteriol. 1988 Mar;170(3):1227–1234. doi: 10.1128/jb.170.3.1227-1234.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waag D. M., Williams J. C. Immune modulation by Coxiella burnetii: characterization of a phase I immunosuppressive complex differentially expressed among strains. Immunopharmacol Immunotoxicol. 1988;10(2):231–260. doi: 10.3109/08923978809014335. [DOI] [PubMed] [Google Scholar]
- Wiater L. A., Grindley N. D. Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. EMBO J. 1988 Jun;7(6):1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. C., Hoover T. A., Waag D. M., Banerjee-Bhatnagar N., Bolt C. R., Scott G. H. Antigenic structure of Coxiella burnetii. A comparison of lipopolysaccharide and protein antigens as vaccines against Q fever. Ann N Y Acad Sci. 1990;590:370–380. doi: 10.1111/j.1749-6632.1990.tb42243.x. [DOI] [PubMed] [Google Scholar]
- Winter R. B., Berg O. G., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981 Nov 24;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- Woods S. A., Cole S. T. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol Lett. 1989 Dec;53(3):305–309. doi: 10.1016/0378-1097(89)90235-8. [DOI] [PubMed] [Google Scholar]