Abstract
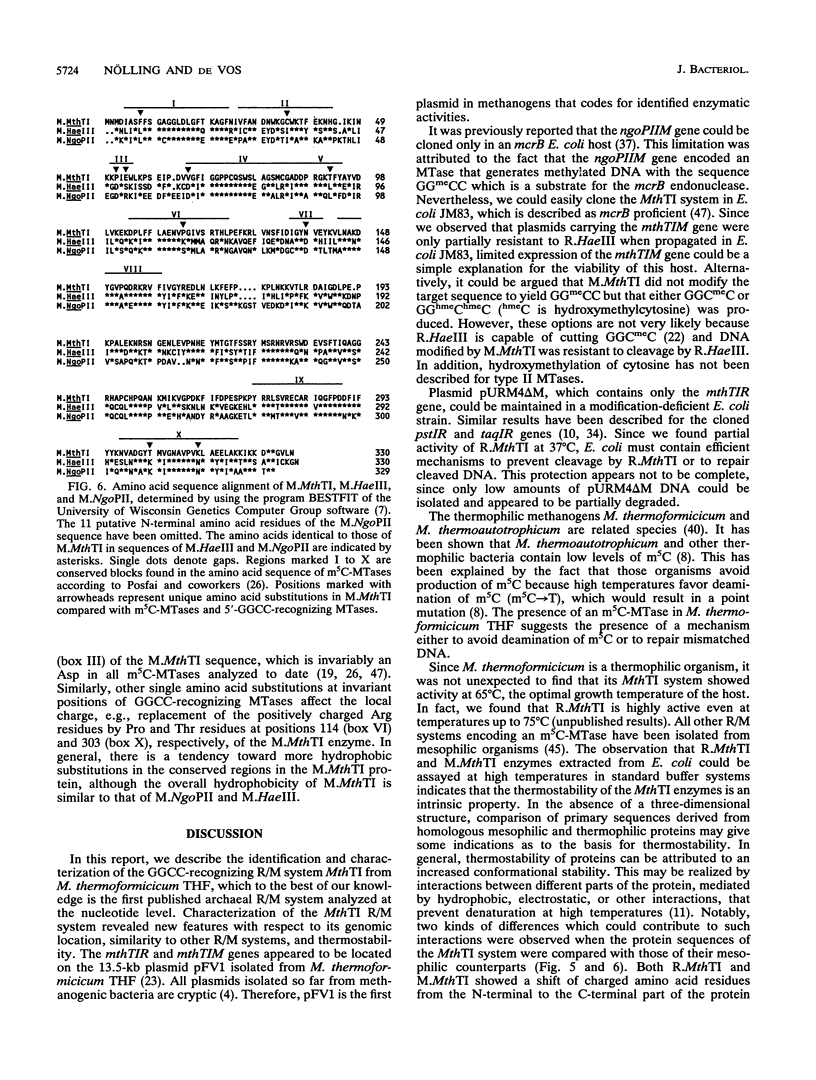
A restriction-modification system, designated MthTI, was localized on plasmid pFV1 from the thermophilic archaeon Methanobacterium thermoformicicum THF. The MthTI system is a new member of the family of GGCC-recognizing restriction-modification systems. Functional expression of the archaeal MthTI genes was obtained in Escherichia coli. The mthTIR and mthTIM genes are 843 and 990 bp in size and code for proteins of 281 (32,102 Da) and 330 (37,360 Da) amino acids, respectively. The deduced amino acid sequence of M.MthTI showed high similarity with that of the isospecific methyltransferases M.NgoPII and M.HaeIII. In addition, extensive sequence similarity on the amino acid level was observed for the endonucleases R.MthTI and R.NgoPII. Moreover, the endonuclease and methyltransferase genes of the thermophilic MthTI system and those of the Neisseria gonorrhoeae NgoPII system show identical organizations and high (54.5%) nucleotide identity. This finding suggests horizontal transfer of restriction-modification systems between members of the domains Bacteria and Archaea.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bron S., Murray K. Restriction and modification in B. subtilis. Nucleotide sequence recognised by restriction endonuclease R. Bsu R from strain R. Mol Gen Genet. 1975 Dec 30;143(1):25–33. doi: 10.1007/BF00269417. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Cannon J. G., Sparling P. F. The genetics of the gonococcus. Annu Rev Microbiol. 1984;38:111–133. doi: 10.1146/annurev.mi.38.100184.000551. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Carreira L. H., Ljungdahl L. G., Kuo K. C., Gehrke C. W. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985 Feb 25;13(4):1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Brooks J. E. Cloned restriction/modification system from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1983 Jan;80(2):402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke R., Závodszky P. Proteins under extreme physical conditions. FEBS Lett. 1990 Aug 1;268(2):344–349. doi: 10.1016/0014-5793(90)81283-t. [DOI] [PubMed] [Google Scholar]
- Kapfer W., Walter J., Trautner T. A. Cloning, characterization and evolution of the BsuFI restriction endonuclease gene of Bacillus subtilis and purification of the enzyme. Nucleic Acids Res. 1991 Dec 11;19(23):6457–6463. doi: 10.1093/nar/19.23.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Manta V. Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3). Gene. 1990 Aug 16;92(1-2):1–248. doi: 10.1016/0378-1119(90)90486-b. [DOI] [PubMed] [Google Scholar]
- Kiss A., Posfai G., Keller C. C., Venetianer P., Roberts R. J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985 Sep 25;13(18):6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Jugel A., Walter J., Noyer-Weidner M., Trautner T. A. 'Pseudo' domains in phage-encoded DNA methyltransferases. Nature. 1991 Aug 15;352(6336):645–648. doi: 10.1038/352645a0. [DOI] [PubMed] [Google Scholar]
- Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J Mol Biol. 1989 Mar 20;206(2):313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Lunnen K. D., Morgan R. D., Timan C. J., Krzycki J. A., Reeve J. N., Wilson G. G. Characterization and cloning of MwoI (GCN7GC), a new type-II restriction-modification system from Methanobacterium wolfei. Gene. 1989 Apr 15;77(1):11–19. doi: 10.1016/0378-1119(89)90354-5. [DOI] [PubMed] [Google Scholar]
- Mazodier P., Davies J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Effect of site-specific methylation on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1989;17 (Suppl):r389–r415. doi: 10.1093/nar/17.suppl.r389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N. H., Pauling C. Evidence for two restriction-modification systems in Halobacterium cutirubrum. J Bacteriol. 1985 Aug;163(2):783–784. doi: 10.1128/jb.163.2.783-784.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prangishvili D. A., Vashakidze R. P., Chelidze M. G., Gabriadze IYu A restriction endonuclease SuaI from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1985 Nov 11;192(1):57–60. doi: 10.1016/0014-5793(85)80042-9. [DOI] [PubMed] [Google Scholar]
- Pósfai G., Kiss A., Erdei S., Pósfai J., Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J Mol Biol. 1983 Nov 5;170(3):597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Hüdepohl U., Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K., Thomm M., Laminet A., Laue F. G., Kessler C., Stetter K. O., Schmitt R. Three new restriction endonucleases MaeI, MaeII and MaeIII from Methanococcus aeolicus. Nucleic Acids Res. 1984 Mar 26;12(6):2619–2628. doi: 10.1093/nar/12.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko B. E., Croft R., Moran L. S., Wilson G. G. Cloning and analysis of the HaeIII and HaeII methyltransferase genes. Gene. 1988 Dec 25;74(1):45–50. doi: 10.1016/0378-1119(88)90248-x. [DOI] [PubMed] [Google Scholar]
- Stephenson F. H., Ballard B. T., Boyer H. W., Rosenberg J. M., Greene P. J. Comparison of the nucleotide and amino acid sequences of the RsrI and EcoRI restriction endonucleases. Gene. 1989 Dec 21;85(1):1–13. doi: 10.1016/0378-1119(89)90458-7. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Saunders J. R. Nucleotide sequence and genetic organization of the NgoPII restriction-modification system of Neisseria gonorrhoeae. Mol Gen Genet. 1989 Apr;216(2-3):380–387. doi: 10.1007/BF00334379. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Saunders J. R. Sequence analysis of the NgoPII methyltransferase gene from Neisseria gonorrhoeae P9: homologies with other enzymes recognizing the sequence 5'-GGCC-3'. Nucleic Acids Res. 1988 May 25;16(10):4369–4387. doi: 10.1093/nar/16.10.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzel J. P., Conway de Macario E., Nölling J., De Vos W. M., Zhilina T., Lysenko A. M. DNA relatedness among some thermophilic members of the genus Methanobacterium: emendation of the species Methanobacterium thermoautotrophicum and rejection of Methanobacterium thermoformicicum as a synonym of Methanobacterium thermoautotrophicum. Int J Syst Bacteriol. 1992 Jul;42(3):408–411. doi: 10.1099/00207713-42-3-408. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Behrens B., Noyer-Weidner M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: comparison of their nucleotide sequences. Gene. 1986;42(1):89–96. doi: 10.1016/0378-1119(86)90153-8. [DOI] [PubMed] [Google Scholar]
- Walter J., Noyer-Weidner M., Trautner T. A. The amino acid sequence of the CCGG recognizing DNA methyltransferase M.BsuFI: implications for the analysis of sequence recognition by cytosine DNA methyltransferases. EMBO J. 1990 Apr;9(4):1007–1013. doi: 10.1002/j.1460-2075.1990.tb08203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wilson G. G. Organization of restriction-modification systems. Nucleic Acids Res. 1991 May 25;19(10):2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]