Abstract
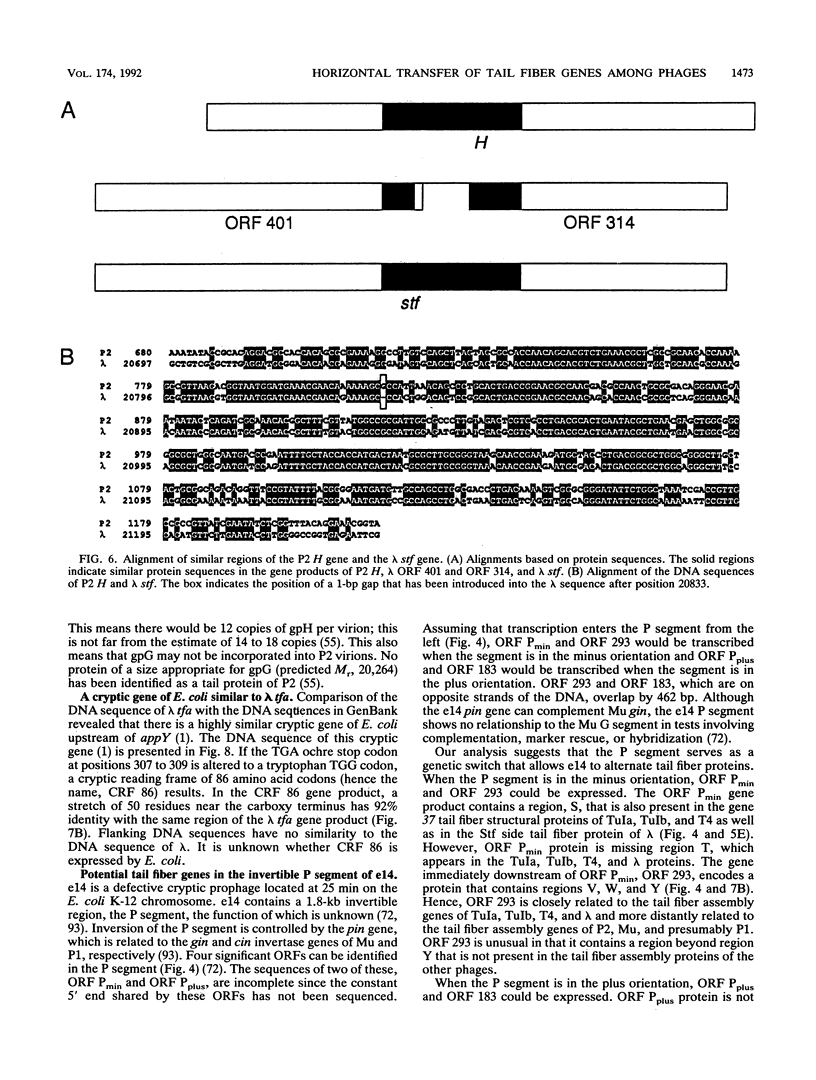
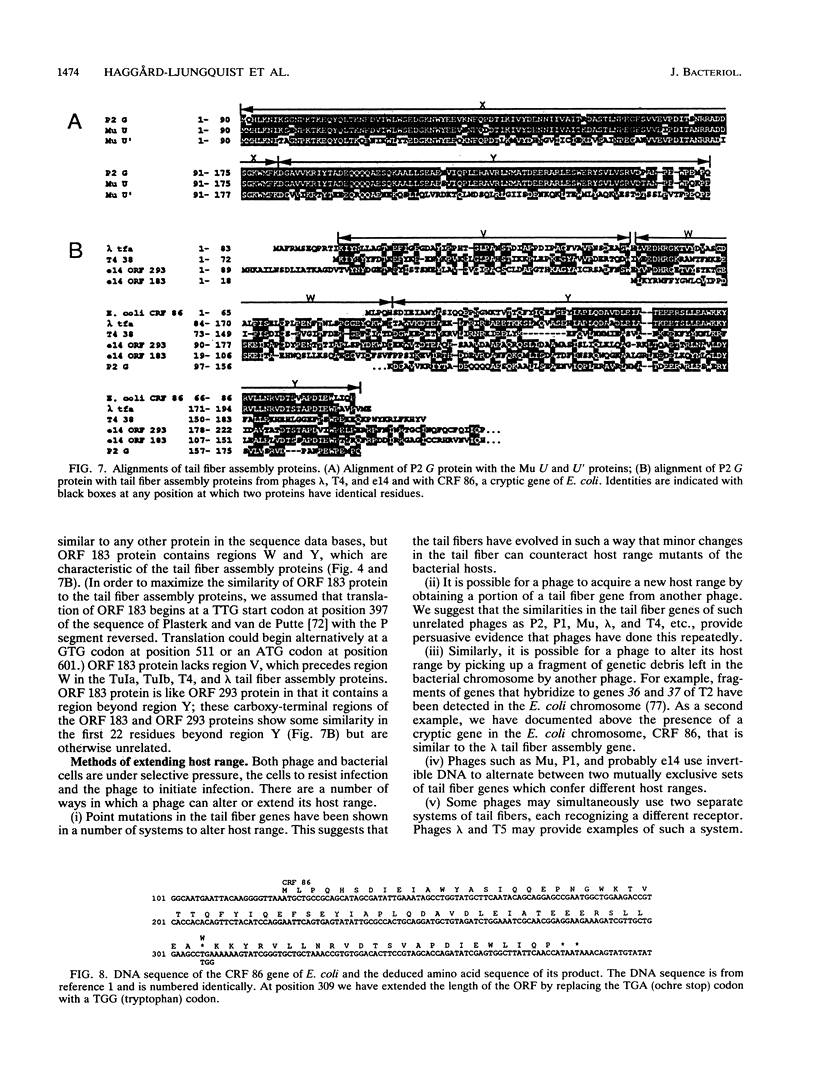
We have determined the DNA sequence of the bacteriophage P2 tail genes G and H, which code for polypeptides of 175 and 669 residues, respectively. Gene H probably codes for the distal part of the P2 tail fiber, since the deduced sequence of its product contains regions similar to tail fiber proteins from phages Mu, P1, lambda, K3, and T2. The similarities of the carboxy-terminal portions of the P2, Mu, ann P1 tail fiber proteins may explain the observation that these phages in general have the same host range. The P2 H gene product is similar to the products of both lambda open reading frame (ORF) 401 (stf, side tail fiber) and its downstream ORF, ORF 314. If 1 bp is inserted near the end of ORF 401, this reading frame becomes fused with ORF 314, creating an ORF that may represent the complete stf gene that encodes a 774-amino-acid-long side tail fiber protein. Thus, a frameshift mutation seems to be present in the common laboratory strain of lambda. Gene G of P2 probably codes for a protein required for assembly of the tail fibers of the virion. The entire G gene product is very similar to the products of genes U and U' of phage Mu; a region of these proteins is also found in the tail fiber assembly proteins of phages TuIa, TuIb, T4, and lambda. The similarities in the tail fiber genes of phages of different families provide evidence that illegitimate recombination occurs at previously unappreciated levels and that phages are taking advantage of the gene pool available to them to alter their host ranges under selective pressures.
Full text
PDF













Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atlung T., Nielsen A., Hansen F. G. Isolation, characterization, and nucleotide sequence of appY, a regulatory gene for growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1989 Mar;171(3):1683–1691. doi: 10.1128/jb.171.3.1683-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckendorf S. K., Kim J. S., Lielausis I. Structure of bacteriophage T4 genes 37 and 38. J Mol Biol. 1973 Jan;73(1):17–35. doi: 10.1016/0022-2836(73)90156-3. [DOI] [PubMed] [Google Scholar]
- Beckendorf S. K. Structure of the distal half of the bacteriophage T4 tail fiber. J Mol Biol. 1973 Jan;73(1):37–53. doi: 10.1016/0022-2836(73)90157-5. [DOI] [PubMed] [Google Scholar]
- Bertani G. Deletions in bacteriophage P2. Circularity of the genetic map and its orientation relative to the DNA denaturation map. Mol Gen Genet. 1975;136(2):107–137. doi: 10.1007/BF00272034. [DOI] [PubMed] [Google Scholar]
- Bertani G., Ljungquist E., Jagusztyn-Krynicka K., Jupp S. Defective particle assembly in wild type P2 bacteriophage and its correction by the lg mutation. J Gen Virol. 1978 Feb;38(2):251–261. doi: 10.1099/0022-1317-38-2-251. [DOI] [PubMed] [Google Scholar]
- Bertani L. E., Bertani G. Genetics of P2 and related phages. Adv Genet. 1971;16:199–237. doi: 10.1016/s0065-2660(08)60359-4. [DOI] [PubMed] [Google Scholar]
- Bertani L. E., Bertani G. Preparation and characterization of temperate, non-inducible bacteriophage P2 (host: Escherichia coli). J Gen Virol. 1970 Feb;6(2):201–212. doi: 10.1099/0022-1317-6-2-201. [DOI] [PubMed] [Google Scholar]
- Beumer J., Beumer-Jochmans M. P., Dirkx J., Dekegel D. Etat actuel des connaissances concernant la nature et la localisation des récepteurs des bactériophages dans la parol cellulaire des Shigella et des Escherichia. Bull Acad R Med Belg. 1965;5(9):749–790. [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun-Breton C., Hofnung M. Explanations accounting for transduction by bacteriophage lambda in maltose negative bacteriophage lambda resistant mutants of Escherichia coli K-12. Mol Gen Genet. 1978 Feb 16;159(2):143–149. doi: 10.1007/BF00270887. [DOI] [PubMed] [Google Scholar]
- Braun V., Schaller K., Wolff H. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim Biophys Acta. 1973 Sep 27;323(1):87–97. doi: 10.1016/0005-2736(73)90433-1. [DOI] [PubMed] [Google Scholar]
- Buchwald M., Siminovitch L. Production of serum-blocking material by mutants of the left arm of the lambda chromosome. Virology. 1969 May;38(1):1–7. doi: 10.1016/0042-6822(69)90121-4. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Ambrosio L. The invertible segment of bacteriophage Mu DNA determines the adsorption properties of Mu particles. Nature. 1978 Feb 9;271(5645):575–577. doi: 10.1038/271575a0. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Bukhari A. I. The invertible DNA segments of coliphages Mu and P1 are identical. Virology. 1976 Oct 1;74(1):242–248. doi: 10.1016/0042-6822(76)90148-3. [DOI] [PubMed] [Google Scholar]
- Christie G. E., Calendar R. Bacteriophage P2 late promoters. II. Comparison of the four late promoter sequences. J Mol Biol. 1985 Feb 5;181(3):373–382. doi: 10.1016/0022-2836(85)90226-8. [DOI] [PubMed] [Google Scholar]
- Dickson R. C. Assembly of bacteriophage T4 tail fibers. IV. Subunit composition of tail fibers and fiber precursors. J Mol Biol. 1973 Oct 5;79(4):633–647. doi: 10.1016/0022-2836(73)90068-5. [DOI] [PubMed] [Google Scholar]
- Dove W. F. Action of the lambda chromosome. I. Control of functions late in bacteriophage development. J Mol Biol. 1966 Aug;19(1):187–201. doi: 10.1016/s0022-2836(66)80060-8. [DOI] [PubMed] [Google Scholar]
- Drexler K., Riede I., Montag D., Eschbach M. L., Henning U. Receptor specificity of the Escherichia coli T-even type phage Ox2. Mutational alterations in host range mutants. J Mol Biol. 1989 Jun 20;207(4):797–803. doi: 10.1016/0022-2836(89)90245-3. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Lielausis I. Serological studies with mutants of phage T4D defective in genes determining tail fiber structure. Genetics. 1965 Dec;52(6):1187–1200. doi: 10.1093/genetics/52.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKLIN N. C. Serological study of tail structure and function in coliphages T2 and T4. Virology. 1961 Aug;14:417–429. doi: 10.1016/0042-6822(61)90333-6. [DOI] [PubMed] [Google Scholar]
- George D. G., Yeh L. S., Barker W. C. Unexpected relationships between bacteriophage lambda hypothetical proteins and bacteriophage T4 tail-fiber proteins. Biochem Biophys Res Commun. 1983 Sep 30;115(3):1061–1068. doi: 10.1016/s0006-291x(83)80043-6. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Plasterk R. H., van de Putte P. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature. 1982 May 27;297(5864):339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Involvement of the invertible G segment in bacteriophage mu tail fiber biosynthesis. Virology. 1984 Apr 30;134(2):296–317. doi: 10.1016/0042-6822(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Guidolin A., Zingg J. M., Arber W. Organization of the bacteriophage P1 tail-fibre operon. Gene. 1989;76(2):239–243. doi: 10.1016/0378-1119(89)90164-9. [DOI] [PubMed] [Google Scholar]
- Heller K., Braun V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J Bacteriol. 1979 Jul;139(1):32–38. doi: 10.1128/jb.139.1.32-38.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller K., Braun V. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J Virol. 1982 Jan;41(1):222–227. doi: 10.1128/jvi.41.1.222-227.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking S. M., Egan J. B. Genetic characterization of twelve P2-186 hybrid bacteriophages. Mol Gen Genet. 1982;187(1):174–176. doi: 10.1007/BF00384403. [DOI] [PubMed] [Google Scholar]
- Iida S. Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology. 1984 Apr 30;134(2):421–434. doi: 10.1016/0042-6822(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Iida S., Meyer J., Kennedy K. E., Arber W. A site-specific, conservative recombination system carried by bacteriophage P1. Mapping the recombinase gene cin and the cross-over sites cix for the inversion of the C segment. EMBO J. 1982;1(11):1445–1453. doi: 10.1002/j.1460-2075.1982.tb01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp D., Kahmann R., Zipser D., Broker T. R., Chow L. T. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978 Feb 9;271(5645):577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kardas E., Ritthaler W., Sandulache R., Schmucker R., Stern B. Comparative analysis of invertible DNA in phage genomes. Cold Spring Harb Symp Quant Biol. 1984;49:301–311. doi: 10.1101/sqb.1984.049.01.036. [DOI] [PubMed] [Google Scholar]
- Krauel V., Heller K. J. Cloning, sequencing, and recombinational analysis with bacteriophage BF23 of the bacteriophage T5 oad gene encoding the receptor-binding protein. J Bacteriol. 1991 Feb;173(3):1287–1297. doi: 10.1128/jb.173.3.1287-1297.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel J. A., Goldstein R. N., Marsh M., Calendar R. Structure of the bacteriophage P2 tail. Virology. 1974 Nov;62(1):161–174. doi: 10.1016/0042-6822(74)90312-2. [DOI] [PubMed] [Google Scholar]
- Lindahl G. Characterization of conditional lethal mutants of bacteriophage P2. Mol Gen Genet. 1974 Feb 6;128(3):249–260. doi: 10.1007/BF00267114. [DOI] [PubMed] [Google Scholar]
- Lindahl G. Genetic map of bacteriophage P2. Virology. 1969 Dec;39(4):839–860. doi: 10.1016/0042-6822(69)90021-x. [DOI] [PubMed] [Google Scholar]
- Lindahl G. On the control of transcription in bacteriophage P2. Virology. 1971 Dec;46(3):620–633. doi: 10.1016/0042-6822(71)90065-1. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A. Bacteriophage receptors. Annu Rev Microbiol. 1973;27:205–241. doi: 10.1146/annurev.mi.27.100173.001225. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bertani L. E. Properties and products of the cloned int gene of bacteriophage P2. Mol Gen Genet. 1983;192(1-2):87–94. doi: 10.1007/BF00327651. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel C. J., Jacq B., Arquès D. G., Bickle T. A. A remarkable amino acid sequence homology between a phage T4 tail fibre protein and ORF314 of phage lambda located in the tail operon. Gene. 1986;44(1):147–150. doi: 10.1016/0378-1119(86)90055-7. [DOI] [PubMed] [Google Scholar]
- Montag D., Hashemolhosseini S., Henning U. Receptor-recognizing proteins of T-even type bacteriophages. The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J Mol Biol. 1990 Nov 20;216(2):327–334. doi: 10.1016/S0022-2836(05)80324-9. [DOI] [PubMed] [Google Scholar]
- Montag D., Henning U. An open reading frame in the Escherichia coli bacteriophage lambda genome encodes a protein that functions in assembly of the long tail fibers of bacteriophage T4. J Bacteriol. 1987 Dec;169(12):5884–5886. doi: 10.1128/jb.169.12.5884-5886.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag D., Riede I., Eschbach M. L., Degen M., Henning U. Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution. J Mol Biol. 1987 Jul 5;196(1):165–174. doi: 10.1016/0022-2836(87)90519-5. [DOI] [PubMed] [Google Scholar]
- Montag D., Schwarz H., Henning U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J Bacteriol. 1989 Aug;171(8):4378–4384. doi: 10.1128/jb.171.8.4378-4384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W., Harris A. W., Fuerst C. R., Siminovitch L. Mutations in bacteriophage lambda affecting particle morphogenesis. Virology. 1968 May;35(1):134–149. doi: 10.1016/0042-6822(68)90313-9. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Crowther R. A. DNA sequence of the tail fibre genes 36 and 37 of bacteriophage T4. J Mol Biol. 1981 Dec 15;153(3):545–568. doi: 10.1016/0022-2836(81)90407-1. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall-Hazelbauer L., Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973 Dec;116(3):1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riede I., Drexler K., Eschbach M. L., Henning U. DNA sequence of genes 38 encoding a receptor-recognizing protein of bacteriophages T2, K3 and of K3 host range mutants. J Mol Biol. 1987 Mar 5;194(1):31–39. doi: 10.1016/0022-2836(87)90713-3. [DOI] [PubMed] [Google Scholar]
- Riede I., Drexler K., Eschbach M. L., Henning U. DNA sequence of the tail fiber genes 37, encoding the receptor recognizing part of the fiber, of bacteriophages T2 and K3. J Mol Biol. 1986 Sep 20;191(2):255–266. doi: 10.1016/0022-2836(86)90262-7. [DOI] [PubMed] [Google Scholar]
- Riede I., Drexler K., Schwarz H., Henning U. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J Mol Biol. 1987 Mar 5;194(1):23–30. doi: 10.1016/0022-2836(87)90712-1. [DOI] [PubMed] [Google Scholar]
- Riede I., Eschbach M. L., Henning U. Presence of DNA, encoding parts of bacteriophage tail fiber genes, in the chromosome of Escherichia coli K-12. J Bacteriol. 1985 Sep;163(3):832–836. doi: 10.1128/jb.163.3.832-836.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Saigo K. Isolation of high-density mutants and identification of nonessential structural proteins in bacteriophage T5; dispensability of L-shaped tail fibers and a secondary major head protein. Virology. 1978 Apr;85(2):422–433. doi: 10.1016/0042-6822(78)90449-x. [DOI] [PubMed] [Google Scholar]
- Sandulache R., Prehm P., Kamp D. Cell wall receptor for bacteriophage Mu G(+). J Bacteriol. 1984 Oct;160(1):299–303. doi: 10.1128/jb.160.1.299-303.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Snustad D. P. Dominance interactions in Escherichia coli cells mixedly infected with bacteriophage T4D wild-type and amber mutants and their possible implications as to type of gene-product function: catalytic vs. stoichiometric. Virology. 1968 Aug;35(4):550–563. doi: 10.1016/0042-6822(68)90285-7. [DOI] [PubMed] [Google Scholar]
- Snyder M., Wood W. B. Genetic definition of two functional elements in a bacteriophage T4 host-range "cassette". Genetics. 1989 Jul;122(3):471–479. doi: 10.1093/genetics/122.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine M. G., Thorn M., Gibbs W., Calendar R., Kelly B. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology. 1971 Dec;46(3):691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- Symonds N., Coelho A. Role of the G segment in the growth of phage Mu. Nature. 1978 Feb 9;271(5645):573–574. doi: 10.1038/271573a0. [DOI] [PubMed] [Google Scholar]
- Temple L. M., Forsburg S. L., Calendar R., Christie G. E. Nucleotide sequence of the genes encoding the major tail sheath and tail tube proteins of bacteriophage P2. Virology. 1991 Mar;181(1):353–358. doi: 10.1016/0042-6822(91)90502-3. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Lefebvre N., Scott J. R., Cowan J. A., de Bruijn F., Bukhari A. I. Relationships between temperate phages Mu and P1. Virology. 1978 Aug;89(1):146–161. doi: 10.1016/0042-6822(78)90048-x. [DOI] [PubMed] [Google Scholar]
- Vanderslice R. W., Yegian C. D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974 Jul;60(1):265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- Walker J. T., Walker D. H., Jr Coliphage P1 morphogenesis: analysis of mutants by electron microscopy. J Virol. 1983 Mar;45(3):1118–1139. doi: 10.1128/jvi.45.3.1118-1139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westö A., Ljungquist E. A restriction endonuclease cleavage map of bacteriophage P2. Mol Gen Genet. 1979 Mar 9;171(1):91–102. doi: 10.1007/BF00274019. [DOI] [PubMed] [Google Scholar]
- Yamada M., Fujisawa H., Kato H., Hamada K., Minagawa T. Cloning and sequencing of the genetic right end of bacteriophage T3 DNA. Virology. 1986 Jun;151(2):350–361. doi: 10.1016/0042-6822(86)90055-3. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Cramer S., Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage mu. Nature. 1980 Jul 17;286(5770):218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Plasterk R., Kuijpers A. A Mu gin complementing function and an invertible DNA region in Escherichia coli K-12 are situated on the genetic element e14. J Bacteriol. 1984 May;158(2):517–522. doi: 10.1128/jb.158.2.517-522.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


