Abstract
We describe tandem mass spectrometric approaches, including multiple stage ion-trap and source collisionally activated dissociation (CAD) tandem mass spectrometry with electrospray ionization (ESI) to characterize inositol phosphorylceramide (IPC) species seen as [M − H]- and [M − 2H + Li]- ions in the negative-ion mode as well as [M + H]+, [M + Li]+, and [M − H + 2Li]+ ions in the positive-ion mode. Following CAD in an ion-trap or a triple-stage quadrupole instrument, the [M − H]- ions of IPC yielded fragment ions reflecting only the inositol and the fatty acyl substituent of the molecule. In contrast, the mass spectra from MS3 of [M − H − Inositol]- ions contained abundant ions that are readily applicable for assignment of the fatty acid and long-chain base (LCB) moieties. Both the product-ion spectra from MS2 and MS3 of the [M − 2H + Alk]-, [M + H]+, [M + Alk]+, and [M − H + 2Alk]+ ions also contained rich fragment ions informative for unambiguous assignment of the fatty acyl substituent and the LCB. However, the sensitivity of the ions observed in the forms of [M − 2H + Alk]-, [M + H]+, [M + Alk]+, and [M − H + 2Alk]+ (Alk = Li, Na) is nearly 10 times less than that observed in the [M − H]- form. In addition to the major fragmentation pathways leading to elimination of the inositol or inositol monophosphate moiety, several structurally informative ions resulting from rearrangement processes were observed. The fragmentation processes are similar to those previously reported for ceramides. While the tandem mass spectrometric approach using MSn (n = 2, 3) permits the structures of the Leishmania major IPCs consisting of two isomeric structures to be unveiled in detail, tandem mass spectra from constant neutral loss scans may provide a simple method for detecting IPC in mixtures.
Inositol phosphorylceramide (IPC) consists of a ceramide moiety, to which an inositol phosphate residue is linked via the 1-OH of the long-chain base (LCB) (see IPC structure in Schemes). The identification of IPCs in the membrane of Saccharomyces cerevisiae was first reported by Smith and Lester [1]. However, the glycosylated IPC (glycosylinositol phosphorylceramides or GIPCs) was first found in the membrane of Saccharomyces cerevisiae and Candida utilis by Wagner and Zofcsik, earlier [2]. In plants, GIPCs also constitute an abundant group of membrane sphingolipids [3, 4]. Different from plants and fungi, the primary sphingolipid species in Leishmania spp. and Trypanosoma spp. is unglycosylated IPC [5–7], which accounts for 5% to 10% of total cellular lipids in Leishmania [5] and are enriched in raft-associated membrane fractions [8].
IPC is synthesized through the transfer of inositol phosphate from phosphatidylinositol to the 1-OH group of ceramide or phytoceramide [9, 10]. This transfer reaction is catalyzed by the phosphatidylinositol: ceramide phosphoinositol transferase (IPC synthase), a membrane-bound enzyme [11, 12]. In S. cerevisiae and C. albicans, phytoceramide is the preferred receptor for phosphoinositol. These fungi synthesize a family of sphingolipids (SLs) composed of unglycosylated IPC, mannose inositol phosphoceramide (MIPC), and mannose di-inositol phosphoceramide (M[IP]2C) [11, 13]. Available evidence indicates that AUR1, an essential gene, encodes the IPC synthase or a subunit of the enzyme in S. cerevisiae [14]. In contrast to fungi and plants, mammalian cells do not synthesize IPC or GIPCs; instead, choline phosphate is transferred from phosphatidylcholine to the 1-OH group of ceramide, leading to the synthesis of sphingomyelin (SM). This difference in SL metabolism between mammalian cells and fungi represents an attractive target for antifungal drugs such as khafrefungin [15] and aureobasidin A [14, 16], which specifically inhibit the IPC synthase.
In addition to being membrane component, IPC species regulate virulence in pathogenic fungi. In Cryptococcus neoformans, IPC-derived diacylglycerol induces the transcription of virulence factor App1, which inhibits phagocytosis by alveolar macrophages [17], as well as activating protein kinase C1, which promotes cell-wall stability and melanin production in C. neoformans [18]. Recent work indicates Leishmania parasites synthesize high levels of IPC as both promastigotes (the stage residing extracellularly in the midgut of sandfly vectors) and amastigotes (the stage residing intracellularly within phagolysosomes of mammalian macrophages), suggesting this lipid plays important roles in both stages [19–21]. Mutants defective in the synthesis of sphingoid bases (via inactivation of serine palmitoyltransferase; spt2-) or degradation of sphingosine-1-phosphate (via inactivation of sphingosine-1-phosphate lyase; spl-) exhibited severe defects in vesicular trafficking upon entry into stationary phase and failed to differentiate to infective metacyclic form, suggesting sphingolipid metabolism is crucial for the virulence of Leishmania parasites [19, 21, 22]. Remarkably, both mutants were rescued by ethanolamine, a downstream metabolite of the de novo SL pathway, a finding with profound implications to role of SLs in the promastigote stage [22]. However, despite disruptions in the de novo synthesis of SLs, spt2- amastigotes retain high levels of IPC, suggesting that this stage salvages one or more IPC precursors from the mammalian host for conversion into the parasite specific IPC [20]. In this regard, the SL mutant amastigotes resemble WT amastigotes, which normally down-regulate the de novo SL and SPL (sphingosine-1-phosphate lyase)-dependent ethanolamine synthetic pathways [19, 21, 22]. Genetic analysis of the role of amastigote IPC thus awaits the development of parasite mutants deficient in IPC synthesis and/or SL salvage from the host.
The structure of IPC is well established. Tandem sector mass spectrometric approach to characterize Leishmania Mexicana IPC as the [M + H]+ ions desorbed by FAB was previously described by Singh et al. [23]. Levery et al. also described tandem mass spectrometric approaches to characterize glycosylinositol phosphorylceramides (GIPCs), the complex derivatives of IPC, using the lithiated adduct ions desorbed by ESI in the positive-ion mode [24–26]. IPC species from Trypanosomatids (Leishmania spp. and Trypanosoma spp.) have been previously analyzed by ESI tandem mass spectrometry in the negative-ion mode [6, 7, 19]. Here we report more detailed structural studies towards the identification of the long-chain base and the fatty acid substituent of IPC using tandem mass spectrometry. ESI tandem quadrupole and ion-trap mass spectrometric approaches are used to characterize the IPC species in the promastigote stage of L. major, a pathogen responsible for cutaneous leishmaniasis. These approaches afford the structures of IPC species in the forms of [M − H]- and [M − 2H + Li]- ions in the negative-ion mode, as well as in the forms of [M + H]+, [M + Li]+, and [M − H + 2Li]+ ions in the positive-ion mode to be unveiled in detail. The mechanisms underlying the fragmentation processes of IPC revealed by tandem mass spectrometry will be presented.
Materials
All chemicals used are in spectroscopic grade and were purchased from Sigma Chemical Co. (St. Louis, MO). Total lipids were extracted from L. major as previously described [19]. Briefly, wild type L. major LV39 clone 5 (Rho/SU/59/P) cells were grown in M199 medium with supplements and 10% heat inactivated fetal bovine serum at 26 °C. Cells were harvested, washed and resuspended in phosphate buffered saline (PBS, 137 mM NaCl, 10 mM Phosphate, 2.7 mM KCl, pH 7.4) at 2 × 108 cells/mL, followed by sonication on ice (15 W, 5 s). Afterwards lysates were extracted with 3.75 vol of CHCl3/methanol (1:2), followed by 1.25 vol of CHCl3, and 1.25 vol of water. After centrifugation (1000 g, 10 min), the upper aqueous phase was removed and the organic phase was washed once with water. The organic phase was then dried under nitrogen and dissolved in CHCl3/methanol (1:2) at the equivalent of 2 × 109 cells/ml and stored at −20 °C.
Methods
Mass spectrometry
Low-energy CAD tandem mass spectrometry experiments were conducted both on a Finnigan (San Jose, CA) LCQ DECA ion-trap (IT) mass spectrometer (MS) with the Xcalibur operating system and a Finnigan TSQ-7000 triple-stage quadrupole (TSQ) mass spectrometer with ICIS operating system. Lipid extract was infused (3 μL/min) to the ESI source, where the skimmer was set at ground potential, the electrospray needle was set at 4.5 kV, and temperature of the heated capillary was 260 °C. The automatic gain control of the ion trap was set to 5 × 107, with a maximum injection time of 400 ms. Helium was used as the buffer and collision gas at a pressure of 1 × 10−3 mbar (0.75 mTorr). The MSn experiments were carried out with a relative collision energy ranging from 30% to 40% and with an activation q value at 0.25. The activation time was set at 100 ms. Mass spectra were accumulated in the profile mode, typically for 3 to 5 min for MS2- and MS3-spectra. The mass resolution of the instrument was tuned to 0.6 Da at half peak height.
For product-ion spectra obtained with a triple quadrupole instrument, the precursor ions were selected in the first quadrupole (Q1), collided with Ar (2.3 mTorr) in the rf-only second quadrupole (Q2) and analyzed in the third quadrupole (Q3). The collision energies were set at 32 to 45 eV. Both Q1 and Q3 were tuned to unit mass resolution and scanned at a rate of 3 s/scan. The mass spectra were accumulated in the profile mode, typically for 5 to 10 min for a tandem mass spectrum. For source collisionally activated dissociation (CAD) product-ion spectra, the skimmer voltage was set 40 eV to yield product ions that were selected for further MS/MS.
Nomenclature
The nomenclature previously described for sphingomyelin and ceramide was used [27–29]. Therefore, the designation of, for example, d18:1/16:0-IPC signifies the IPC consists of a sphingosine-LCB and a palmitoyl (16:0-fatty acyl) substituent; while t16:0/18:0-IPC signifies that IPC contains a 16:0-phytosphingosine LCB and a stearoyl moiety.
Results and Discussion
IPC yielded abundant [M − H]- ions, when subjected to ESI in the negative-ion mode. In the presence of alkali metal ion (Alk+, Alk = Li, Na, 1 nm/uL), IPC also yielded [M − 2H + Alk]- ions in the negative-ion mode, as well as the [M + Alk]+ and [M − H + 2Alk]+ ions in the positive ion mode. To obtain the [M + H]+ ion, methanolic solution of NH4OAc was added to a final concentration of 1 nm/uL. In general, the sensitivity observed as the [M − H]- ion is at least 10 times higher than that observed as the [M + Li]+, [M − H + 2Li]+, [M + H]+, and [M − 2H + Li]- ions. This may be attributable to the fact that IPC possesses a phosphate residue that is readily ionizable. The structures of the Leishmania major IPC deduced from the tandem mass spectra of the various molecular species in this study are listed in Table 1.
Table 1.
Composition of IPC from Leishmania majora
| [M - H]− | [M - 2H + Li]− | [M - 2H + Na]− | [M + H]+ | [M + Li]+ | [M + Na]+ | [M - H + 2Li]+ | [M - H + 2Na]+ | Structure | Relative abundance (% of base peak) |
|---|---|---|---|---|---|---|---|---|---|
| 750.6 | *756.6 | *772.6 | 752.6 | *758.6 | *774.6 | *764.6 | *796.6 | d14:1/18:0-IPC & d16:1/16:0-IPC | 1 |
| 764.6 | *770.6 | *786.6 | 766.6 | *772.6 | *788.6 | *778.6 | *810.6 | d16:1/17:0-IPC & d15:1/18:0-IPC | 1 |
| 776.6 | *782.6 | *798.6 | 778.6 | *784.6 | *800.6 | *790.6 | *822.6 | d16:1/18:1-IPC | 1 |
| 778.6 | 784.6 | 800.6 | 780.6 | 786.6 | 802.6 | 792.6 | 824.6 | d16:1/18:0-IPC | 100 |
| 792.6 | 798.6 | 804.6 | 794.6 | *800.6 | *816.6 | *806.6 | *832.6 | d17:1/18:0-IPC & d16:1/19:0-IPC | 2 |
| 796.6 | 802.6 | 818.6 | 798.6 | 804.6 | 820.6 | 810.6 | *842.6 | t16:0/18:0-IPC | 10 |
| 804.6 | *810.6 | *826.6 | 806.6 | *812.6 | *828.6 | *818.6 | *850.6 | d18:2/18:0-IPC | 1 |
| 806.6 | 812.6 | *828.6 | 808.6 | 814.6 | 830.6 | *820.6 | *852.6 | d18:1/18:0-IPC | 9 |
| 820.6 | *826.6 | *842.6 | 822.6 | *828.6 | *844.6 | *834.6 | *866.6 | d19:1/18:0-IPC & d18:1/19:0-IPC | 1 |
| 822.6 | *828.6 | *844.6 | 824.6 | *830.6 | *846.6 | *836.6 | *868.6 | d19:0/18:0-IPC | 1 |
Data are derived from product-ion analysis with IT and TSQ mass spectrometers.
Not studied.
Structural Characterization with the [M − H]− Ions
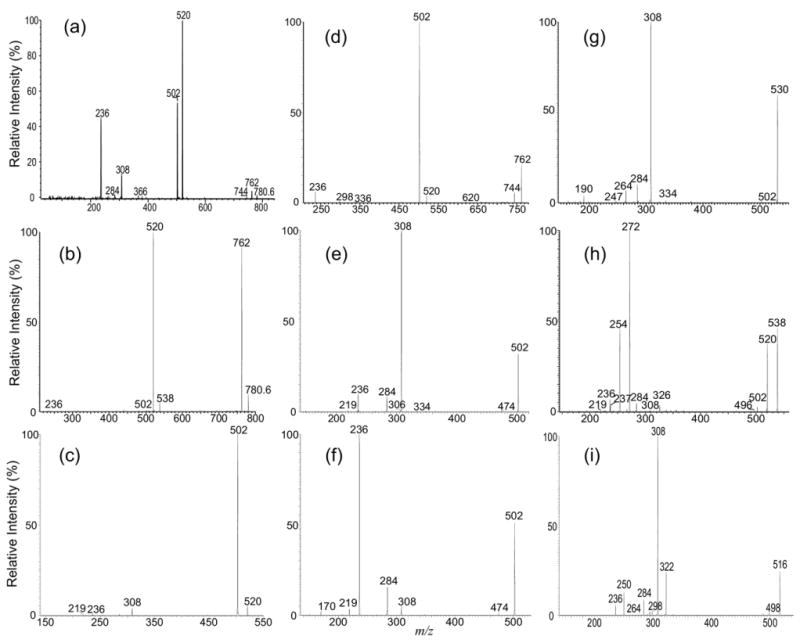
The ESI-MS profile of the [M − H]- ions of IPC isolated from Leishmania major is shown in Figure 1a, which contains major ions at m/z 778, 780, 796, and 806, consistent with the results previously reported by Zhang et al. [20]. The MS2 spectra of the ion at m/z 778 obtained with an ITMS (Figure 2a) and a TSQ (Figure 2b) instruments are nearly identical. The phosphoinositol residue of the molecule is seen by the presence of the prominent ions at m/z 241, corresponding to a inositol-1,2-cyclic phosphate anion, and at m/z 259 corresponding to a inositol monophosphate anion, along with the ion at m/z 223 arising from m/z 241 by further loss of H2O. These ions are characteristic to molecules of inositol phosphorylceramides [7], and are similar to those observed for phosphatidylinositol [30]. The spectrum also contains the ions at m/z 616 (778 − 162) and 598 (778 − 180), arising from losses of a dehydrated inositol and a inositol residues, respectively, as well as the ion at m/z 512 (778 − 266), corresponding to loss of a 18:0-fatty acyl substituent as a ketene (Scheme 1a). The presence of the ion at m/z 512 suggested that the molecule is a d16:1/18:0-IPC rather than a d18:1/16:0-IPC structure as previously reported by Serrano et al. [7]. The structural assignment is further supported by the IT MS3 spectrum of the ion at m/z 598 (778 → 598, Figure 2c), which contains ions at m/z 332 (598 − 266) corresponding to loss of an 18:0-fatty acyl ketene and at m/z 314 (332 − H2O) from further loss of H2O. The spectrum also contains the ion at m/z 388, arising from loss of the d16:1-LCB as an aldehyde and the ion at m/z 386, arising from the simultaneous losses of the aldehyde and H2 residues (Scheme 1a, Routes a and b) [31]. The 18:0-fatty acyl substituent is recognized by the presence of the ion at m/z 283, which probably arises from the pathway involving a rearrangement process (Scheme 1a, Route c) and represents a 18:0-carboxylate anion. This unique formation of carboxylate anion was previously observed for ceramides [31] and provides information for assignment of the fatty acyl substituent. Similarly, the IT MS2 product-ion spectrum of the ion at m/z 806 (Figure 2d) contains the common ions at m/z 259, 241, 223, 644 (806 − 162) and 626 (644 − 180) seen for an IPC, along with the ion at m/z 540, corresponding to loss of a 18:0-fatty acyl ketene, suggesting the compound is a d18:1/18:0-IPC. This structural assignment is further confirmed by the IT MS3 spectrum of the ion at m/z 626 (806 → 626, Figure 2e), in which the prominent ions at m/z 360 (626 − 266) from loss of the 18:0-fatty acyl ketene and at m/z 342 (360 − H2O) from further loss of H2O, along with the ion at m/z 283, representing a 18:0-carboxylate anion are present. The spectrum also contains the analogous ions at m/z 388 and 366 arising from the similar losses of the d18:1-LCB as described earlier, consistent with the assigned d18:1/18:0-IPC structure.
Figure 1.
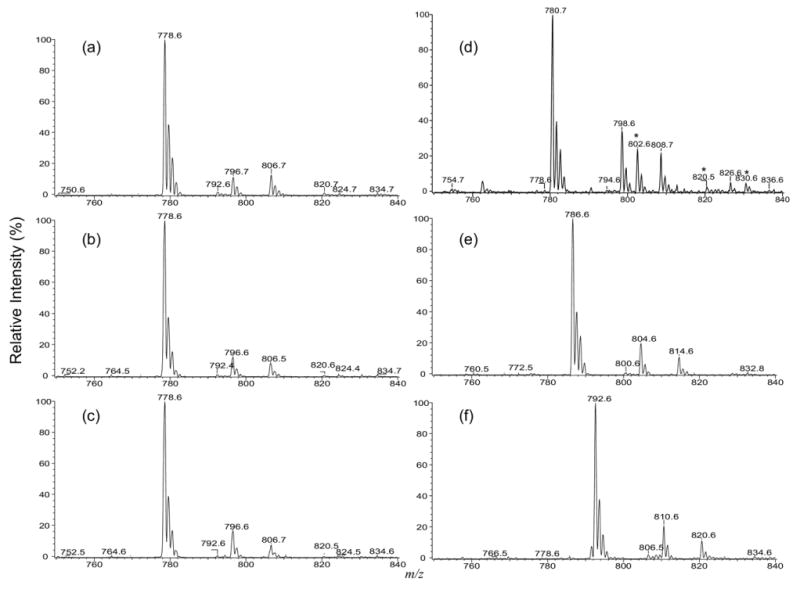
(a) The ESI/MS spectrum of the [M − H]- ion of IPC extract from Leishmania major, and the MS2 spectra of the same extract obtained by precursor-ion scans of 241 (b), of 259 (c), by neutral loss scans of 260 (d), of 266 (e), and of 248 (f) that detect IPCs as the [M − H]- (b), [M − H]- (c), [M + H]+ (d), [M + Li]+ (e), and the [M − H + 2Li]+ (f) species, respectively. The ions labeled with an asterisk in (d) are the corresponding [M + Na]+ ions.
Figure 2.
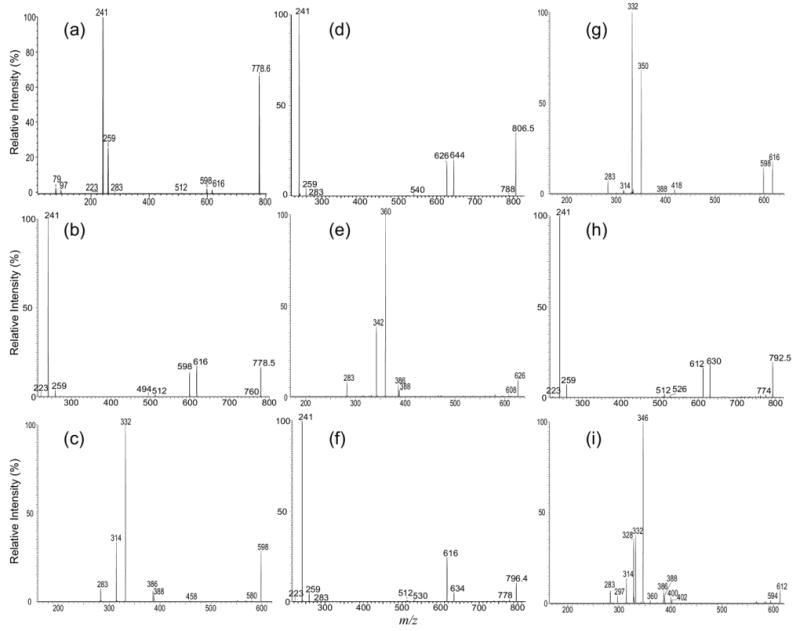
The MS2 product-ion spectra of the [M − H]- ion of d16:1/18:0-IPC at m/z 778 obtained with an ITMS (a), and a TSQ (b) instruments are similar and contain limited structural information. However, the IT MS3 spectrum of the ion at m/z 598 (778 → 598) (c) contains ions readily applicable for structural assignment. Similar results were also observed for the IT MS2 spectrum of the [M − H]- ion of d18:1/18:0-IPC at m/z 806 (d) and its IT MS3 spectrum of the ion at m/z 626 (806 → 626) (e), for the IT MS2 spectrum of the [M − H]- ion of t16:0/18:0-IPC m/z 796 (f) and its MS3 spectrum of the ion at m/z 616 (796 → 616) (g), and for the IT MS2 spectrum of the ion at m/z 792 (h) and its IT MS3 spectrum of the ion at m/z 612 (792 → 612) (i), from which two isomeric structures were found.
Scheme 1.
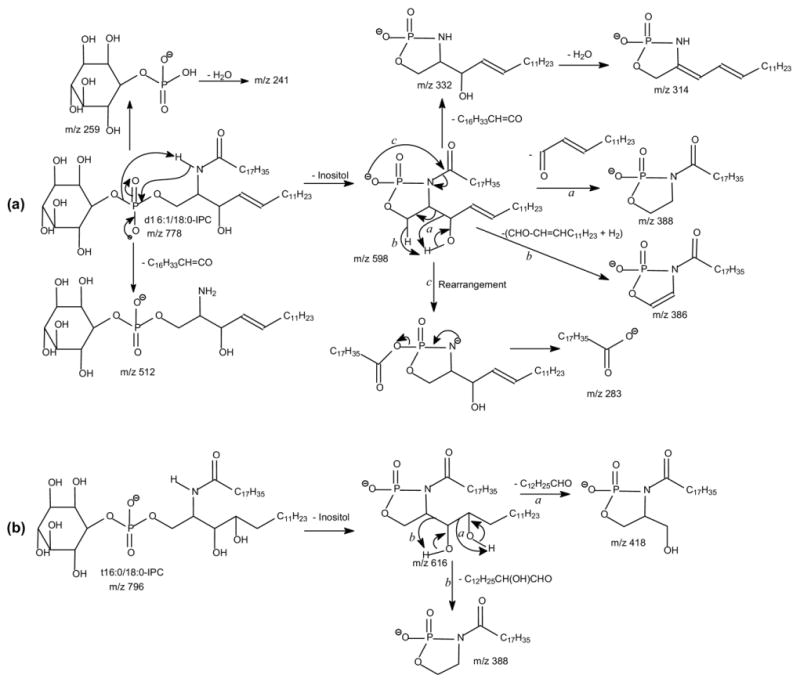
In Figure 2f, the IT MS2 spectrum of m/z 796 also contains the ion at m/z 530, arising from loss of a 18:0-fatty acyl ketene, along with the ions at m/z 259, 241, 223, 634 (796 − 162), and 616 (796 − 180) that are characteristic to IPC. However, the ion at m/z 616 ([M − H − 180]-) is more abundant than the ion at m/z 634 ([M − H − 162]-). By contrast, the abundances of the analogous ions at m/z 598 ([M − H − 180]-) and 616 ([M − H − 162]-) observed in Figure 2a and b and of the ions at m/z 626 ([M − H − 180]-) and 644 ([M − H − 162]-) observed in Figure 2d are nearly of equal abundance. These differences in the abundances of the [M − H − 162]- and the [M − H − 180]- ions indicate that the m/z 796 ion may consist of a phytosphingosine LCB rather than a sphingosine LCB. This notion is further revealed by the MS3 spectrum of the ion at m/z 616 (796 → 616, Figure 2g), in which the ions at m/z 418, arising from elimination of a tridecaldehyde (198 Da) residue by cleavage of the 3,4-dihydroxy bond (Scheme 1b, Route a) and at m/z 388 by loss of a 2-hydroxy-tetradecaldehyde (Scheme 1b, Route b) are present. The spectrum is also dominated by the ions at m/z 350 (616 − 266) arising from loss of a 18:0-fatty acyl ketene and at m/z 332 (350 − H2O) arising from further loss of H2O. The observation of these ions along with the ion at m/z 283 reflecting a 18:0-fatty acyl residue, clearly demonstrated that the molecule is, indeed, a t16:0/18:0-IPC. The distinction in the production spectra between, for example, d16:1/18:0-IPC and t16:0/18:0-IPC that are differed by their LCB moieties are also seen in the MS2 and MS3 spectra of the [M − 2H + Li]-, [M + Li]+, [M − H + 2Li]+, and [M + H]+ ions (discuss later).
Several isobaric structures were observed for the ions at m/z 750, 764, and 792 (Table 1). The IT MS2 spectrum of the ion at m/z 792 (Figure 2h), for example, contains the ions at m/z 259, 241, 223, 630 (792 − 162), and 612 (792 − 180) that are characteristic to an IPC. The spectrum also contains the ions at m/z 526 and 512, corresponding to losses of 18:0- and 19:0-fatty acyl substituents, respectively. The structural assignment is further deduced from the IT MS3 spectrum of the ion at m/z 612 (792 → 612) (Figure 2i), which contains two sets of the ion-pair at m/z 346 and 328, arising from loss of 18:0-fatty acyl substituent, and at m/z 332 and 314, arising from the analogous losses of the 19:0-fatty acyl substituent, along with the ions at m/z 297 and 283, corresponding to the 19:0- and 18:0-carboxylate anions, respectively. The spectrum also contains two sets of the ion-pair at m/z 402 and 400, arising from elimination of d16:1-LCB, and at m/z 388 and 386, arising from elimination of d17:1-LCB. The results demonstrated that the ion at m/z 792 is composed of both a d17:1/18:0-IPC and a d16:1/19:0-IPC structures.
Structural Characterization with the [M − 2H + Li]− Ions
Both the MS2 spectra of the [M − 2H + Li]- ion at m/z 784 (Table 1) obtained with IT (Figure 3a) and TSQ (not shown) instruments are dominated by the ion at m/z 622, corresponding to loss of a dehydrated inositol residue. The presence of the ions at m/z 518 (784 − 266) arising from loss of the 18:0-fatty acyl ketene and at m/z 500 (518 − H2O) from further loss of H2O indicate that the molecule contains a 18:0-fatty acyl substituent. Further dissociation of the ion at m/z 622 (784 → 622) (Figure 3b), gives rise to ions at m/z 604 (622 − H2O) and 592 (622 − HCHO) via losses of H2O and HCHO, respectively [31]. The fragmentation process leading to formation of the ion at m/z 592 may involve rearrangement of the phosphate residue to C3 from C1 position of the LCB, followed by loss of a HCHO residue (Scheme 2a, Route a). The MS4 product-ion spectrum of the ion at m/z 592 (784 → 622 → 592) (Figure 3c) is dominated by the ion at m/z 504, corresponding to loss of a P(OH)2(OLi) (88 Da) residue (Scheme 2a), consistent with the proposed mechanism that involves rearrangement of the phosphate group. Similar rearrangement of phosphate group has been previously observed for phosphatidylinositol phosphate [30], phosphatidylserine [32], and plasmenylethanolamine [33].
Figure 3.
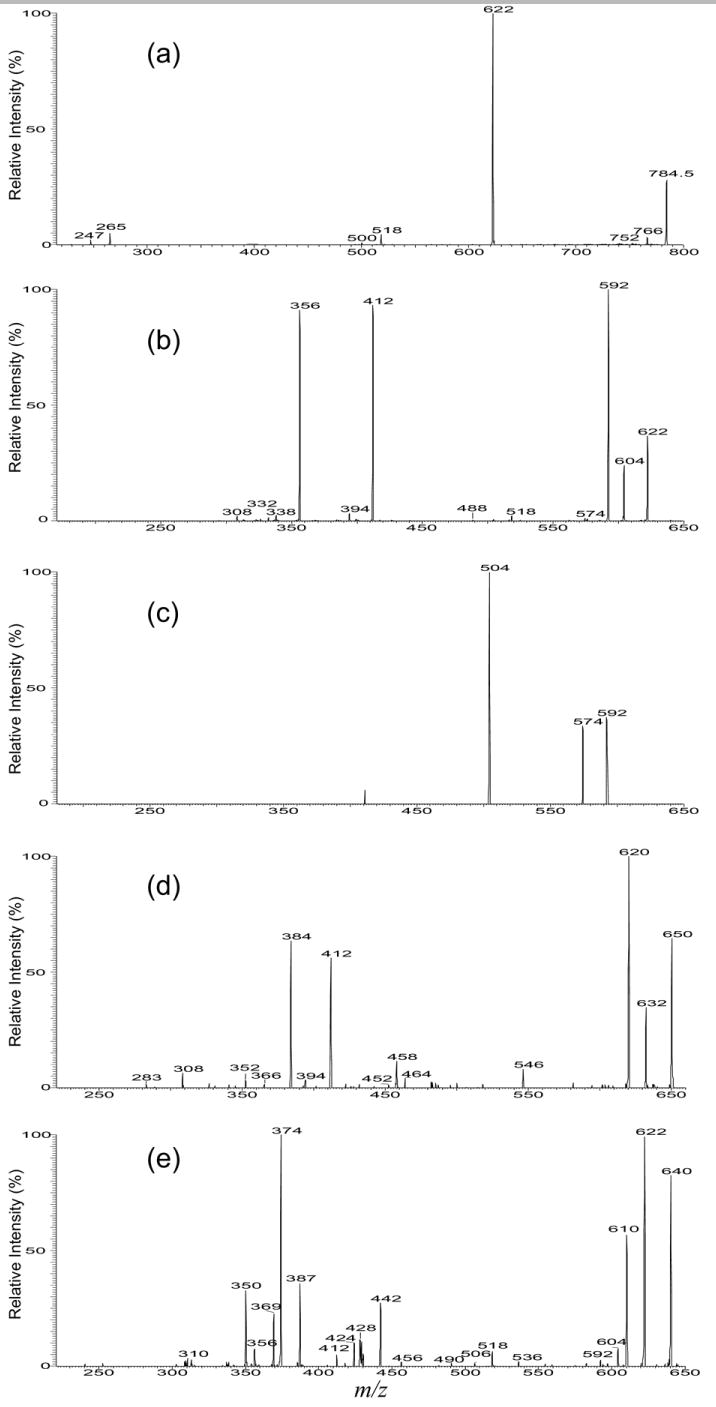
The IT MS2 spectra of the [M −2H + Li]- ion at m/z 784 (a), its IT MS3 spectrum of the ion at m/z 622 (784 → 622) (b), and its MS4 spectrum of the ion at m/z 592 (784 → 622 → 592) (c); (d) is the IT MS3 spectrum of m/z 650 (812 → 650) from d18:1/18:0-IPC; (e) is the IT MS3 spectrum of m/z 640 (802 → 640) from t16:0/18:0-IPC.
Scheme 2.
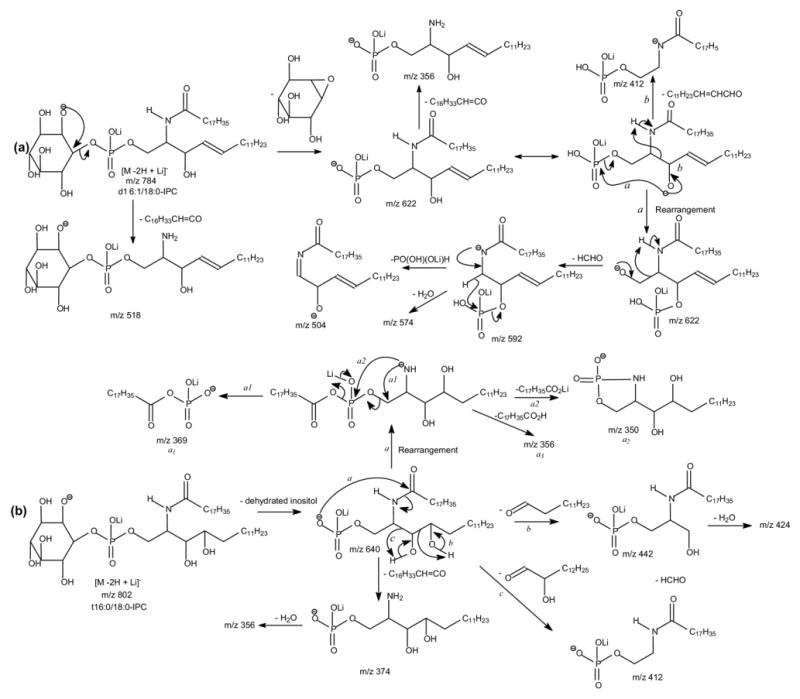
In Figure 3b, the ion at m/z 412 probably arises from elimination of the LCB as an α,β-unsaturated aldehyde as seen before (Scheme 2a, Route b); the ion at m/z 356 corresponds to loss of the 18:0-fatty acyl substituent. These results combined with the results from MS2 (Figure 3a) indicate that the compound is a d16:1/18:0-IPC. Similarly, the IT MS2 spectrum of the [M − 2H + Li]- ion at m/z 812 contains a major ion at m/z 650 (not shown) from loss of the inositol residue and the IT MS3 spectrum of m/z 650 (812 → 650) (Figure 3d) also contains the analogous ions at m/z 412 and 384, arising from loss of a 18:1-LCB as an α,β-unsaturated aldehyde and from loss of a 18:0-fatty acyl substituent, respectively. The observation of these ions, along with the ions at m/z 632 and 620, arising from further losses of H2O and HCHO, respectively, suggests that the molecule is a d18:1/18:0-IPC, which is consistent with the structural assignment obtained from the [M − H]- ion (Table 1).
The IT MS2 spectrum of the [M − 2H + Li]- ion at m/z 802 (not shown) is, again, dominated by the ion at m/z 640 (802 − 162), which gives rise to the prominent ions at m/z 622 (640 − H2O) and 610 (640 − HCHO) as seen earlier (Figure 3e, 802 → 640). Nevertheless, the spectrum (Figure 3e) also contains the ion at m/z 442, probably arising from cleavage of the 3,4-dihydroxy bond of the t16:0-LCB to yield an alcohol by elimination of a tridecaldehyde (Scheme 2b), and the ion at m/z 412, arising from loss of a 2-hydroxy-tetradecaldehyde, similar to the fragmentation processes as seen for the corresponding [M − H]- ion of m/z 796 (Scheme 1b). The 18:0-fatty acyl substituent is recognized by the observation of the prominent ion at m/z 374 (640 –266), arising from loss of 18:0-fatty acyl ketene. The ion at m/z 369 may represent a stearoylphosphoric anion (a1 ion,Scheme 2b), arising from a lipophosphoryl LCB via a prior rearrangement process (Scheme 2b, Route a). This fragmentation process is consistent with the observation of the ions at m/z 350 (a2) and 356 (a3), arising from further losses of the 18:0-fatty acid substituent as a lithium salt and as a free acid, respectively. These combined information lead to the assignment of a t16:0/18:0-IPC structure. By contrast, the MS3 spectrum from the sphingosine-containing IPC as shown in Figure 3b is simple. The striking differences among the MS3 spectra arising from the IPCs with various LCBs are consistent with the distinction among the production spectra arising from their [M − H]- ions as seen earlier.
Structural Characterization with the [M + H]+, [M + Alk]+, and [M − H +2 Alk]– Ions
Among the most useful features in the product-ion spectra arising from the protonated and the alkali adduct ions of ceramide and sphingomyelin is that multiple sets of the fragment ions reflecting the identities of LCB and of fatty acyl substituent are present, leading to unequivocal structural determination [27–29]. The structurally informative ions as described below are similar to those observed for the [M + H]+ and alkali adduct ions of ceramide and of sphingomyelin.
The [M + H]+
The MS2 product-ion spectra of the [M + H]+ ion at m/z 780 obtained with a TSQ (Figure 4a) and an IT (Figure 4b) instruments are dominated by the ion at m/z 520, arising from loss of the inositol monophosphate residue. The m/z 520 ion is equivalent to a protonated [Ceramide – H2O] ion, which gives rise to the ion at m/z 502 by elimination of H2O. This is evidenced by observation of a prominent ion at m/z 502 in the IT MS3 spectrum of the ion at m/z 520 (Figure 4c, 780 → 520). The m/z 502 ion can also arise from m/z 762 by elimination of the inositol monophosphate residue. This fragmentation process is supported by the IT MS3 spectrum of m/z 762 (780 → 762) (Figure 4d), in which the ion at m/z 502 is prominent. Both the IT MS4 spectra of the ion at m/z 502 originated from m/z 520 (780 → 520 → 502, Figure 4e) and from m/z 762 (780 → 762 → 502, Figure 4f) contain ions at m/z 308 and 284, identifying the 18:0-fatty acyl substituent, along with the ions at m/z 236 and 219 reflecting the d16:1-LCB moiety [23, 27, 28]. However, the ion at m/z 308 is the most prominent in Figure 4e; whereas the ion at m/z 236 is the most prominent in Figure 4f. The apparent differences between the two IT MS3 spectra from the m/z 502 ions may be attributable to the fact that the mechanism leading to dissociation of m/z 502 as seen in Figure 4e involved in a prior formation of 5-member cyclic ring intermediate as previously proposed (Scheme 3a) [29], while the mechanism leading to dissociation of m/z 502 as seen in Figure 4f involved a prior formation of an aziridine intermediate (Scheme 3b). Similar results were also observed for the [M + H]+ ion at m/z 806, which undergoes consecutive losses of inositol monophosphate and H2O to yield ions at m/z 548 and 530, respectively (data not shown). The IT MS4 spectrum of the ion at m/z 530 (808 → 548 → 530) (Figure 4g) contains the ions at m/z 308 and 284, reflecting the 18:0-fatty acyl substituent, along with the ions at m/z 264 and 247 that are 28 Da higher than the analogous ions observed for d16:1/18:0-IPC (i.e., the m/z 236 and 219 ions) and are signifying the presence of d18:1-LCB residue.
Figure 4.
The MS2 product-ion spectra of the [M + H]+ ion at m/z 780 obtained with (a) a TSQ and (b) an IT instruments. The IT MS3 spectra of the ion at m/z 520 (780 → 520) (c), and of the ion at m/z 762 (780 → 762) (d) provide limited structural information but the IT MS4 spectrum of m/z 502 originated from m/z 520 (780 → 520 → 502) (e) and from m/z 762 (780 → 762 → 502) (f) give abundant ions readily applicable for structure identification; (g) is the IT MS4 spectrum of the ion at m/z 530 (808 → 548 → 530) from the [M + H]+ ion of d18:1/18:0-IPC; (h) is the IT MS3 spectrum of the ion at m/z 538 (798 →538) from the [M + H]+ ion of t16:0/18:0-IPC; (i) is the IT MS4 spectrum of the ion at m/z 516 (794 → 534 → 516) arising from both a d17:1/18:0-IPC and a d16:1/19:0-IPC isomers.
Scheme 3.
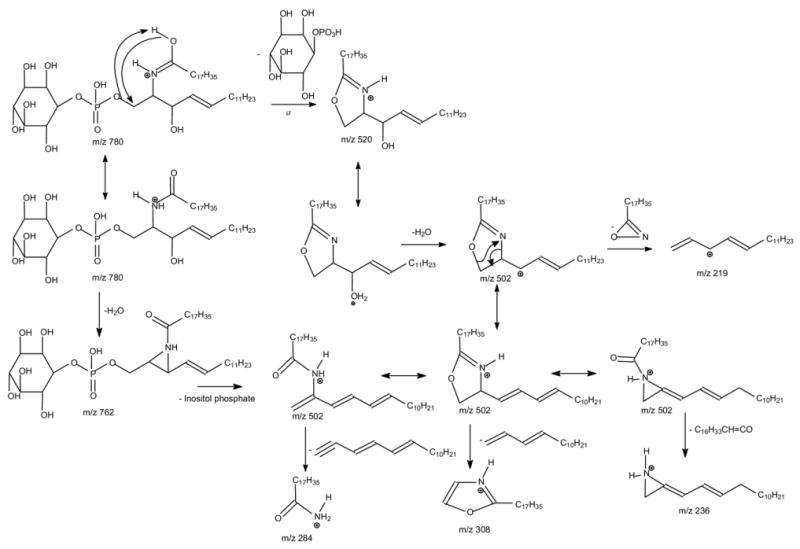
Both the IT and TSQ MS2 product-ion spectra of the [M + H]+ ion at m/z 798 are dominated by the ion at m/z 538 (not shown), arising from loss of inositol monophosphate. The IT MS3 spectrum of the ion at m/z 538 (798 → 538, Figure 4h) contains the ions at m/z 520 and 272 from further losses of H2O and 18:0-fatty acyl ketene, respectively. However, the ions at m/z 308 and 284, reflecting the 18:0-fatty acyl residue, along with the ions at m/z 254 and 237, reflecting the t16:0-LCB, residue were also seen. Similar ions (i.e., the m/z 308, 282, 254, and 237 ions) were also observed in the MS4-spectrum of the ion at m/z 520 (798 → 538 → 520) (data not shown). The results are consistent with the assignment of the t16:0/18:0-IPC structure deduced from the [M − H]- ion at m/z 796 as described earlier. The MS3 spectrum is readily distinguishable from that arising from the IPC consisting of sphingosine, such as d16:1/18:0- IPC (Figure 4c), which would require MS4 spectrum for its structural identification.
The tandem mass spectrometric approach that reveals the multiple isobaric structures of IPCs in the [M + H]+ form is exemplified by the ion at m/z 794, which gives rise to a prominent ion at m/z 534 (794 − 260) (not shown), which further dissociated to m/z 516 (534 − H2O) by a water loss (not shown). The MS4 spectrum of the ion at m/z 516 (794 → 534 → 516) (Figure 4i) contains two sets of the ion-pair at m/z 322/298 and 308/284, indicating the presence of 19:0- and 18:0-fatty acyl substituents, respectively. The spectrum (Figure 4g) also contains two sets of the ion-pair at m/z 250/233 and 236/219 that signify the presence of 17:1-LCB and 16:1-LCB, respectively. The major set of the m/z 308/284 ion-pair is formed concurrently with the m/z 250/237 ion-pair, arising from the major d17:1/18:0-IPC structure; while the minor m/z 322/298 ion-pair is formed together with the set of m/z 236/219 ion-pair, arising from a minor d16:1/19:0-IPC species. Again, the structural assignment is consistent with that deduced from the [M − H]- ion at m/z 792 (Figure 2i) (Table 1).
The [M + Alk]+ (Alk = Li, Na) Ions
The major fragmentation process observed for the [M + Alk]+ (Alk = Li, Na) ion of IPC also arises from cleavage of the inositol monophosphate residue. This is seen in the tandem quadrupole product-ion spectrum of the [M+ Li]+ ion at m/z 786 (Figure 5a), which contains prominent ions at m/z 526 (786 − 260) arising from elimination of inositol monophosphate residue and at m/z 267 ([C6H11O5OPO(OH)2 + Li]+), corresponding to a lithiated adduct ion of inositol monophosphate. The ion at m/z 544 (Figure 5a) is equivalent to a lithiated ceramide ion and gives rise to ions at m/z 526, 520, 502, and 496, as well as the ions at m/z 308 and 236, reflecting the 18:0-fatty acyl and 16:1-LCB moieties, respectively [27–29]. Similar results were also observed for the [M + Na]+ ion at m/z 802 (Figure 5b), which is dominated by the analogous ions at m/z 542 (802 − 260) and m/z 283 ([C6H11O5OPO(OH)2 + Na]+), along with ions at m/z 308 and 236 that reflect the 18:0-fatty acyl and 16:1-LCB, respectively. The IT MS2 spectra of the ions at m/z 786 (Figure 5c) and at m/z 802 (Figure 5d) are similar to those obtained with TSQ, but the ions reflecting the fatty acyl and LCB moieties are not present. However, the IT MS4 spectra of the ions at m/z 502 (786 → 520 → 502) (not shown) are identical to that shown in Figure 4e, which gives assignment of the 18:0-fatty acyl and 16:1-LCB moieties.
Figure 5.
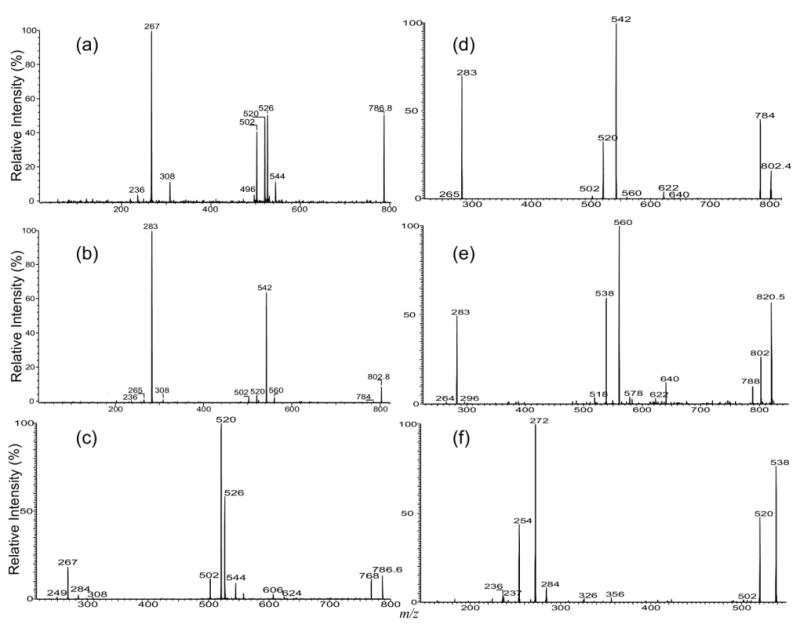
The MS2 product-ion spectra of (a) the [M + Li]+ ion at m/z 786, (b) of the [M+ Na]+ ion at m/z 802 (b) obtained with a TSQ, and of the [M + Li]+ ion at m/z 786 (c), and of the [M + Na]+ ion at m/z 802 (d) obtained with an IT instrument. The IT MS2 spectrum of the [M + Na]+ ion of t16:0/18:0-IPC at m/z 820 (e) and its IT MS3-spectrum of the ion at m/z 538 (820 → 538) (f) are also shown.
The IT MS2 spectrum of the [M + Na]+ ion at m/z 820 (Figure 5e) is also dominated by the ions at m/z 560 (820 − 260) and 538 (820 − 282), arising from losses of the inositol monophosphate as an acid and as a sodium salt, respectively, along with the ion at m/z 283 ([C6H11O5OPO(OH)2 + Na]+). The ions informative to the identities of the 18:0-fatty acyl and t16:0-LCB residues are not present in the spectrum, but the IT MS3-spectrum of the ion at m/z 538 (820 → 538) (Figure 5f) is identical to that shown in Figure 4h, which represents the t16:0/18:0-IPC structure. Similar results were also observed for the [M + Li]+ ion at m/z 804 (data not shown).
The [M − H + 2 Alk]- (Alk = Li, Na) Ions
The major ions observed for the [M − H + 2 Alk]+ ion of IPC arises from cleavage of the P–OC1 bond, similar to that previously observed for the [M − H + 2Alk]+ of phosphatidylserine [32]. As seen in Figure 6, the MS2 product-ion spectra of the [M − H + 2 Li]+ ion at m/z 792 obtained with an TSQ (Figure 6a) and with an IT (Figure 6b) instruments are dominated by the ions at m/z 544 and 255, corresponding to a lithiated ceramide and a dilithiated inositol-1,2-cyclic phosphate ions, respectively (Scheme 4). In contrast, the product-ion spectrum of the corresponding [M − H + 2 Na]+ ion at m/z 824 (Figure 6c) is dominated by a disodiated inositol-1,2-cyclic phosphate ion at m/z 287, and the ion at m/z 560, corresponding to a sodiated ceramide cation, is of low abundance. This drastic rise in the abundance of the alkali phosphate cations upon substitution of Li+ with Na+ was previously seen for the [M − H + 2Alk]+ of phosphatidylserine [32].
Figure 6.
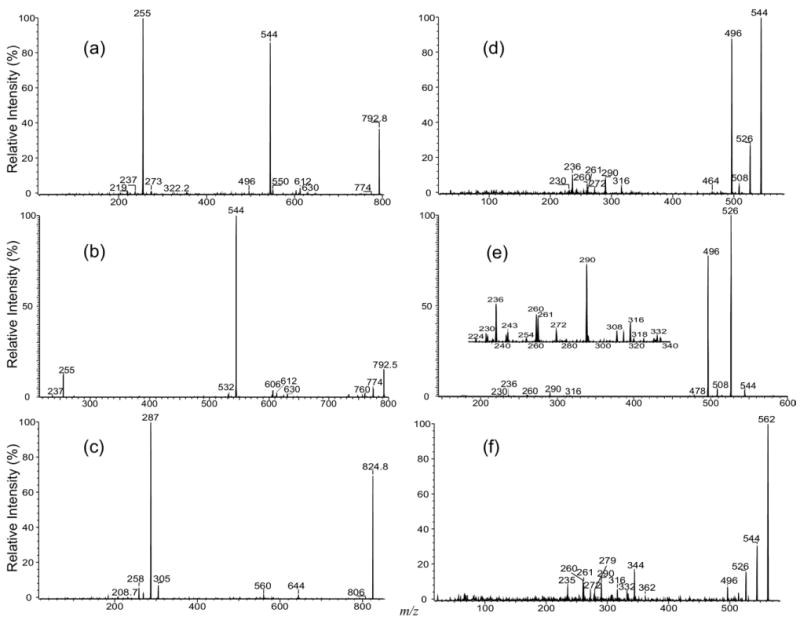
The MS2 product-ion spectra of the [M − H + 2Li]+ ion of d16:1/18:0-IPC at m/z 792 obtained with (a) a TSQ, (b) an IT instruments, and the tandem quadrupole product-ion spectrum of the corresponding [M − H + 2Na]+ ion at m/z 824 (c). The product-ion spectrum of the ion at m/z 544 from source CAD (d), and from MS3 (792 → 544) with IT (e) are also shown; (f) is the product-ion spectrum of the ion at m/z 562, generated by skimmer CAD on the [M − H + 2Li]+ ion of t16:0/18:0-IPC at m/z 810.
Scheme 4.
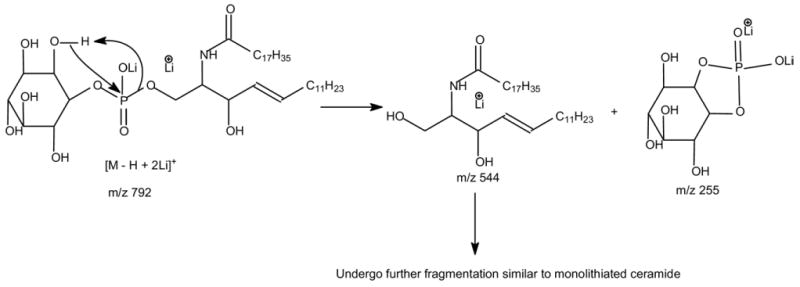
The fragment ions informative for the fatty acyl substituent and LCB are not present in the MS2 spectra (Figure 6a-c). However, the product-ion spectrum of the ion at m/z 544 (Figure 6d) generated by source CAD and the IT MS3 spectrum of the ion at m/z 544 (792 → 544) (Figure 6e) contains the ions at m/z 236, 260, and 261, signifying the 16:1-LCB moiety, along with the ions at m/z 272, 290, and 316, identifying the 18:0-fatty acyl substituent. These ions are similar to those observed for the [M + Li]+ ion of ceramide with a sphingosine moiety [27–29], supporting the assignment of the d16: 1/18:0-IPC structure. These results are consistent with the notion that the ions informative for structural identification of the [M + Li]+ ion of the d16:1/18:0-IPC arise from further fragmentation of m/z 544, a lithiated d16:1/18:0-ceramide. Similarly, the product-ion spectrum of the ion at m/z 562 (Figure 6f), generated by source CAD on the [M − H + 2Li]+ ion of t16:0/18:0-IPC at m/z 810, contains analogous ions arising from the [M + Li]+ ion of t16:0/18:0-ceramide at m/z 562. These ions (Figure 6f) were seen at m/z 279, 261, 260, and 235 that identify the t16:0-LCB, as well as the ions at m/z 344, 332, 316, 290, and 272 that identify the 18:0-fatty acyl moiety [28].
Profiling the IPC Species by Linked-Scanning
As shown earlier, the ions at m/z 259 and 241 are common ions observed in the MS2 spectra of the [M − H]- ions of IPCs and are among the most prominent (Figure 2). The MS2 spectra obtained by parent-ion scans of 241 (Figure 1b) and of 259 (Figure 1c) are similar to the MS spectrum shown in Figure 1a, suggesting that the MS2 spectra arising from these scans may be applicable for profiling the IPCs in mixtures, similar to that previously described for phosphatidylinositol [30]. The profiles of the MS2 spectra acquired by neutral losses of 260 (loss of C6H11O6PO[OH]2) (Figure 1d), of 266 (loss of C6H11O6PO[OH][OLi]) (Figure 1e), and of 248 (loss of C6H10O6PO[H][OLi]) (Figure 1f), from the IPCs observed in the forms of [M + H]+, [M + Li]+, and [M − H + Li]+, respectively, are also similar, indicating the MS2 spectra from these neutral loss (NL) scans may be also useful for detecting IPC molecules. However, the sensitivities of IPCs detected by ESI as the [M + H]+, [M + Li]+, and [M − H + Li]+ forms are at least 10 times less than the [M − H]- form, resulting in decrease in sensitivities using the corresponding NL scannings (i.e., NL scannings of 260, 266, and 248), regardless of the fact that the [M + H - 260]+ (NL of 260), [M + Li - 266]+ (NL of 266), and [M − H + 2Li - 248]+ (NL of 248) ions are among the most prominent in the product-ion spectra of IPC seen as the [M + H]+ (e.g., Figure 4a), [M + Li]+ (e.g., Figure 5a) and as the [M − H + 2Li]+ (e.g., Figure 6a) ions, respectively.
Conclusions
Multiple-stage IT mass spectrometry with subsequent applications of MS3 and MS4 and tandem quadrupole mass spectrometry with application of source CAD afford the fragmentation processes of IPC under low-energy CAD to be unveiled in detail, leading to its structural characterization. While multiple-stage mass spectrometry, in particular with IT, provides information that is valuable to the understanding of the mechanism(s) underlying the fragmentation processes, tandem quadrupole mass spectrometry with application of precursor-ion or NL scanning is useful for specific detection of IPC species in mixture from biological specimens. The observation of the d16:1/18:0-IPC as the most abundant IPC from this study (Table 1) suggests that the majority of LCB in Leishmania major is synthesized through the condensation of serine and myristoyl-CoA (14:0). This unique requirement for myristoyl-CoA in the IPC synthesis in Leishmania major is similar to the synthesis of lipid anchor for the variant surface glycoprotein in a related protozoan parasite, Trypanosoma brucei [34].
Acknowledgments
Research at the Mass Spectrometry Resource of Washington University was supported by U.S. Public Health Service grants P41-RR-00954, R37-DK-34388, P60-DK-20579, P01-HL-57278, P30-DK56341, and AI31078 (SMB).
References
- 1.Smith SW, Lester RL. Inositol Phosphorylceramide, a Novel Substance and the Chief Member of a Major Group of Yeast Sphingolipids Containing a Single Inositol Phosphate. J Biol Chem. 1974;249:3395–3405. [PubMed] [Google Scholar]
- 2.Wagner H, Zofcsik W. On New Sphingolipids of Yeast. Biochem Zeitschrift. 1966;344:314–316. [PubMed] [Google Scholar]
- 3.Carter HE, Celmer WD, Lands WE, Mueller KL, Tomizawa HH. Biochemistry of the Sphingolipids. VIII Occurrence of a Long Chain Base in Plant Phosphatides. J Biol Chem. 1954;206:613–623. [PubMed] [Google Scholar]
- 4.Hetherington AM, Drobak BK. Inositol-Containing Lipids in Higher Plants. Prog Lipid Res. 1992;31:53–63. doi: 10.1016/0163-7827(92)90015-b. [DOI] [PubMed] [Google Scholar]
- 5.Kaneshiro ES, Jayasimhulu K, Lester RL. Characterization of Inositol Lipids from Leishmania donovani Promastigotes: Identification of an Inositol Sphingophospholipid. J Lipid Res. 1986;27:1294–1303. [PubMed] [Google Scholar]
- 6.Guther ML, Lee S, Tetley L, Acosta-Serrano A, Ferguson MA. GPI-Anchored Proteins and Free GPI Glycolipids of Procyclic Form Trypanosoma brucei are Nonessential for Growth, are Required for Colonization of the Tsetse Fly, and are not the Only Components of the Surface Coat. Mol Biol Cell. 2006;17:5265–5274. doi: 10.1091/mbc.E06-08-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serrano AA, Schenkman S, Yoshida N, Mehlert A, Richardson JM, Ferguson MA. The Lipid Structure of the Glycosylphosphatidylinositol-Anchored Mucin-Like Sialic Acid Acceptors of Trypanosoma cruzi Changes During Parasite Differentiation from Epimastigotes to Infective Metacyclic Trypomastigote Forms. J Biol Chem. 1995;270:27244–27253. doi: 10.1074/jbc.270.45.27244. [DOI] [PubMed] [Google Scholar]
- 8.Denny PW, Field MC, Smith DF. GPI-Anchored Proteins and Glycoconjugates Segregate into Lipid Rafts in Kinetoplastida. FEBS Lett. 2001;491:148–153. doi: 10.1016/s0014-5793(01)02172-x. [DOI] [PubMed] [Google Scholar]
- 9.Lester RL, Dickson RC. Sphingolipids with Inositolphosphate-Containing Head Groups. Adv Lipid Res. 1993;26:253–274. [PubMed] [Google Scholar]
- 10.Becker GW, Lester RL. Biosynthesis of Phosphoinositol-Containing Sphingolipids from Phosphatidylinositol by a Membrane Preparation from Saccharomyces cerevisiae. J Bacteriol. 1980;142:747–754. doi: 10.1128/jb.142.3.747-754.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson RC, Lester RL. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426:347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- 12.Figueiredo JM, Dias WB, Mendonca-Previato L, Previato JO, Heise N. Characterization of the Inositol Phosphorylceramide Synthase Activity from Trypanosoma cruzi. Biochem J. 2005;387:519–529. doi: 10.1042/BJ20041842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells GB, Dickson RC, Lester RL. Isolation and Composition of Inositolphosphorylceramide-Type Sphingolipids of Hyphal Forms of Candida albicans. J Bacteriol. 1996;178:6223–6226. doi: 10.1128/jb.178.21.6223-6226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid Synthesis as a Target for Antifungal Drugs. Complementation of the Inositol Phosphorylceramide Synthase Defect in a Mutant Strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 15.Mandala SM, Thornton RA, Rosenbach M, Milligan J, Garcia-Calvo M, Bull HG, Kurtz MB. Khafrefungin, a Novel Inhibitor of Sphingolipid Synthesis. J Bio Chem. 1997;272:32709–32714. doi: 10.1074/jbc.272.51.32709. [DOI] [PubMed] [Google Scholar]
- 16.Zhong W, Jeffries MW, Georgopapadakou NH. Inhibition of Inositol Phosphorylceramide Synthase by Aureobasidin A in Candida and Aspergillus Species. Antimicrob Agents Chemother. 2000;44:651–653. doi: 10.1128/aac.44.3.651-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mare L, Iatta R, Montagna MT, Luberto C, Del Poeta M. APP1 Transcription is Regulated by Inositol-Phosphorylceramide Synthase 1-Diacylglycerol Pathway and is Controlled by ATF2 Transcription Factor in Cryptococcus neoformans. J Biol Chem. 2005;280:36055–36064. doi: 10.1074/jbc.M507285200. [DOI] [PubMed] [Google Scholar]
- 18.Heung LJ, Luberto C, Plowden A, Hannun YA, Del Poeta M. The Sphingolipid Pathway Regulates Pkc1 Through the Formation of Diacylglycerol in Cryptococcus neoformans. J Biol Chem. 2004;279:21144–21153. doi: 10.1074/jbc.M312995200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Showalter M, Revollo J, Hsu FF, Turk J, Beverley SM. Sphingolipids are Essential for Differentiation but not Growth in Leishmania. EMBO J. 2003;22:6016–6026. doi: 10.1093/emboj/cdg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K, Hsu FF, Scott DA, Docampo R, Turk J, Beverley SM. Leishmania Salvage and Remodeling of Host Sphingolipids in Amastigote Survival and Acidocalcisome Biogenesis. Mol Microbiol. 2005;55:1566–1578. doi: 10.1111/j.1365-2958.2005.04493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny PW, Goulding D, Ferguson MA, Smith DF. Sphingolipid-Free Leishmania are Defective in Membrane Trafficking, Differentiation, and Infectivity. Mol Microbiol. 2004;A52:313–327. doi: 10.1111/j.1365-2958.2003.03975.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang K, Pompey JM, Hsu FF, Key P, Bandhuvula P, Saba JD, Turk J, Beverley SM. Redirection of Sphingolipid Metabolism Toward de Novo Synthesis of Ethanolamine in Leishmania. EMBO J. 2007;26:1094–1104. doi: 10.1038/sj.emboj.7601565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh BN, Costello CE, Beach DH, Holz GG., Jr Di-O Alkylglycerol, mono-O-Alkylglycerol and Ceramide Inositol Phosphates of Leishmania Mexicana Mexicana promastigotes. Biochem Biophys Res Commun. 1988;157:1239–1246. doi: 10.1016/s0006-291x(88)81007-6. [DOI] [PubMed] [Google Scholar]
- 24.Levery SB, Toledo MS, Straus AH, Takahashi HK. Comparative Analysis of Glycosylinositol Phosphorylceramides from Fungi by Electrospray Tandem Mass Spectrometry with Low-Energy Collision-Induced Dissociation of Li(+) Adduct Ions. Rapid Commun Mass Spectrom. 2001;15:2240–2258. doi: 10.1002/rcm.505. [DOI] [PubMed] [Google Scholar]
- 25.Toledo MS, Levery SB, Glushka J, Straus AH, Takahashi HK. Structure Elucidation of Sphingolipids from the Mycopathogen Sporothrix schenckii: Identification of Novel Glycosylinositol Phosphorylceramides with Core Manalpha1–>6I ns Linkage. Biochem Biophys Res Commun. 2001;280:19–24. doi: 10.1006/bbrc.2000.4091. [DOI] [PubMed] [Google Scholar]
- 26.Bennion B, Park C, Fuller M, Lindsey R, Momany M, Jennemann R, Levery SB. Glycosphingolipids of the Model Fungus Aspergillus nidulans: Characterization of GIPCs with Oligo-α-Mannose-Type Glycans. J Lipid Res. 2003;44:2073–2088. doi: 10.1194/jlr.M300184-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Hsu FF, Turk J. Structural Determination of Sphingomyelin by Tandem Mass Spectrometry with Electrospray Ionization. J Am Soc Mass Spectrom. 2000;11:437–449. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- 28.Hsu FF, Turk J, Stewart ME, Downing DT. Structural Studies on Ceramides as Lithiated Adducts by Low Energy Collisional-Activated Dissociation Tandem Mass Spectrometry with Electrospray Ionization. J Am Soc Mass Spectrom. 2002;13:680–695. doi: 10.1016/S1044-0305(02)00362-8. [DOI] [PubMed] [Google Scholar]
- 29.Hsu FF, Turk J. Tandem Mass Spectrometry with Electrospray Ionization of Sphingomyelins. In: Caprioli R, Gross ML, editors. The Encyclopedia of Mass Spectrometry, Vol III. Applications in Biochemistry, Biology, and Medicine, Part A. Elsevier Science; New York: 2005. pp. 430–447. [Google Scholar]
- 30.Hsu FF, Turk J. Characterization of Phosphatidylinositol, Phosphatidylinositol-4-Phosphate, and Phosphatidylinositol-4,5-Bisphosphate by Electrospray Ionization Tandem Mass Spectrometry: A Mechanistic Study. J Am Soc Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 31.Hsu FF, Turk J. Characterization of Ceramides by Low Energy Collisional-Activated Dissociation Tandem Mass Spectrometry with Negative-Ion Electrospray Ionization. J Am Soc Mass Spectrom. 2002;13:558–570. doi: 10.1016/S1044-0305(02)00358-6. [DOI] [PubMed] [Google Scholar]
- 32.Hsu FF, Turk J. Structural Studies on Phosphatidylserine by Tandem Quadrupole and Multiple Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. J Am Soc Mass Spectrom. 2005;16:1510–1522. doi: 10.1016/j.jasms.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Zemski Berry KA, Murphy RC. Electrospray Ionization Tandem Mass Spectrometry of Glycerophosphoethanolamine Plasmalogen Phospholipids. J Am Soc Mass Spectrom. 2004;15:1499–1508. doi: 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Doering TL, Pessin MS, Hoff EF, Hart GW, Raben DM, Englund PT. Trypanosome Metabolism of Myristate, the Fatty Acid Required for the Variant Surface Glycoprotein Membrane Anchor. J Biol Chem. 1993;268:9215–9222. [PubMed] [Google Scholar]


