Abstract
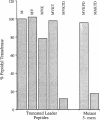
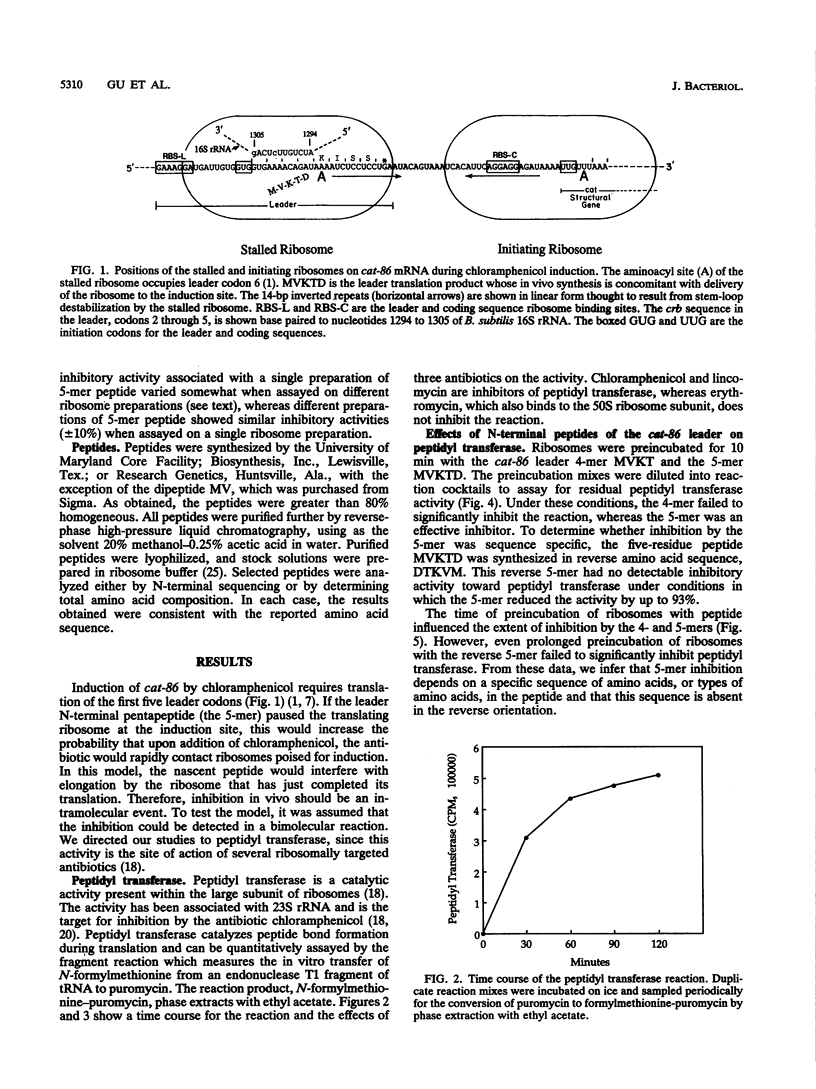
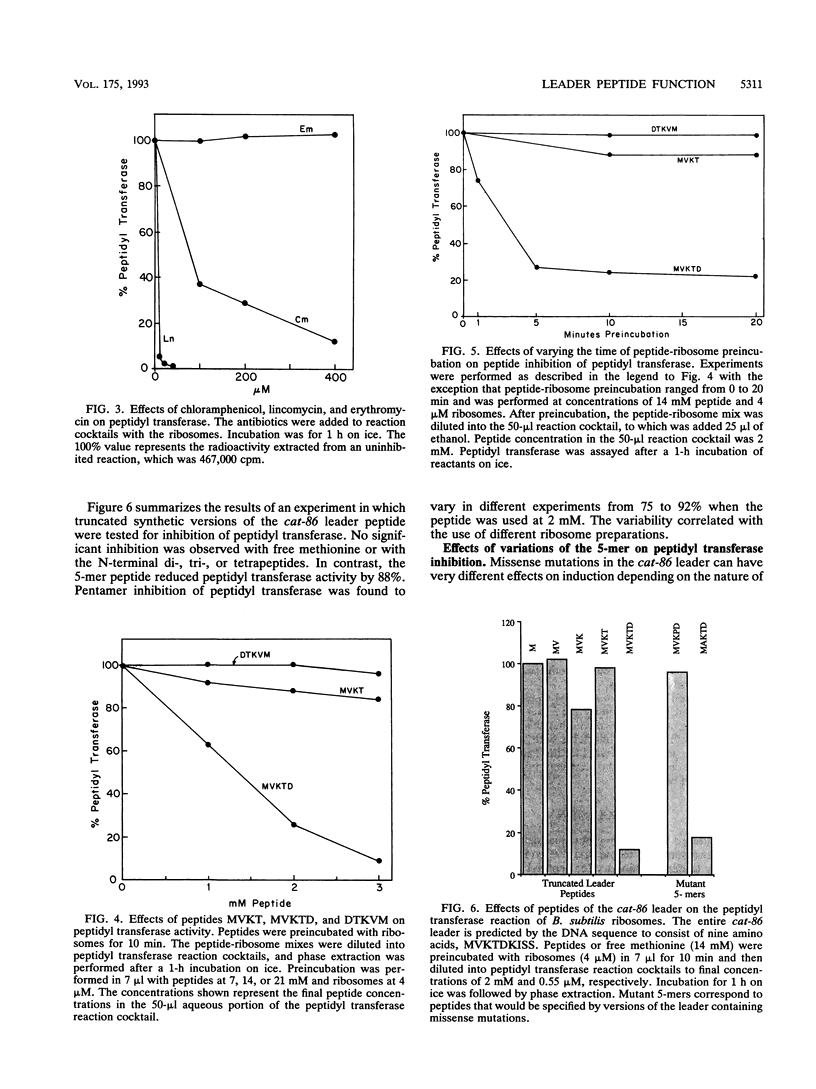
The site of ribosome stalling in the leader of cat transcripts is critical to induction of downstream translation. Site-specific stalling requires translation of the first five leader codons and the presence of chloramphenicol, a sequence-independent inhibitor of ribosome elongation. We demonstrate in this report that a synthetic peptide (the 5-mer) corresponding to the N-terminal five codons of the cat-86 leader inhibits peptidyl transferase in vitro. The N-terminal 2-, 3-, and 4-mers and the reverse 5-mer (reverse amino acid sequence of the 5-mer) are virtually without effect on peptidyl transferase. A missense mutation in the cat-86 leader that abolishes induction in vivo corresponds to an amino acid replacement in the 5-mer that completely relieves peptidyl transferase inhibition. In contrast, a missense mutation that does not interfere with in vivo induction corresponds to an amino acid replacement in the 5-mer that does not significantly alter peptidyl transferase inhibition. Our results suggest that peptidyl transferase inhibition by the nascent cat-86 5-mer peptide may be the primary determinant of the site of ribosome stalling in the leader. A model based on this concept can explain the site specificity of ribosome stalling as well as the response of induction to very low levels of the antibiotic inducer.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexieva Z., Duvall E. J., Ambulos N. P., Jr, Kim U. J., Lovett P. S. Chloramphenicol induction of cat-86 requires ribosome stalling at a specific site in the leader. Proc Natl Acad Sci U S A. 1988 May;85(9):3057–3061. doi: 10.1073/pnas.85.9.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Duvall E. J., Lovett P. S. Analysis of the regulatory sequences needed for induction of the chloramphenicol acetyltransferase gene cat-86 by chloramphenicol and amicetin. J Bacteriol. 1986 Sep;167(3):842–849. doi: 10.1128/jb.167.3.842-849.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner R., Matzura H. Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. EMBO J. 1985 Sep;4(9):2295–2300. doi: 10.1002/j.1460-2075.1985.tb03929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick T., Matzura H. Positioning ribosomes on leader mRNA for translational activation of the message of an inducible Staphylococcus aureus cat gene. Mol Gen Genet. 1988 Sep;214(1):108–111. doi: 10.1007/BF00340187. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Induction of ermC requires translation of the leader peptide. EMBO J. 1985 Feb;4(2):533–537. doi: 10.1002/j.1460-2075.1985.tb03661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Ambulos N. P., Jr, Lovett P. S. Drug-free induction of a chloramphenicol acetyltransferase gene in Bacillus subtilis by stalling ribosomes in a regulatory leader. J Bacteriol. 1987 Sep;169(9):4235–4241. doi: 10.1128/jb.169.9.4235-4241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D. A., Sisodia S. S., Cleveland D. W. Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Lovett P. S. Perturbing highly conserved spatial relationships in the regulatory domain that controls inducible cat translation. Mol Microbiol. 1992 Oct;6(19):2769–2776. doi: 10.1111/j.1365-2958.1992.tb01456.x. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. Translational attenuation as the regulator of inducible cat genes. J Bacteriol. 1990 Jan;172(1):1–6. doi: 10.1128/jb.172.1.1-6.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcker K. The formation of N-formyl-methionyl-sRNA. J Mol Biol. 1965 Nov;14(1):63–70. doi: 10.1016/s0022-2836(65)80230-3. [DOI] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989 Mar 5;206(1):69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- Monro R. E., Marcker K. A. Ribosome-catalysed reaction of puromycin with a formylmethionine-containing oligonucleotide. J Mol Biol. 1967 Apr 28;25(2):347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- Monro R. E., Vazquez D. Ribosome-catalysed peptidyl transfer: effects of some inhibitors of protein synthesis. J Mol Biol. 1967 Aug 28;28(1):161–165. doi: 10.1016/s0022-2836(67)80085-8. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992 Jun 5;256(5062):1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Rogers E. J., Ambulos N. P., Jr, Gu Z., Lovett P. S. Parallel induction strategies for cat-86: separating chloramphenicol induction from protein synthesis inhibition. Mol Microbiol. 1993 Jun;8(6):1063–1069. doi: 10.1111/j.1365-2958.1993.tb01651.x. [DOI] [PubMed] [Google Scholar]
- Rogers E. J., Ambulos N. P., Jr, Lovett P. S. Complementarity of Bacillus subtilis 16S rRNA with sites of antibiotic-dependent ribosome stalling in cat and erm leaders. J Bacteriol. 1990 Nov;172(11):6282–6290. doi: 10.1128/jb.172.11.6282-6290.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E. J., Kim U. J., Ambulos N. P., Jr, Lovett P. S. Four codons in the cat-86 leader define a chloramphenicol-sensitive ribosome stall sequence. J Bacteriol. 1990 Jan;172(1):110–115. doi: 10.1128/jb.172.1.110-115.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Huang W. M., Dunn D. M. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990 Jul 13;62(1):117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]