Abstract
Transcription during the bacteriophage Mu lytic cycle occurs in three phases: early, middle, and late. Middle transcription requires the early gene product Mor for its activation. Mor protein overproduction was accomplished by fusing the mor gene to an efficient phage T7 promoter and translation initiation region. A protein fraction highly enriched for Escherichia coli RNA polymerase (E sigma 70) from the Mor-overproducing strain was able to activate transcription from both the tac promoter (Ptac) and the Mu middle promoter (Pm) in vitro. Transcription initiation from Pm was Mor dependent, and the RNA 5' end was identical to that of in vivo RNA. Addition of anti-sigma 70 antibody to transcription reactions containing Ptac and Pm resulted in inhibition of transcription from both promoters; addition of purified sigma 70 restored transcription. These results indicate that Mor-dependent activation requires sigma 70 and therefore imply that Mor is not an alternate sigma factor. This conclusion was further substantiated by a reconstitution experiment with purified proteins in which all three components, Mor, sigma 70, and core RNA polymerase, were required for Pm-dependent transcription in vitro. The sigma 70 dependence of Mor-specific transcription and the amino acid sequence similarity between Mor and C (an activator for Mu late transcription) both support the hypothesis that Mor functions mechanistically as an activator protein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barron C., Bade E. G. Transcriptional mapping of the bacteriophage Mu DNA. J Gen Virol. 1988 Feb;69(Pt 2):385–393. doi: 10.1099/0022-1317-69-2-385. [DOI] [PubMed] [Google Scholar]
- Bölker M., Wulczyn F. G., Kahmann R. Role of bacteriophage Mu C protein in activation of the mom gene promoter. J Bacteriol. 1989 Apr;171(4):2019–2027. doi: 10.1128/jb.171.4.2019-2027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987 Jun 11;15(11):4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBow M. S., Bukhari A. I. The proteins of bacteriophage mu: composition of the virion and biosynthesis in vivo during lytic growth. Prog Clin Biol Res. 1981;64:47–67. [PubMed] [Google Scholar]
- Giusti M., Di Lallo G., Ghelardini P., Paolozzi L. The bacteriophage Mu gem gene: a positive regulator of the C operon required for normal levels of late transcription. Virology. 1990 Dec;179(2):694–700. doi: 10.1016/0042-6822(90)90136-f. [DOI] [PubMed] [Google Scholar]
- Gross C., Engbaek F., Flammang T., Burgess R. Rapid micromethod for the purification of Escherichia coli ribonucleic acid polymerase and the preparation of bacterial extracts active in ribonucleic acid synthesis. J Bacteriol. 1976 Oct;128(1):382–389. doi: 10.1128/jb.128.1.382-389.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Heisig P., Kahmann R. The sequence and mom-transactivation function of the C gene of bacteriophage Mu. Gene. 1986;43(1-2):59–67. doi: 10.1016/0378-1119(86)90008-9. [DOI] [PubMed] [Google Scholar]
- Ho Y. S., Wulff D. L., Rosenberg M. Bacteriophage lambda protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature. 1983 Aug 25;304(5928):703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Jaurin B., Cohen S. N. Streptomyces contain Escherichia coli-type A + T-rich promoters having novel structural features. Gene. 1985;39(2-3):191–201. doi: 10.1016/0378-1119(85)90313-0. [DOI] [PubMed] [Google Scholar]
- Kaufman M. R., Seyer J. M., Taylor R. K. Processing of TCP pilin by TcpJ typifies a common step intrinsic to a newly recognized pathway of extracellular protein secretion by gram-negative bacteria. Genes Dev. 1991 Oct;5(10):1834–1846. doi: 10.1101/gad.5.10.1834. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Krause H. M., Rothwell M. R., Higgins N. P. The early promoter of bacteriophage Mu: definition of the site of transcript initiation. Nucleic Acids Res. 1983 Aug 25;11(16):5483–5495. doi: 10.1093/nar/11.16.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Margolin W., Howe M. M. Activation of the bacteriophage Mu lys promoter by Mu C protein requires the sigma 70 subunit of Escherichia coli RNA polymerase. J Bacteriol. 1990 Mar;172(3):1424–1429. doi: 10.1128/jb.172.3.1424-1429.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W., Howe M. M. Localization and DNA sequence analysis of the C gene of bacteriophage Mu, the positive regulator of Mu late transcription. Nucleic Acids Res. 1986 Jun 25;14(12):4881–4897. doi: 10.1093/nar/14.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W., Rao G., Howe M. M. Bacteriophage Mu late promoters: four late transcripts initiate near a conserved sequence. J Bacteriol. 1989 Apr;171(4):2003–2018. doi: 10.1128/jb.171.4.2003-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs C. F., Howe M. M. Kinetics and regulation of transcription of bacteriophage Mu. Virology. 1990 Jan;174(1):192–203. doi: 10.1016/0042-6822(90)90068-3. [DOI] [PubMed] [Google Scholar]
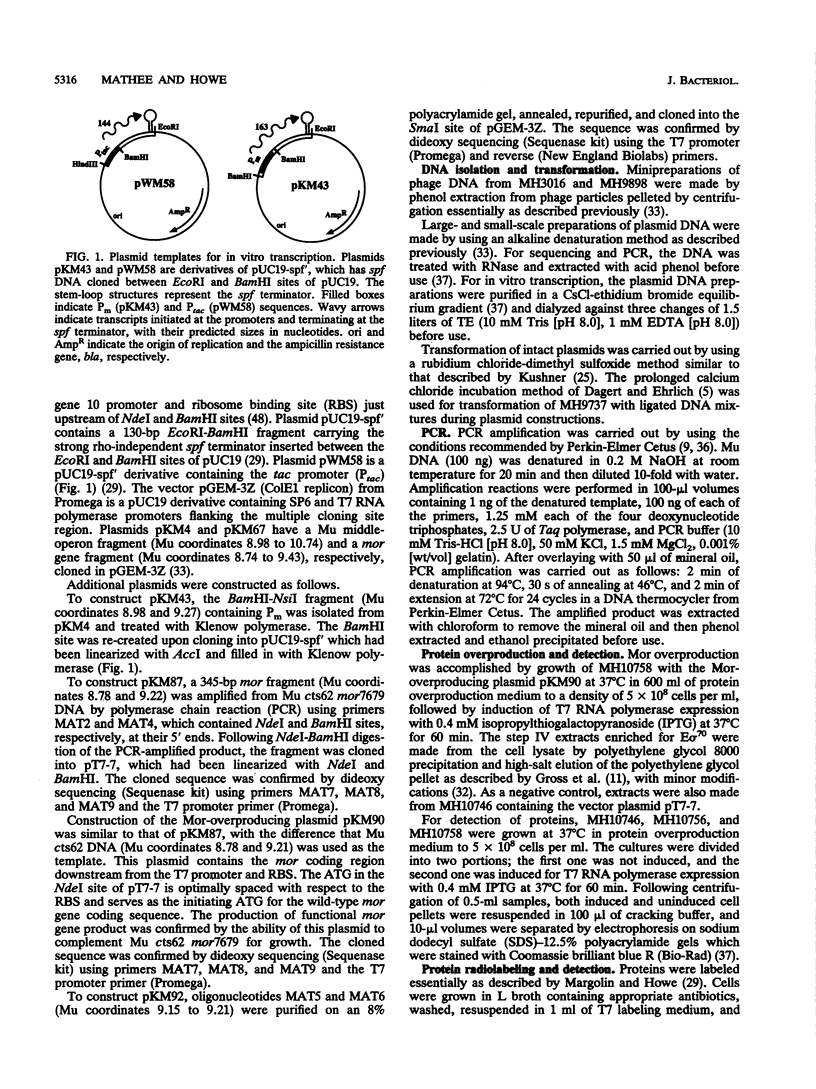
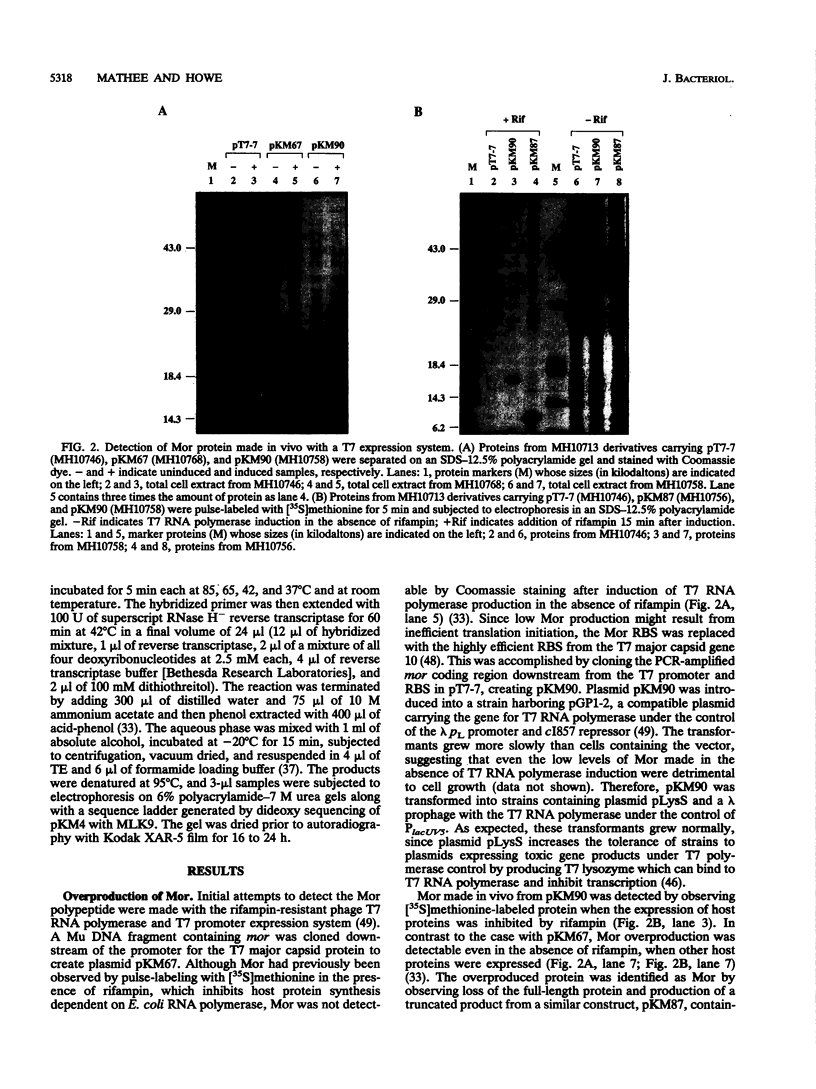
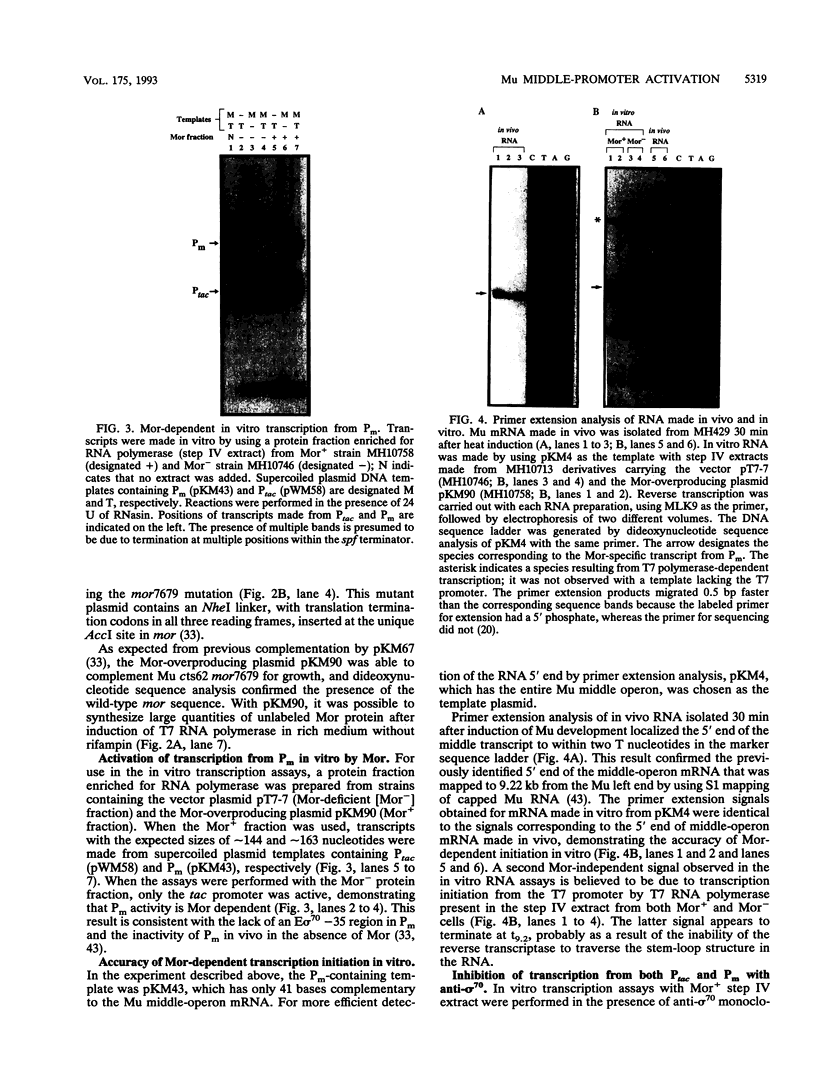
- Mathee K., Howe M. M. Identification of a positive regulator of the Mu middle operon. J Bacteriol. 1990 Dec;172(12):6641–6650. doi: 10.1128/jb.172.12.6641-6650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess H., Kamp D., Kahmann R., Bräuer B., Delius H. Nucleotide sequence of the immunity region of bacteriophage Mu. Mol Gen Genet. 1982;186(3):315–321. doi: 10.1007/BF00729448. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumm J. W., Moore D. D., Blattner F. R., Howe M. M. Correlation of the genetic and physical maps in the central region of the bacteriophage Mu genome. Virology. 1980 Aug;105(1):185–195. doi: 10.1016/0042-6822(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Shore S. H., Howe M. M. Bacteriophage Mu T mutants are defective in synthesis of the major head polypeptide. Virology. 1982 Jul 15;120(1):264–268. doi: 10.1016/0042-6822(82)90026-5. [DOI] [PubMed] [Google Scholar]
- Stoddard S. F., Howe M. M. Characterization of the C operon transcript of bacteriophage Mu. J Bacteriol. 1990 Jan;172(1):361–371. doi: 10.1128/jb.172.1.361-371.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard S. F., Howe M. M. DNA sequence within the Mu C operon. Nucleic Acids Res. 1987 Sep 11;15(17):7198–7198. doi: 10.1093/nar/15.17.7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard S. F., Howe M. M. Localization and regulation of bacteriophage Mu promoters. J Bacteriol. 1989 Jun;171(6):3440–3448. doi: 10.1128/jb.171.6.3440-3448.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland M. S., Thompson N. E., Burgess R. R. Structure and function of the sigma-70 subunit of Escherichia coli RNA polymerase. Monoclonal antibodies: localization of epitopes by peptide mapping and effects on transcription. Biochemistry. 1988 Jul 26;27(15):5755–5762. doi: 10.1021/bi00415a054. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., van de Putte P. Transcription of bacteriophage mu. An analysis of the transcription pattern in the early phase of phage development. Mol Gen Genet. 1974;135(4):327–337. doi: 10.1007/BF00271147. [DOI] [PubMed] [Google Scholar]
- van Meeteren R., Giphart-Gassler M., van de Putte P. Transcription of bacteriophage Mu. II. Transcription of the repressor gene. Mol Gen Genet. 1980;179(1):185–189. doi: 10.1007/BF00268462. [DOI] [PubMed] [Google Scholar]








