SUMMARY
The Endosomal Sorting Complex Required for Transport-I (ESCRT-I) complex, which is conserved from yeast to humans, directs the lysosomal degradation of ubiquitinated transmembrane proteins and the budding of the HIV virus. Yeast ESCRT-I contains four subunits, Vps23, Vps28, Vps37 and Mvb12. The crystal structure of the heterotetrameric ESCRT-I complex reveals a highly asymmetric complex of 1:1:1:1 subunit stoichiometry. The core complex is nearly 18 nm long, and consists of a headpiece attached to a 13 nm stalk. The stalk is important for cargo sorting by ESCRT-I, and is proposed to serve as a spacer regulating the correct disposition of cargo and other ESCRT components. Hydrodynamic constraints and crystallographic structures were used to generate a model of intact ESCRT-I in solution. The results show how ESCRT-I uses a combination of a rigid stalk and flexible tethers to interact with lipids, cargo, and other ESCRT complexes over a span of ~25 nm.
INTRODUCTION
The Endosomal Sorting Complexes Required for Transport (ESCRTs) direct transmembrane proteins into inwardly-budding vesicles at the multivesicular body (MVB) (Babst, 2005; Conibear, 2002; Hurley and Emr, 2006; Slagsvold et al., 2006). Sorting into the MVB pathway is a key step in the regulated degradation of transmembrane proteins in the lysosome or yeast vacuole. The ESCRT network is required for the downregulation of many cell surface receptors, for the delivery of resident hydrolases during lysosome and vacuole biogenesis, and for the production of exosomes from human cells (Katzmann et al., 2002). ESCRT complexes participate in the budding of HIV-1 and a number of other viruses (Demirov and Freed, 2004; Morita and Sundquist, 2004).
The ESCRT proteins were discovered as the products of class E vacuolar protein sorting (VPS) genes in the budding yeast Saccharomyces cerevisiae (Bowers and Stevens, 2005). Class E vps mutants have a characteristic enlarged cargo-rich compartment adjacent to the vacuole. Yeast class E VPS genes encode the subunits of at least four hetero-oligomeric protein complexes: the Vps27/Hse1 complex and ESCRT-I, II, and III (Bowers and Stevens, 2005; Hurley and Emr, 2006). Whereas monomeric ESCRT-III components are believed to assemble at the endosomal membrane, the other three complexes are assembled in the cytosol and cycle on and off membranes. Each of these soluble complexes contains specific ubiquitin binding domains that recognize ubiquitinated cargo proteins (Bilodeau et al., 2002; Hicke et al., 2005; Hurley et al., 2006; Katzmann et al., 2001; Shih et al., 2002; Slagsvold et al., 2005). Vps27/Hse1 (Katzmann et al., 2003) and ESCRT-II (Slagsvold et al., 2005; Teo et al., 2006) contain lipid binding domains that target these complexes to endosomal membranes. P(S/T)XP sequences in Vps27 bind to the UEV domain of the ESCRT-I subunit Vps23 (Bilodeau et al., 2003; Katzmann et al., 2003), whereas the C-terminal domain of the ESCRT-I subunit Vps28 binds to ESCRT-II (Kostelansky et al., 2006; Teo et al., 2006). Collectively, these interactions direct the assembly of the ESCRT network on endosomal membranes.
ESCRT-I contains the subunits Vps23, Vps28, and Vps37, and plays a central role in the MVB pathway (Katzmann et al., 2001). Human Vps23 (Tsg101) is one of a handful of ESCRT proteins whose knockdown by RNA interference almost completely blocks HIV budding (Garrus et al., 2001). The structure and function of ESCRT-I has come under intense scrutiny, and the structure of many of its components has been determined. The N-terminal UEV domains of yeast and human Vps23 have been determined in complex with ubiquitin (Sundquist et al., 2004; Teo et al., 2004b), and for the human case, with a PTAP motif peptide from HIV-1 (Pornillos et al., 2002). The C-terminal domain of Vps28, responsible for binding to ESCRT-II, is a four-helix bundle (Pineda-Molina et al., 2006).
The structure of a globular core complex comprising the C-terminal steadiness box of Vps23, the N-terminal half of Vps28, and the C-terminal half of Vps37 showed how the three subunits assemble together (Kostelansky et al., 2006; Teo et al., 2006). However, the structures and functions of the central region of Vps23 and the N-terminal half of Vps37, both conserved from yeast to man, have yet to be characterized. Furthermore, ESCRT-I isolated from yeast has an apparent molecular weight of 350 kDa as judged by gel filtration (Katzmann et al., 2001), yet near-full length recombinant ESCRT-I purified from E. coli has an apparent molecular weight of only ~200 kDa (Kostelansky et al., 2006; Teo et al., 2006). This discrepancy led to the question as to whether ESCRT-I in yeast oligomerizes or contains additional subunits.
Here we identify a fourth ESCRT-I subunit, Mvb12, and characterize the structure and function of the quaternary Vps23:Vps28:Vps37:Mvb12 ESCRT-I complex. The complete recombinant ESCRT-I complex migrates on gel filtration in a manner identical to native yeast ESCRT-I and has a 1:1:1:1 subunit stoichiometry. We determined the crystal structure of the core of the complete heterotetrameric complex, which includes a novel 130 Å-long stalk. The integrity of the stalk is essential for sorting MVB cargo. Hydrodynamic analysis of full-length and multiple truncation constructs was used in combination with crystal structures to determine the conformation of intact ESCRT-I in solution. The physical interactions of ESCRT-I with membranes were inventoried and used to model the structure of ESCRT-I bound to an endosomal membrane.
RESULTS
Mvb12 is the Fourth Subunit of ESCRT-I
Genome-wide phenotypic profiling was used to identify mutants defective in the sorting of carboxypeptidase Y (CPY) (Conibear, unpublished data). Known ESCRT-associated factors, including the VPS4-regulatory genes VTA1 and DID2, and the Vps27/Hse1 component HSEI, were found in a cluster enriched for genes that encode endosome-localized proteins, including the ORF YGR206w (Conibear, unpublished data), which was also identified in another screen for CPY sorting mutants (Bonangelino et al., 2002). Co-clustering with ESCRT regulatory factors suggests this endosome-associated ORF, which is now known as Mvb12, could play a role in sorting at the MVB. mvb12Δ mutants have a weak CPY missorting phenotype compared to that of vps4Δ strains (Fig. 1A). In wild type cells, Mvb12-GFP was diffusely localized, with faint punctate staining (Fig. 1B); however, in vps4Δ mutants, Mvb12-GFP was found in brighter puncta that co-localized with FM4-64, a characteristic of ESCRT components. mvb12Δ mutants also exhibited defects in MVB cargo sorting similar to those of ESCRT mutants (Fig. 1C). The cargo proteins Ste3 and Sna3 largely failed to reach the vacuole lumen in mvb12Δ mutants and instead accumulated in aberrant MVB structures and at the vacuole limiting membrane. Each mvb12Δ mutant phenotype was fully complemented by a MVB12-encoding plasmid. The strong Ste3 and Sna3 mislocalization phenotypes suggest that Mvb12 is important for sorting MVB cargo.
Figure 1. Identification of the Novel ESCRT-I Subunit Mvb12.
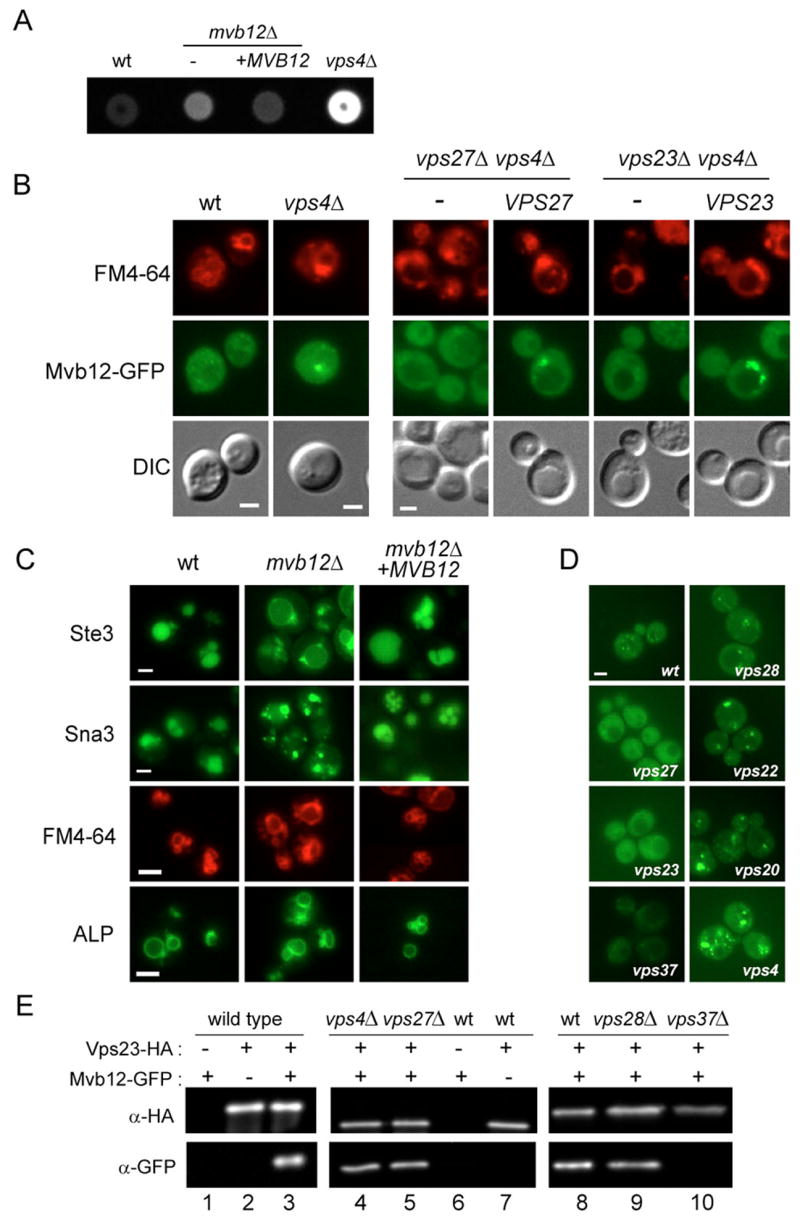
(A) mvb12 mutants exhibit a weak CPY sorting defect. CPY missorting from wild type, vps4Δ or mvb12Δ strains with and without a MVB12-expressing plasmid (+pMVB12) was detected by colony overlay assay (Conibear and Stevens, 2002). (B) Mvb12 is recruited to the endosome in vps4Δ mutants. vps27Δ vps4Δ and vps23Δ vps4Δ strains containing integrated Mvb12-GFP and plasmid-expressed VPS27 and VPS23, or empty vector, were incubated with FM4-64 and viewed by double-label fluorescence microscopy. (C) Fluorescence microscopy of wild type and mvb12Δ strains containing Ste3-GFP, Sna3-GFP, or ALP-GFP plasmids together with a complementing MVB12 plasmid or empty vector. (D) Mvb12 recruitment does not require ESCRT-II or -III. The chromosomal copy of MVB12 was tagged with GFP in wild type, vps27Δ, vps23Δ, vps37Δ, vps28Δ, vps22Δ and vps20Δ strains and visualized in live cells. (E) Mvb12 is associated with ESCRT-I. Detergent extracts prepared from 20 OD600 units (lanes 1–7) or 100 OD600 (lanes 8–10) of wild type or mutant strains expressing GFP-tagged Mvb12 and/or HA-tagged Vps23 were immunoprecipitated with anti-HA antiserum and analyzed by western blotting with anti-GFP and anti-HA mAbs. Loading of lanes 9–10 was 7.5X greater than lane 8 to compare relative levels of co-purifying Mvb12-GFP despite differences in Vps23 stability. Bar = 2 μM.
Mvb12 is a Component of the ESCRT-I Complex
Because ESCRT complexes are recruited to the MVB in a stepwise fashion, the localization of Mvb12 in representative mutants provides a way to evaluate its association with a particular ESCRT complex. Mvb12 was found at the enlarged MVB in ESCRT-II (vps22Δ and vps36Δ) and ESCRT-III (vps20Δ) mutants, indicating that recruitment of Mvb12 is independent of these ESCRT complexes (Fig. 1D and Fig. S1A). Mvb12 was diffusely localized in Vps27/Hse1 (vps27Δ) mutants and in mutants lacking two different components of ESCRT-I (vps23Δ and vps3Δ7). The recruitment of Mvb12-GFP to FM4-64-labeled endosomes was completely blocked in either vps27Δ vps4Δ or vps23Δ vps4Δ double mutants, and restored upon introduction of a complementing plasmid-borne copy of VPS27 or VPS23, respectively (Fig. 1B).
The physical interaction between Mvb12 and the ESCRT-I subunit Vps23 was tested by co-immunoprecipitation of tagged proteins from cell lysates. Mvb12 and Vps23 co-purified efficiently, with immunoprecipitation of 80% of the cellular Vps23 resulting in the co-precipitation of a nearly equal proportion (70%) of cellular Mvb12 (Fig. 1E lane 3). These findings are consistent with the physical association of Mvb12 with ESCRT-I components in proteome-wide screens of yeast protein complexes (Gavin et al., 2006; Krogan et al., 2006). Unexpectedly, loss of the ESCRT-I subunit Vps28 resulted in an enhanced MVB recruitment of Mvb12, similar to that seen in ESCRT-II and –III mutants (Fig. 1D). The co-purification of Mvb12 and Vps23 required Vps37, but not Vps28 (Fig. 1E lanes 8–10). Mvb12 thus appears to associate with a stable, MVB-localized Vps23/37 subcomplex in vps28Δ mutants. Although ESCRT-I is found both in cytosol and membrane-bound pools, the amount of Mvb12 that co-precipitated with Vps23 was unchanged in vps27Δ mutants, in which ESCRT-I is entirely cytosolic, compared to vps4Δ strains, where it is primarily endosome-localized (Fig. 1E lanes 4–5). This indicates that Mvb12 does not preferentially associate with a specific subcellular fraction of ESCRT-I. Taken together, these data support the model that Mvb12 is an integral component of the ESCRT-I complex.
Structure of the Heterotetrameric ESCRT-I Core Complex
The structure of a heterotetrameric core complex of ESCRT-I was determined at a nominal resolution of 2.7Å (Fig. 2A, Table S1). The heterotetrameric core ESCRT-I structure consists of a globular headpiece attached to an extended stalk (Fig. 2B). The overall structure is 176 Å long. The headpiece is a flattened blade of 25 × 55 × 60 Å that corresponds to the ESCRT-I ternary core complex (Fig. 2C–E) (Kostelansky et al., 2006; Teo et al., 2006). The stalk is a cylinder of 20Å diameter bent gently into a 130 Å long S-shaped curve. The headpiece includes portions of all four subunits, while the stalk contains regions of Vps23, Vps37, and Mvb12, but not Vps28 (Fig. 2F–H). Consistent with the finding that Mvb12 forms a subcomplex in vivo with Vps23 and Vps37, but not Vps28, the Mvb12:Vps28 interface buries a negligible amount of solvent accessible surface area, 68 Å2 (Fig. 4). By contrast, Mvb12 buries 1470 and 2318 Å2, respectively in its interfaces with Vps23 and Vps37. In total, Mvb12 buries a remarkable 42 % of its solvent accessible surface area in the complex, leaving no doubt that it is an integral core component of ESCRT-I.
Figure 2. Structure of the ESCRT-I Heterotetramer Core.
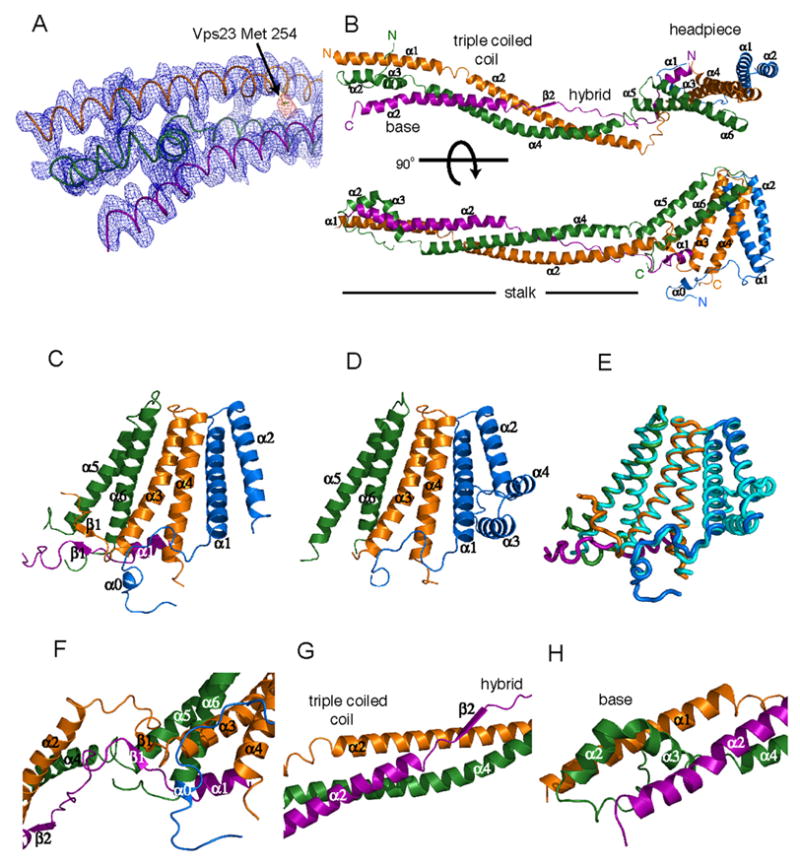
(A) Electron density from the density-modified experimental map (blue) contoured at 1.0 σ and from a Se anomalous difference Fourier contoured at 4.0 σ (yellow) overlaid on the refined structure in the region of the helix bundle at the distal end of the stalk. (B) Structure of ESCRT-I, Vps23, orange, Vps28, blue, Vps37, green, Mvb12, purple. (C) Structure of the headpiece in the heterotetramer, shown with (D) the previously determined trimeric core structure in the same orientation. The two structures are overlaid in (E) with the heterotetramer headpiece colored as in (C) and the trimer colored cyan. (F) The headpiece contains a small β-sheet near its junction with the stalk.
(G) The triple coiled coil is continued and stabilized by an unusual hybrid between a two-stranded coiled-coil and an extended region of Mvb12. (H) The helix bundle at the base of the stalk. Structural figures were generated with Pymol (www.pymol.org).
Figure 4. Stoichiometry and Solution Structure of the Complete ESCRT-I Complex.
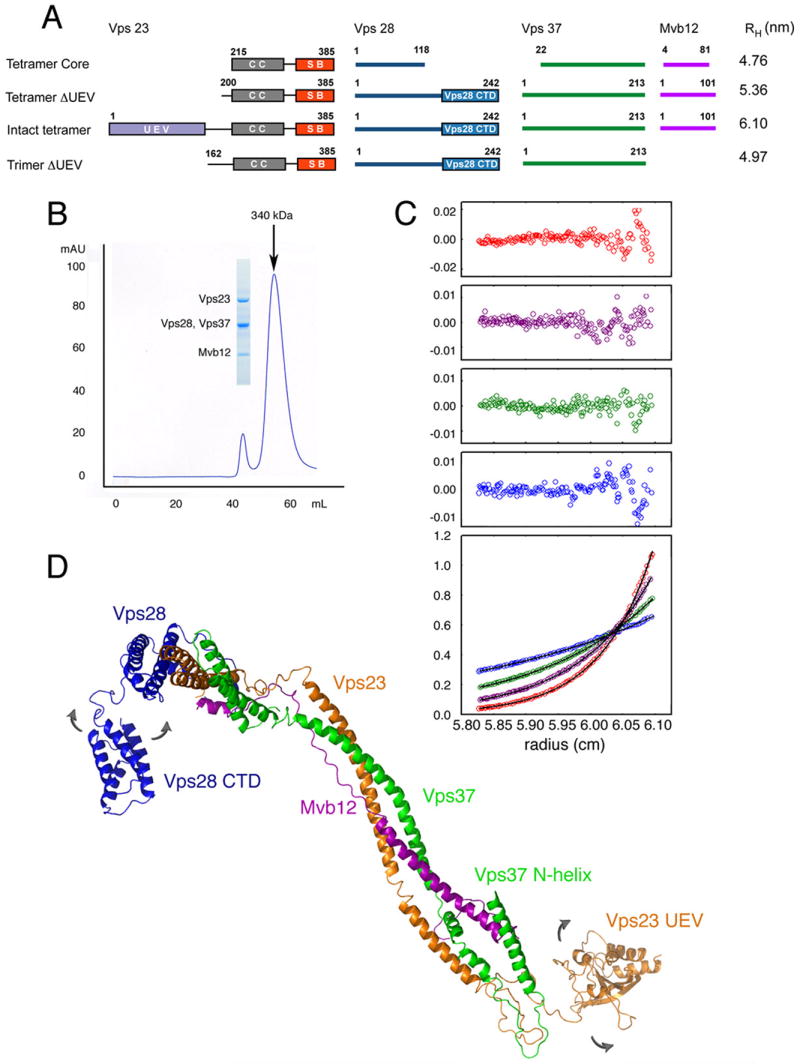
(A) Domain structure of full-length ESCRT-I and other constructs used in this study, and their Stokes radii RH as determined by analytical ultracentrifugation. (B) Gel filtration of full-length ESCRT-I, monitored at 280 nm. The inset shows a Coomassie blue-stained SDS-PAGE gel of the peak fraction indicated. (C) Sedimentation equilibrium profiles of full-length ESCRT-I plotted as a distribution of A280 vs. r at equilibrium. Data were collected at 6 (blue), 8 (green), 10 (purple) and 12 (red) krpm at a loading A280 of 0.38 (alternate data points are shown). The solid lines show the best-fit global analysis in terms of a single ideal solute, with the corresponding residuals shown in the panels above the plot. (D) Solution structural model of intact ESCRT-I. The curved arrows indicate that the Vps23 UEV and Vps28 C-terminal domain (CTD) are conformationally dynamic.
The headpiece is built around three antiparallel two-helix hairpins (Fig. 2C–E), and superimposes well with the ternary core (r.m.s.d. of 0.8 Å for 193 Cα positions). Residues of Vps28 that correspond to α3 in the ternary core are disordered in this structure, while Vps28 helix α4 is in a completely different position than in the ternary core. Mvb12 contributes its N-terminal α1 helix to the headpiece. The headpiece contains a two-stranded antiparallel β-sheet (Fig. 2F), not present in the ternary complex. Mvb12-β1 (residues 14-16) is antiparallel to Vps23 residues 317-319. This miniature β-sheet connects the headpiece to the stalk.
The stalk is centered on four long α-helices, two from Vps23, and one each from Vps37 and Mvb12 (Fig. 2B, S2, S3). The Vps23 α1- and α2-helices are nearly co-linear and run the entire length of the stalk (Fig. 2B). A short 310-helix (residues 247-253) separates these two α-helices. The α4 helix of Vps37 is nearly as long. Mvb12 contributes its α2 helix to the stalk (Fig. 3A). These three long helices come together to form a triple coiled coil over part of their lengths (Fig. 2G). Vps23 residues 254-265, Vps37 residues 89-106, and Mvb12 residues 43-61 form a triple coiled coil in which Vps23 and Vps37 are parallel, and Mvb12 antiparallel. The triple coiled coil comprises the center of the stalk. Proximal to the headpiece, Vps23 α2 and Vps37 α4 form an unusual partnering (the “hybrid”) with an extended portion of Mvb12, residues 18-39 (Fig. 3C,D). Here Mvb12 contributes a series of hydrophobic residues to stabilize the groove between Vps23 and Vps37 (Fig. 3D). At the base of the stalk, distal to the headpiece, helices α2 and α3 of Vps37 form a small four helix bundle with the N-terminus of Vps23 α1 and the C-terminus of Mvb12 α2 (Fig. 2H, 3A). The very N-terminal helix α1 of Vps37 is not part of the stalk at all. It is not directly associated with the rest of the protein, and is apparently trapped in an ordered conformation due to lattice contacts. A well-ordered sulfate ion is bound in the stalk region by Vps37 His-71, His-73 and Gln-74, suggestive of a potential phospholipid interaction site. However, the overall electronegative character of the stalk argues against a direct interaction with membranes.
Figure 3. Interactions Within the ESCRT-I Heterotetramer.
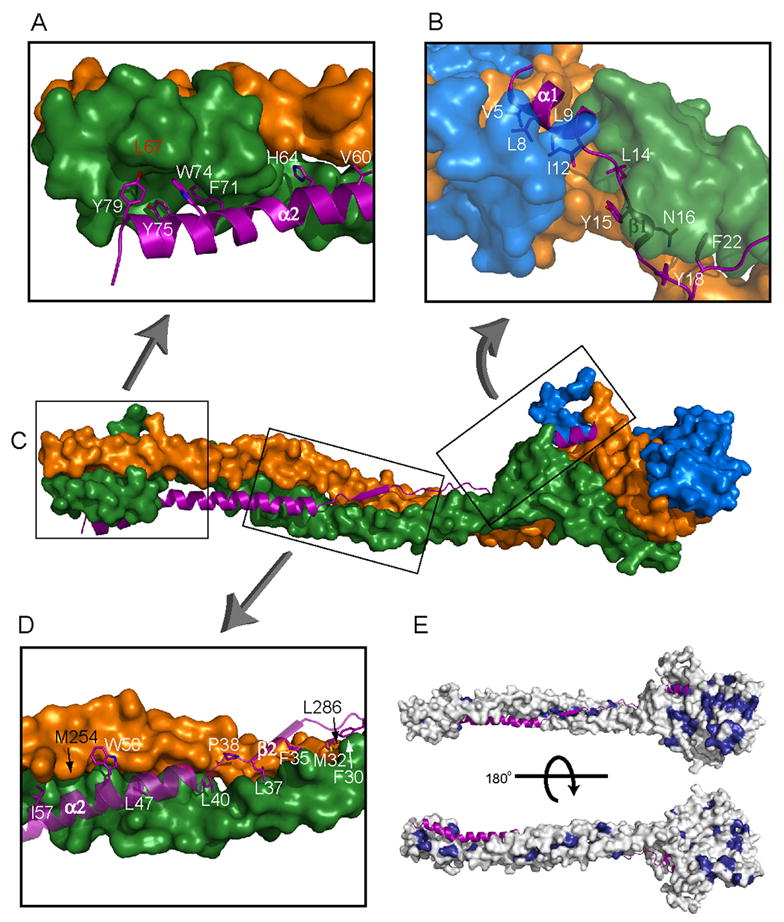
(A, B, D) Close-ups of the interface of Mvb12 (purple ribbon and stick model) with Vps23, Vps28 and Vps37 (orange, blue, and green surfaces, respectively). (C) Shows locations of the regions of the ESCRT-I heterotetramer highlighted in panels A, B and D. Mvb12 residues are labeled in white, while significant Vps23 residues are labeled in black and Vps37 residues in red. (E) Surface depiction of the ESCRT-I heterotetramer showing in dark blue residues highly conserved in orthologs of Vps23, Vps28 and Vps37. Mvb12 is depicted as a ribbon. The conservation of the Mvb12 interaction surface on the rest of the stalk suggests that there is a structural counterpart of Mvb12 in non-fungal species.
Stoichiometry and Solution Conformation of ESCRT-I
The full-length heterotetrameric ESCRT-I complex consisting of intact Vps23, Vps28, Vps37 and Mvb12 runs on gel filtration chromatography with an apparent molecular weight of 340 kDa (Fig. 4B), identical within error to the 350 kDa previously reported for native ESCRT-I isolated from S. cerevisiae (Katzmann et al., 2001). Sedimentation equilibrium centrifugation analysis revealed that full length ESCRT-I sample has an experimental molecular mass of 117 ± 4.6 kDa (Fig. 4C, Table S2), as compared to a calculated molecular mass for a 1:1:1:1 heterotetramer of 108 kDa. The experimental molecular mass is consistent with a “monomeric” complex of n = 1.08 ± 0.04, and is inconsistent with any higher oligomeric structures.
The stoichiometry and solution conformation of intact ESCRT-I was characterized by sedimentation velocity experiments on intact heterotetrameric ESCRT-I and two truncated constructs (Fig. 4A, Table S2). The frictional ratio of intact ESCRT-I, f/fo, is 1.9 ± 0.1. Globular proteins have f/fo values ranging from 1.05 to 1.35, whereas rod-like particles have f/fo values of 3 and larger. The elongated shape of ESCRT-I in solution quantitatively predicts its elution volume on gel filtration chromatography (Fig. S2). The solution conformations of the Vps23 UEV domain, the Vps28 C-terminal domain, and the Vps37 N-terminal helix were assigned on the basis of iterative modeling of the three experimental values for the Stokes radii RH (Fig. 4A). The best fit for the double deletion construct was obtained by modeling the N-terminus of Vps37 in a partially extended conformation (Fig. 4D). The experimental Stokes radii rule out a “closed” conformation in which the UEV or Vps28 C-terminal domain contacts the core in solution. In the absence of Mvb12, the heterotrimeric UEV-domain deletion of ESCRT-I has an experimental Stokes radius RH = 4.97 nm and a frictional ratio of 1.8, indicating ESCRT-I retains an extended structure. On the other hand, if the length and stability of the stalk were fully maintained in the absence of Mvb12, this would yield a calculated RH = 5.45 nm for the heterotrimeric UEV-domain deletion construct. Thus when Mvb12 is not present, the solution conformation of ESCRT-I is only modestly less extended than the intact complex.
The Stalk is Important for the Function and Assembly of ESCRT-I
To test the role of the stalk in ESCRT-I function, mutations were engineered in Vps23, Vps37, and Mvb12. Residues in the base, triple coiled coil, and hybrid regions of the stalk were altered alone and in combination. A spectrum of effects was observed in various mutants (Table S3). Some mutants, such as the combined triple coiled coil and hybrid region mutant Vps23M254D/L286D (Fig. S5), severely disrupted all ESCRT-I functions by blocking the assembly of the complex and destabilizing the subunits. Other mutants produced less drastic effects. We chose three representative alleles of this latter class for further study, Vps23M254D and Mvb12L47D/I57D (triple coiled coil) and Vps37L67D (base). With the exception of reduced stability of Mvb12 in vps37L67D strains, the different mutations had little if any effect on steady-state levels of the other subunits (Fig. S1C). However, all three gave rise to defects in the sorting of the MVB cargos Ste3 and Sna3 (Fig. 5A). Some labeling of both cargos was observed in the vacuolar lumen of the mutants, similar to that seen in mvb12Δ cells. Vps37L67D and Vps23M254D mutations also resulted in a CPY sorting defect at least as severe as that of mvb12Δ mutants, whereas little CPY was secreted from mvb12L47D/I57D mutants (Fig. S1B, Table S3).
Figure 5. The Stalk is Essential for ESCRT-I Function.
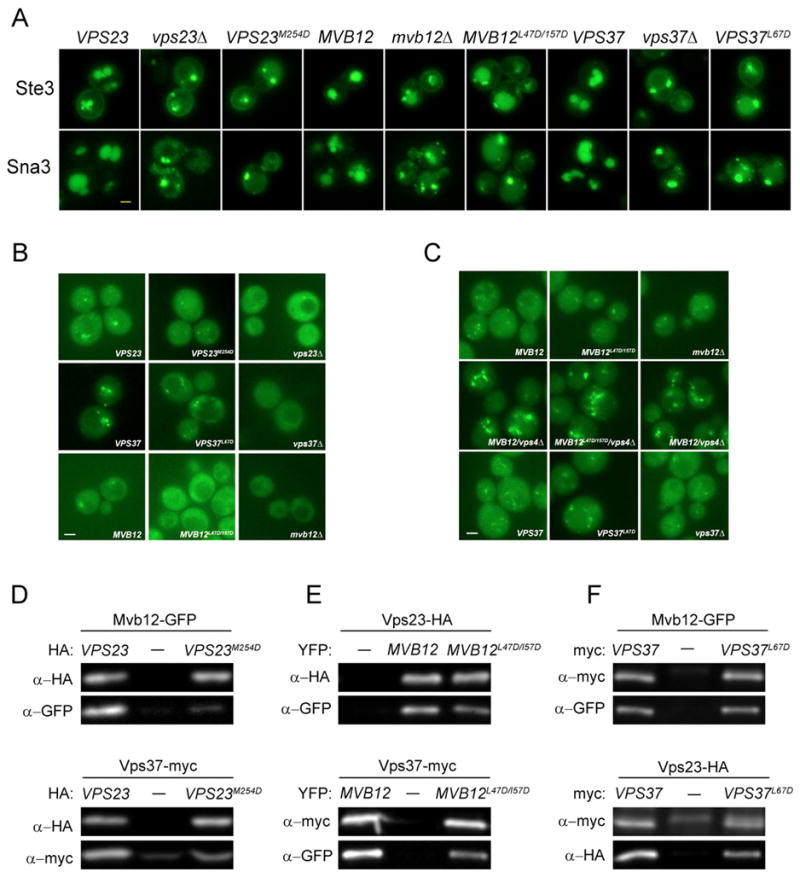
(A) Ste3 and Sna3 sorting defects in stalk mutants. Plasmids expressing wild type or mutant forms of VPS23, MVB12 or VPS37 were introduced into vps23Δ, mvb12Δ or vps37Δ strains expressing plasmid-encoded Ste3-GFP or Sna3-GFP, as indicated. (B) Mvb12 localization depends on the integrity of the stalk. Wild type or mutant forms of VPS23 or VPS37 were expressed from plasmids in vps23Δ vps4Δ or vps37 vps4Δ strains containing chromosomally integrated Mvb12-GFP, whereas plasmid-encoded wild type and mutant forms of Mvb12-YFP were expressed in mvb12Δ vps4Δ strains. (C) Stalk mutants do not prevent Vps23 localization to the MVB. Wild type or mutant forms of VPS37 or MVB12 were expressed from plasmids in vps37Δ, mvb12Δ, or mvb12Δ vps4Δ strains containing chromosomally integrated Vps23-GFP. (D-F) Stalk mutations have differential effects on ESCRT-I assembly. (D) Detergent extracts from vps23Δ mutants containing chromosomally tagged Mvb12-GFP (upper panels) or Vps37-13myc (lower panels) and plasmids for the expression of HA-tagged wild type or mutant forms of Vps23 were subjected to immunoprecipitation with anti-HA antiserum (E) mvb12Δ mutants containing chromosomally tagged Vps23-6HA (upper panels) or Vps37-13myc (lower panels) and plasmid-borne wild type or mutant forms of Mvb12-YFP were immunoprecipitated with either anti-HA (upper panels) or anti-myc (lower panels) antiserum. (F) vps37Δ mutants containing chromosomally tagged Mvb12-GFP (upper panels) or Vps23-6HA (lower panels) and plasmids for the expression of wild type or mutant forms of Vps37-myc were immunoprecipitated with anti-myc antiserum. Co-precipitating proteins were resolved by SDS-PAGE and analyzed by western blotting with mAbs to HA, myc or GFP as indicated. Immunoprecipitates from cells expressing wild type VPS23 (D) or VPS37 (F) were loaded at 5X (D) or 2X (F) reduced levels relative to immunoprecipitates from cells expressing empty vector or mutant proteins to compare relative levels of co-purifying proteins despite differences in the stability of the mutant complexes. Bar = 2 μM.
We examined the effect of each mutation on ESCRT-I membrane recruitment. Both Vps23M254D and Mvb12L47D/I57D greatly reduced Mvb12 localization to the MVB in vps4Δ strains (Fig. 5B). However, Mvb12L47D/I57D had little effect on the MVB localization of Vps23 in either wild type cells or vps4Δ mutants (Fig. 5C), suggesting this mutation reduces the assembly of Mvb12 into the ESCRT-I complex without disrupting the membrane association of ESCRT-I. In contrast, Vps37L67D did not block the MVB association of either Mvb12 or Vps23 (Fig. 5B,C).
Co-immunoprecipitation experiments were used to test the effect of each mutation on ESCRT-I assembly (Fig. 5D–F). The mutant complexes showed varying levels of stability during co-purification from native cell extracts. Consistent with the in vivo localization data, Vps23M254D and Mvb12L47D/I57D mutants had the greatest effect on the incorporation of Mvb12 into the complex. Vps23M254D blocked the Vps23-Mvb12 interaction and reduced, but did not prevent, Vps23 binding to Vps37, whereas Mvb12L47D/I57D showed reduced binding to both Vps23 and Vps37. Vps37L67D-containing complexes contained wild type levels of Mvb12 and Vps23, in keeping with the MVB localization of both proteins.
These results indicate that the stalk contributes to the function of the ESCRT-I complex in vivo. CPY secretion of the Mvb12L47D/I57D mutant was close to wild type (Fig. S1B), suggesting that perturbations of the stalk that do not disrupt CPY sorting can nevertheless lead to significant MVB cargo sorting defects. Vps37L67D assembled at near wild-type efficiency yet caused significant missorting of CPY, Ste3 and Sna3. Thus the base of the stalk is less critical for assembly than the triple coiled coil or hybrid regions of the stalk, yet is equally important for cargo sorting.
Interactions of ESCRT-I with Model Membranes in vitro
To investigate the physical basis for the binding of ESCRT-I to the endosomal membrane, we sought to define the interactions responsible for its binding to membranes in vitro. Full length ESCRT-I bound to brain lipid and bound weakly to synthetic PI(3)P containing vesicles (Fig. 6A, B). No enhancement of binding was observed in PI(3)P-containing vesicles compared to generic brain lipid vesicles. No binding was observed in the Vps37 Δ1-21 construct, indicating that the basic N-terminus of Vps37 is required for binding. ESCRT-II bound weakly to Folch lipids (Fig. 6A), but strongly to PI(3)P-containing synthetic liposomes (Fig. 6B), consistent with previous findings (Teo et al., 2006). ESCRT-I and ESCRT-II were mixed at a 1:1 ratio and found to bind weakly to Folch but strongly to PI(3)P-containing synthetic liposomes. These data show that ESCRT-I can bind weakly and non-specifically to anionic lipids via the basi c N-terminus of Vps37. In contrast, ESCRT-I and ESCRT-II together bind strongly and specifically to a PI(3)P-containing model membrane. The triple complex of Vps23:Vps28:Vps37 Δ1-21 bound to roughly the same extent as intact ESCRT-I in the presence of ESCRT-II, indicating the loss of Mvb12 does not affect the assembly of a membrane-bound ESCRT-I/II complex (Fig. 6C).
Figure 6. Sedimentation of ESCRT-I with Model Liposomes in vitro.
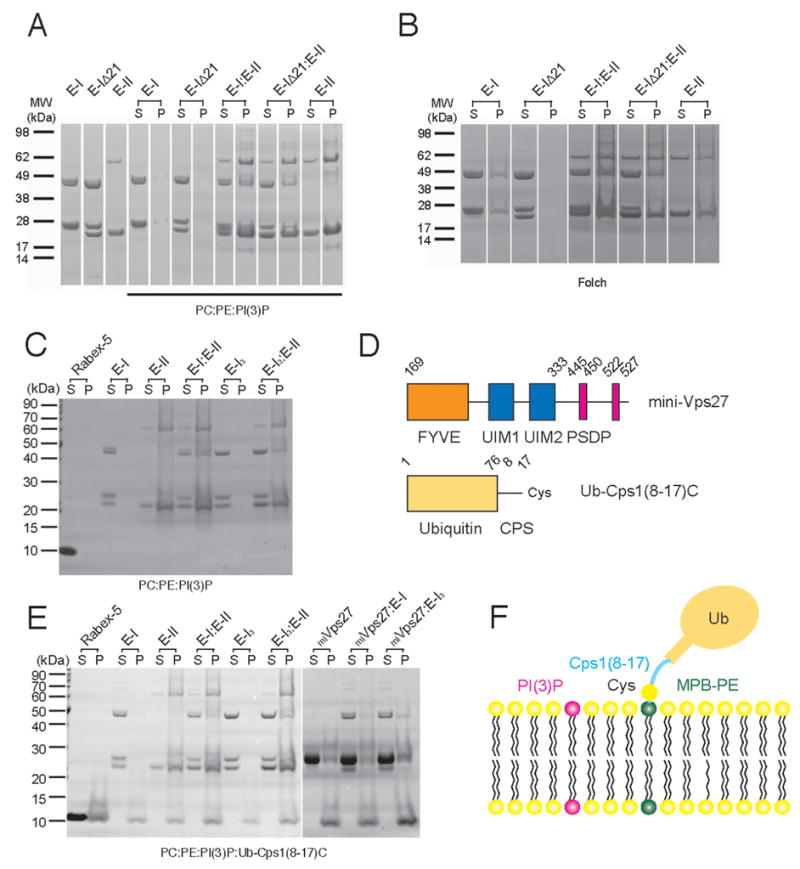
(A) ESCRT-I has a weak intrinsic ability to bind to synthetic PI(3)P-containing liposomes that depends on the N-terminal helix of Vps37. ESCRT-II binds strongly to these liposomes and recruits ESCRT-I in vitro independent of the Vps37 N-terminal helix. (B). ESCRT-I sediments with brain liposomes, indicating the interaction with the N-terminal helix does not require PI(3)P. ESCRT-I Vps37 Δ1-21 does not bind to brain liposomes, showing that the stalk and headpiece do not bind lipids. (C) The triple ESCRT-I complex with the Vps37 Δ1-21 deletion (EI3) is compared to intact ESCRT-I for binding to synthetic PI(3)P-containing liposomes. Both complexes are recruited to the same extent by ESCRT-II. (D) Schematic of mini-Vps27 and Ub-Cps1(8-17)C constructs. (E) Sedimentation with ubiquitin-Cps1 linker-conjugated synthetic PI(3)P-containing liposomes, schematized in (F). ESCRT-II and mini-Vps27 bind to these liposomes. ESCRT-I and triple ESCRT-I do not bind in the absence of other complexes, but bind strongly in the presence of ESCRT-II and more weakly in the presence of mini-Vps27.
We developed an in vitro model MVB cargo based on Cps1, which is ubiquitinated at Lys-8 (Katzmann et al., 2001), 11 residues from the start of its transmembrane helix. A construct in which ubiquitin was fused to Cps1 residues 8-17 followed by a unique C-terminal Cys residue (Ub-Cps1(8-17)C) was covalently conjugated to liposomes (Fig. 6D). These liposomes, but not control liposomes, bound to the ubiquitin binding domains of Rabex-5 (Fig. 6E). The ability of ESCRT-I, II and the ESCRT-I/II complex to bind to PI(3)P-containing vesicles was not substantially enhanced by the model cargo, consistent with the low affinities of the ESCRT-I and II ubiquitin-binding domains. To explore whether Mvb12 alters recruitment by Vps27, a “miniVps27” consisting of the PI(3)P-binding FYVE domain, the ubiquitin binding UIM domains, and the ESCRT-I binding P(S/T)XP sequences was purified. MiniVps27 binds to PI(3)P-containing ubiquitin-Cps1-conjugated liposomes (Fig. 6E), but does not bind to control PC:PE liposomes (data not shown). ESCRT-I binding to these liposomes was markedly enhanced by the presence of miniVps27 (Fig. 6E), independent of the presence or absence of Mvb12. Thus Mvb12 does not appear to regulate the interaction of Vps27 and ESCRT-I in vitro, consistent with the absence of a structural interaction between Mvb12 and the Vps27-binding UEV domain of Vps23.
DISCUSSION
The results presented here essentially complete the structural description of yeast ESCRT-I. The presence of a 130 Å long stalk is the most dramatic new feature of the heterotetramer structure. Known structures of the yeast Vps23 UEV (Teo et al., 2004b) and Vps28 C-terminal domains (Pineda-Molina et al., 2006), together with the heterotetrameric core presented here, represent 88 % of the mass of the complex, missing only flexible loops linking the UEV and Vps28 C-terminal domains, and functionally non-essential extensions from the termini of Vps37 and Mvb12. Hydrodynamic analyses show that the UEV and Vps28 C-terminal domains are in an open conformation, constrained only by their fully flexible tethers to the core. This core-and-appendage organization is familiar in other contexts such as the adaptor protein-2 complex (Collins et al., 2002). The combination of crystallographic analysis of the core and domains separately with hydrodynamic analysis illustrates the power of hybrid structural methodology to address challenging structural problems.
A Structural Model for ESCRT-I on the Membrane
In order to model ESCRT-I bound to an endosomal membrane, we inventoried its physical interactions with the membrane. Consistent with a lack of known phosphoinositide binding domains, we found that ESCRT-I binds acidic phospholipids to a modest extent, but does not bind specifically to PI(3)P (Fig. 6A,B). The highly basic N-terminal 21 amino acids of Vps37, which are absent in the crystallized construct, are required for the weak binding to acidic phospholipids in vitro, although they are not essential for ESCRT-I function and localization in vivo (not shown). When ESCRT-I and full-length ESCRT-II are mixed, ESCRT-I associates tightly with PI(3)P containing liposomes. However, the interaction with ESCRT-II is insufficient to target ESCRT-I in vivo in the absence of Vps27, even though it can do so in vitro. We also found that ESCRT-I can assemble with model cargo and a model Vps27 protein on PI(3)P containing liposomes in the absence of ESCRT-II. The model Vps27 used is less potent than ESCRT-II at recruiting ESCRT-I in vitro, which may reflect the limitations of the model system.
The orientation of ESCRT-I on the endosomal membrane can be inferred from the heterotetramer core structure, the known interactions of the UEV and Vps28 C-terminal domains, and the Vps37 N-terminal helix interaction with membranes. The ubiquitination of the ESCRT-I substrate Cps1 11 residues from the start of its transmembrane helix shows that the UEV domain is capable of functional ubiquitin binding very near the membrane. The NZF1 zinc finger, which is inserted within the split GLUE domain of the ESCRT-II subunit Vps36, binds to the ESCRT-I Vps28 C-terminal domain, while the GLUE domain itself binds to membrane-embedded PI(3)P (Teo et al., 2006). This dictates proximity of the Vps28 C-terminal domain to the membrane. The interaction between the Vps37 basic N-terminal segment and acidic phospholipids suggests that this part of the complex also contacts the membrane. Taken together, these constraints align the stalk roughly parallel to the plane of the membrane (Fig. 7).
Figure 7. Membrane Docked Model for ESCRT-I.
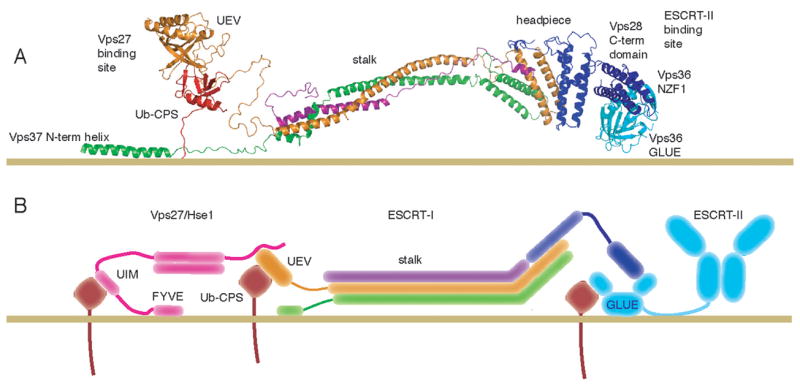
(A) Structural model for yeast ESCRT-I docked to an endosomal membrane. The GLUE domain (PDB code 2CAY)(Teo et al., 2006) and NZF1 domain of ESCRT-II (PDB code 2J9U) (Gill et al., 2007) are cyan, ubiquitinated Cps1 is red, and ESCRT-I subunits are colored as in Fig. 2-4. (B) Schematic diagram of the docked model, incorporating simplified models of the interacting Vps27/Hse1 and ESCRT-II complexes.
Function of the Stalk
The stalk is formed by three of the four subunits of ESCRT-I, and within these three subunits, residues that participate in inter-subunit contacts were selected for mutagenesis. Mutations within the triple-helical stalk region of Mvb12 or Vps23 reduced the assembly of Mvb12 into the ESCRT-I complex and led to sorting defects similar to those of cells lacking Mvb12. Simultaneous mutation of two different regions of the Vps23 stalk appears to destabilize the entire ESCRT-I complex by disrupting the association of Vps23 with both Vps37 and Mvb12, accounting for the abrogation of sorting of all cargo tested. Taken together the Vps23 and Mvb12 mutant data show that the integrity of the stalk is important for the stability and assembly of the complex. However, mutation of Vps37 at the base of the stalk had only mild effects on the stability and assembly of ESCRT-I subunits, yet still disrupted the sorting of CPY, Ste3, and Sna3. This result implicates the stalk in cargo sorting independent of its roles in ESCRT-I stability or assembly.
The postulate that Mvb12 has a central role in organizing ESCRT-I structure begs the question as to why the phenotype of mvb12 is milder than that of other ESCRT-I subunits. Mvb12 is not essential for headpiece assembly (Kostelansky et al., 2006; Teo et al., 2006) and has only one clear-cut function, to increase the stability of the stalk. Vps23 and Vps37 are as important for stalk formation as Mvb12, but in contrast to Mvb12, they are also required for headpiece assembly, which is in turn required to integrate Vps28 into the complex. The solution structure of heterotrimeric ESCRT-I in the absence of Mvb12 is still extended, although it is not as extended as intact ESCRT-I, and it is destabilized as judged by limited proteolysis (Kostelansky et al., 2006). The presence of some residual extended structure in the absence of Mvb12 is consistent with the residual function and stability of ESCRT-I in the absence of Mvb12. The stalk regions of yeast Vps23 and Vps37 are conserved in their human orthologs (Fig. 3E, S3, S4). Human Vps23 (Tsg101) and Vps37B/C are required for HIV budding on the basis of knockdown studies (Garrus et al., 2001) (Eastman et al., 2005), and mutations within the triple coiled-coil and hybrid regions of human Vps23 block budding of HIV (Martin-Serrano et al., 2003). There is thus likely to be a human counterpart to Mvb12, although its identity is unclear. Thus the stalk is a structurally and functionally conserved feature of ESCRT-I from yeast to humans.
Function of Mvb12
While this paper was under review, three other laboratories reported the identification of Mvb12 as an ESCRT-I component (Curtiss et al., 2006; Oestreich et al., 2006). Each of these three reports emphasizes a different biological function for Mvb12. Babst and coworkers note that Mvb12 is required for the efficient recycling of ESCRT-I from membranes in addition to its role in sorting (Curtiss et al., 2006). Katzmann’s group suggests a cargo-selective role for Mvb12 based on their observation that trafficking defects in mvb12Δ cells are more severe for some cargo than others. Two major models have been put forward to account for the differential effects of mvb12 deletion on various cargo (Chu et al., 2006; Curtiss et al., 2006; Oestreich et al., 2006). In the first, Mvb12 has a direct cargo-selective adaptor role, while in the second model, cargo-dependent defects reflect the existence of two or more partially separate MVB pathways. At this stage, there are no data to decisively favor one model over the other, and this is an important area for further study.
Emr and co-workers have proposed that Mvb12 promotes trimerization of soluble ESCRT-I in yeast cytosol and that its biological function is to inhibit the ESCRT-I/ESCRT-II interaction (Chu et al., 2006). However, the soluble form of ESCRT-I is a monomer of 1:1:1:1 subunit stoichiometry, not a trimer (Fig. 2, S2, Table 2). We found that Mvb12 does not affect the binding of ESCRT-I/ESCRT-II to membranes, and Williams and co-workers found that Mvb12 has no effect on the affinity of ESCRT-I for ESCRT-II in solution (Gill et al., 2007). This is consistent with our structural finding that the Vps28 C-terminal domain, which binds to ESCRT-II, does not interact with Mvb12, and is therefore unlikely to be regulated by Mvb12. Taken together, the available data argue against the concept that Mvb12 has as its main function the direct regulation of ESCRT-I/ESCRT-II binding. Instead, the accumulated biological, biochemical, and structural data on Mvb12 are most consistent with a model in which it is an integral component of the core structure of ESCRT-I whose main role is to stabilize the stalk.
Membrane trafficking is an inherently mechanical process. Rigid rod- and stalk-like molecules are omnipresent in trafficking pathways. The coiled-coil stalk structures in proteins such as EEA1 (Dumas et al., 2001) are oriented normal to membranes, and tether vesicles to each other or to organelles. Elongated structures in membrane-binding and coat proteins are oriented parallel to the membrane surface. The elongated BAR domains of proteins such as amphiphysin contact the membrane directly (Peter et al., 2004), while the clathrin leg is part of a scaffold that organizes membrane bound structures without contacting the membrane directly (Brodsky et al., 2001; Kirchhausen, 2000). The docked model of ESCRT-I places the stalk parallel to the plane of the membrane, but not in direct contact with it. In this model, the stalk spatially organizes interactions at the membrane by imposing physical restrictions on their distances relative to each other, yet does not contact the membrane directly.
Lumenal vesicles in yeast multivesicular bodies have a mean diameter of 24 nm (Nickerson et al., 2006). Several ESCRT pathway components have rigid, elongated structures whose lengths are of the same order of magnitude as the lumenal vesicles themselves. These include the ESCRT-I core at 18 nm, ESCRT-II at 15 nm (Hierro et al., 2004; Teo et al., 2004a), and the Bro1 domain at 10 nm (Kim et al., 2005). Strikingly, although perhaps coincidentally, the maximum dimension of intact ESCRT-I in solution is ~25 nm, remarkably close to the mean diameter of a yeast MVB luminal vesicle. A key role for the stalk is most consistent with a function for ESCRT-I as a sophisticated machine acting in coordination with other rigid, elongated ESCRT components.
Conclusion
We have developed a comprehensive structural model for the intact ESCRT-I complex with a number of ramifications for ESCRT function. Our results favor a model in which Mvb12 is an integral component of ESCRT-I. The UEV and Vps28 C-terminal domains have substantial freedom to diffuse in search of their interaction partners, restricted only by their tethers. This structural model is consistent with the lack of any observed effect of Mvb12 on the binding of ESCRT-I to membranes, ubiquitin, Vps27, or ESCRT-II in vitro. ESCRT-I interacts only weakly with membranes and ubiquitin, and is thus not recruited to endosomes in the absence of targeting by Vps27. The observation that all ESCRT ubiquitin binding domains interact with the same Ile-44 patch on ubiquitin led to the “hand-off” model in which ESCRT complexes sequentially bind to cargo (reviewed by Hurley and Emr, 2006). The structure shows that a rigid stalk holds the ESCRT-I and ESCRT-II ubiquitin binding sites too far apart for direct hand-off, strongly arguing against hand-off between ESCRT-I and II. We favor an alternative model in which an assembly of multiple ESCRT complexes simultaneously binds to multiple ubiquitinated cargo molecules. The structure of the trimeric ESCRT-I headpiece (Kostelansky et al., 2006; Teo et al., 2006) revealed a globular 6 nm structure consistent with a simple role as an assembly hub, linking domains that bound cargo and other ESCRT complexes. We now find that the core is nearly 18 nm long, and the intact complex is ~25 nm long. It is difficult to rationalize the evolution of such an elongated structure merely to assemble the subunits. The shape and size of ESCRT-I favor a model in which ESCRT-I directly participates in regulating mechanical aspects of cargo recruitment and membrane remodeling.
EXPERIMENTAL PROCEDURES
Fluorescence Microscopy
Wild type and mutant strains expressing chromosomally integrated Mvb12-GFP or Vps23-GFP were labeled with FM4-64 and viewed by double-label fluorescence microscopy of fixed cells. Uptake of 40 μM FM4-64 by live cells was performed at 30°C for 1 h, after which cells were resuspended in YPD and incubated for 1 h at 30°C. Cells were washed twice in 1M Tris pH 8.0 containing 1% sodium azide and 1% sodium fluoride before imaging. Live and fixed cells were imaged with a 100x oil-immersion objective on a Zeiss Axioplan2 fluorescence microscope, and images were captured with a CoolSnap camera using MetaMorph software and adjusted using Adobe Photoshop.
Co-precipitation Experiments and Western Blotting
Log phase cells were converted to spheroplasts and stored at −70° C (Conibear and Stevens, 2002). For immunoprecipitation of Vps23, spheroplasts from either 20 OD600 or 100 OD600 of cells were resuspended in 1ml of lysis buffer (600 mM sorbitol, 20 mM Hepes pH 7.2, 50 mM KCl, 100 mM potassium acetate, 1% TX-100). Lysates were incubated with 2μl of either rabbit anti-HA antiserum or rabbit anti-myc antiserum for 1h at 4°C, and then with protein G sepharose (Amersham) for 1 h at 4°C. The pellets were washed three times in 20 mM Hepes pH 7.2, 50 mM KCl and subjected to SDS-PAGE. Co-precipitated proteins were detected by Western blotting with monoclonal antibodies to HA (HA.11, Covance Research Products), GFP (Roche scientific) or myc (4A6, Upstate) followed by HRP-labeled anti-mouse secondary antibody (Jackson ImmunoResearch or Sigma). Blots were developed with ECL and either exposed to film (X-OMAT LS, Kodak), or luminescent images were captured with a Fluor S Max Multi-imager and analyzed using Quantity one software.
Crystal Structure Determination
Crystals of the ESCRT-I heterotetramer were grown at 21°C by hanging drop vapor diffusion mixing 2 μL of 4 mg/ml protein in 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM DTT with an equal volume of 100 mM citric acid (pH 4.0), 800 mM ammonium sulfate. Phases were calculated by combining a 3.6 Å two wavelength Se-MAD data set with MIR data from 3.0 Å gold and platinum derivative data sets. The structure was refined against a 2.7 Å native data set, as described in the supplementary methods.
Hydrodynamic Modeling
All calculations of hydrodynamic properties from structural coordinates were carried out using HYDROPRO (Garcia de la Torre et al., 2000). Details of the procedure are described in the supplementary information.
Liposome Binding Experiments
Liposomes used for this study are as follows: Folch fraction (Sigma), 80% POPC:20% POPE (Avanti Polar Lipids), 77% POPC:20% POPE: 3% PI(3)P (Echelon), 77% POPC:15% POPE: 5% MPB-PE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl)butyramide]; Avanti Polar Lipids):3% PI(3)P. The liposomes were prepared at a total lipid concentration of 1 mg/ml by evaporating the solvent from the desired lipid mixture using a nitrogen stream. The dried lipids were resuspended in 150 μl of 0.3 M sucrose, and the solution was incubated at room temperature under nitrogen for 1 h with periodic vortexing; 1 ml of water was added and the sample was sedimented in an ultracentrifuge at 17,000 × g for 30 min at 4°C. The supernatant was removed and the pellet was frozen and thawed 3 times using liquid nitrogen to break up large particles. The pellet was dissolved in 1 ml of buffer A (10 mM Hepes pH 7.4, 150 mM NaCl), and extruded 15 times through a 0.1 μm filter. To chemically conjugate Ub-Cps1(8-17)C to the liposome (77% POPC:15% POPE: 5% MPB-PE: 3% PI(3)P), 450 μg of Ub-Cps1(8-17)C was added to 850 μl of 1 mg/ml solution of total lipids and incubated at room temperature for 3 h. The supernatant was removed by sedimentation at 128,000 × g at 4° C for 1 h, washed with 1 ml of the buffer A, and sedimented for 1h. The pellet was resuspended in 850 μl of the buffer A.
For binding experiments 50 μg of the liposomes were mixed with 50 μg of protein and were brought up to a total volume of 200 μl with buffer A, incubated at room temperature for 30 minutes, and sedimented at 128,000 × g at 4°C for 30 minutes. The pellet was washed once with 200 μl of the buffer A and again sedimented for 30 minutes. Samples of the supernatant (15 μl) and pellet were analyzed by SDS-PAGE. As a control for ubiquitin binding, the tandem A20 ZnF – MIU domain construct of Rabex-5 was used (Lee et al., 2006). This Rabex-5 construct runs on SDS-PAGE at the same position as lipid-conjugated Ub-Cps1(8-17)C, but binding is clearly visualized by an increase in the intensity of the Coomassie blue stained band. To further confirm that the lipid-conjugated Ub-Cps1(8-17)C was functional, surface plasmon resonance experiments were carried out with liposomes bound to an L-1 chip using a Biacore T-100 instrument. Rabex-5 A20 ZnF – MIU bound with ~ 10 μM affinity (Lee and Hurley, unpublished), consistent with previous SPR analysis using immobilized GST-ubiquitin (Lee et al., 2006).
Supplementary Material
Acknowledgments
We thank A. Hierro for preparing ESCRT-II, S. Horte and M. Davey for technical advice and assistance, R. Piper and S. Emr for plasmids, Y. Ye and J. Bonifacino for comments on the manuscript, and G. Hummer for discussions. J. H. H. thanks S. Emr for exchanging unpublished information. Use of the APS was supported by the U. S. DOE, Basic Energy Sciences, Office of Science, under Contract No.W-31-109-Eng-38. This research was supported by NIH intramural support, NIDDK and IATAP (J. H. H.) and funding from the Canadian Institutes of Health Research, the BC Research Institute for Children’s & Women’s Health, and the Michael Smith Foundation for Health Research (E.C.). M. S. K. is a PRAT fellow of the NIGMS, NIH.
Footnotes
Accession numbers
Coordinates have been deposited in the protein data bank with accession code XXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babst M. A Protein’s Final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p-Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochimica Et Biophysica Acta-Molecular Cell Research. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: Formation and function of clathrin- coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Chu T, Sun J, Saksena S, Emr SD. New component of ESCRT-I regulates endosomal sorting complex assembly. J Cell Biol. 2006;175:815–823. doi: 10.1083/jcb.200608053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Conibear E. An ESCRT into the endosome. Mol Cell. 2002;10:215–216. doi: 10.1016/s1097-2765(02)00601-9. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Studying yeast vacuoles. Guide To Yeast Genetics And Molecular And Cell Biology. 2002;Pt C:408–432. doi: 10.1016/s0076-6879(02)51861-9. [DOI] [PubMed] [Google Scholar]
- Curtiss M, Jones C, Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol Biol Cell. 2006;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Research. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Eastman SW, Martin-Serrano J, Chung W, Zang T, Bieniasz PD. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J Biol Chem. 2005;280:628–636. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Emr SD, Williams RL. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. Embo J. 2007;26:600–612. doi: 10.1038/sj.emboj.7601501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, Hurley JH. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004;431:221–225. doi: 10.1038/nature02914. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomolec Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, Hierro A, Emr SD, Hurley JH. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–126. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nature Structural & Molecular Biology. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich AJ, Davies BA, Payne JA, Katzmann DJ. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol Biol Cell. 2006;18:646–657. doi: 10.1091/mbc.E06-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiano D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends In Cell Biology. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: Crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004a;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J Biol Chem. 2004b;279:28689–28696. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- Teo HL, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


