Abstract
Fibroblast growth factor (FGF) signaling mediates reciprocal mesenchymal-epithelial cell interactions in the developing mouse lung and limb. In the gastrointestinal (GI) tract, FGF10 is expressed in the cecal mesenchyme and signals to an epithelial splice form of FGF receptor (FGFR) 2 to regulate epithelial budding. Here, we identify FGF9 as a reciprocal epithelial-mesenchymal signal required for cecal morphogenesis. Fgf9 null (Fgf9−/−) mouse embryos have agenesis of the embryonic cecum, lacking both mesenchymal expansion and an epithelial bud. In the cecal region of Fgf9−/− embryos, mesenchymal expression of Fgf10 and Bmp4 is notably absent, whereas the expression of epithelial markers, such as sonic hedgehog, is not affected. Using epithelial and whole explant cultures, we show that FGF9 signals to mesenchymal FGFRs and that FGF10 signals to epithelial FGFRs. Taken together, these data show that an epithelial FGF9 signal is necessary for the expansion of cecal mesenchyme and the expression of mesenchymal genes that are required for epithelial budding. Thus, these data add to our understanding of FGF-mediated reciprocal epithelial-mesenchymal signaling.
Keywords: Fibroblast growth factor 9 (FGF9), FGF10, Gut development, Cecum, Budding morphogenesis, Epithelial-mesenchymal crosstalk
INTRODUCTION
The development of the gastrointestinal (GI) tract in vertebrates begins with the invagination of embryonic endoderm to form a simple tube with rostral and caudal openings. In mice, this process starts at embryonic day (E) 7 (Grapin-Botton and Melton, 2000; Roberts, 2000; Snell and Stevens, 1966). At E8, the midgut is first visible as a closed tube (Kaufman, 1992). Two sac-like structures, the stomach and cecum, bud from this tube and define functional subdivisions in the gut. The stomach is positioned at the junction of the esophagus and the proximal small intestine, whereas the cecum is located at the junction of the distal small intestine (ileum) and the colon. By E13.5, the entire gut is lined by a pseudostratified, uniformly proliferative endoderm (Kaufman, 1992).
The cecum begins to form at E11.5 as a mesenchymal expansion followed by epithelial budding into the primordial cecal mesenchyme (Burns et al., 2004). This process begins prior to the proximal to distal wave of cytodifferentiation that occurs from E15 to E18 in the small intestinal endoderm, which converts it to a simple columnar epithelium with rudimentary villi (Calvert and Pothier, 1990; Schmidt et al., 1988).
The wall of the cecum, like other parts of the gut tube, contains two types of tissue: an outer layer of mesoderm-derived mesenchyme and an inner layer of endoderm-derived epithelium. Developmental studies have demonstrated that an interaction between the endoderm and mesoderm is required for normal differentiation of region-specific gut epithelium (Haffen et al., 1987; Kedinger et al., 1998; Koike and Yasugi, 1999; Roberts et al., 1998). Reciprocal molecular interactions between epithelium and mesenchyme are crucial for budding morphogenesis in many organ systems (Cardoso, 2001; Shannon and Hyatt, 2004; Tanaka and Gann, 1995). Fibroblast growth factors (FGFs) are candidates for cecal development, as recent studies show that both FGF10 and its receptor, FGFR2b, are required for the formation of this organ (Burns et al., 2004).
FGFs bind to and activate four tyrosine kinase receptors (FGFRs) to regulate intracellular signaling pathways controlling cell proliferation, differentiation and migration. An alternative splice form of FGFR2 (FGFR2b) is expressed in epithelial tissues and is activated by mesenchymally expressed FGFs, such as FGF7 and FGF10. By contrast, FGFR1c and FGFR2c are expressed primarily in mesenchyme, and are activated by FGF ligands expressed in epithelia, such as FGF4, FGF8 and FGF9 (Ornitz and Itoh, 2001; Ornitz et al., 1996). In the developing lung, FGF9 and FGF10 form a reciprocal pair of ligands that regulate branching and budding morphogenesis. FGF10 is expressed at high levels in the distal lung mesenchyme, immediately adjacent to budding airway epithelium. FGF10 signals through its high affinity receptor, FGFR2b, resulting in epithelial migration towards the source of FGF10 (Bellusci et al., 1997; De Moerlooze et al., 2000; Min et al., 1998; Sekine et al., 1999). By contrast, FGF9 is expressed in lung airway epithelium and the mesothelial visceral pleura, where it regulates lung mesenchymal proliferation. Fgf9−/− mice have a significant reduction in lung mesenchyme, and subsequent decreased FGF10 expression, resulting in decreased lung branching (Colvin et al., 1999; Colvin et al., 2001b). Fgf10−/− mice fail to form primary bronchi and thus have complete agenesis of the lung (Min et al., 1998; Sekine et al., 1999). It is not known whether FGF10 is required for the expression of Fgf9 during lung branching morphogenesis. Reciprocal FGF signaling also occurs during limb development. The formation of the limb bud is initiated with the expression of mesenchymal FGF10, which induces formation of the apical ectodermal ridge (AER) (Martin, 1998). FGFs 4, 8, 9 and 17 are subsequently expressed in the AER and signal back to limb mesenchyme.
Although reciprocal FGF signaling pathways have been identified in several developmental systems, it is not known whether reciprocal FGF signals are universally required for organogenesis. For example, in midgestational heart development, FGF9 has been shown to signal from the endocardium and epicardium to the myocardium. However, a reciprocal myocardial to epicardial signal has not been identified (Lavine et al., 2005). We identify FGF9 as a necessary signal to induce expansion of the cecal mesenchyme. However, FGF9 is not sufficient to induce cecal development at other sites along the length of the intestine. Moreover, comparative analysis of both Fgf9−/− and Fgf10−/− mice demonstrates that expansion of cecal mesenchyme precedes FGF10 expression and epithelial budding.
MATERIALS AND METHODS
Mouse embryos
Fgf9 and Fgf10 mutant mice (maintained on a C57/Bl6 background) were generated as described previously (Colvin et al., 2001a; Min et al., 1998). For embryo collection, time-mated Fgf9+/− or Fgf10+/− females were killed at E10.5–E18.5, the embryos were removed and the intestinal tract was isolated, as previously described (Stappenbeck and Gordon, 2000). Wild-type or heterozygous littermates from each line were collected as controls.
Histology and immunohistochemistry
Mouse embryonic GI tracts were fixed overnight in 4% paraformaldehyde (PFA)/PBS, dehydrated through ethanol, embedded in paraffin, and 5 μm serial sections were prepared for Hematoxylin and Eosin (H&E) staining and immunohistochemical staining. To confirm FGFR1 and FGFR2 expression in E12.5 mouse embryonic cecum, the sections were deparaffinized, rehydrated, treated with blocking solution (Histostain-SP kit, Invitrogen, Carlsbad, CA), and incubated overnight at 4°C with rabbit anti-FGFR1 and FGFR2 antibodies (Santa Cruz Biotech, Santa Cruz, CA). The signals were visualized with the Histostain-SP Kit (Invitrogen, Carlsbad, CA), as recommended by the manufacturer.
BrdU labeling
For analysis of cell proliferation, time-mated Fgf9+/− or Fgf10+/− females were given an intraperitoneal injection of bromodeoxyuridine (BrdU, 120 μg/gm body weight; Sigma) 1 hour before sacrifice at E12.5–E18.5. The intestinal tract was isolated and processed for histology. The embedded specimens were cut (5 μm thick sections), and BrdU-labeled cells were identified by immunohistochemistry using a monoclonal antibody to BrdU (BD Biosciences). Horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G was obtained from BioSource. Bound antibodies were visualized using diaminobenzidine (Sigma) in the presence of H2O2. Sections were counterstained with Hematoxylin. Proliferation in the cecal epithelium and mesenchyme was scored as the ratio of BrdU-labeled nuclei to total cell nuclei in fields viewed through a 40× objective.
Whole-mount in situ hybridization
Embryonic intestinal tracts were dissected in cold diethyl pyrocarbonate (DEPC)-treated PBS, fixed overnight in 4% PFA/DEPC-PBS at 4°C, dehydrated through graded methanol in DEPC-PBT (PBS+0.1% Tween 20) and stored at −20°C. Whole-mount in situ hybridization was performed as described (Colvin et al., 2001b). Control wild-type and Fgf9−/− or Fgf10−/− tissues were processed together to ensure identical hybridization conditions. Mouse RNA probes were synthesized from Fgf9 (Colvin et al., 1999), Fgf10 (provided by B. Hogan), Shh and Bmp4 (provided by A. McMahon) cDNA clones. At least three independent hybridizations for each probe were tested at each developmental stage.
Cecal and epithelial explant cultures
Embryos from wild-type C57BL/6J mice were dissected at E12.5. The cecums were removed from the GI tract and embedded in Growth Factor Reduced Matrigel™ (BD Biosciences) diluted 1:1 with culture medium (5% FCS, 50% DMEM:F12, penicillin/streptomycin + L-glutamine) in 24-well tissue culture plates (Burns et al., 2004). To isolate the epithelium, distal intestines were treated with 5 mg/ml collagenase A for 5 minutes on ice, and mesenchyme was removed using tungsten needles. The isolated epithelium was embedded in Growth Factor Reduced Matrigel™, as described above. Human FGF10 (100 ng/ml; PeproTech), mouse FGF9 (100 ng/ml; PeproTech), or BSA (0.1%) soaked heparin-coated beads (Sigma) were placed one bead diameter away from the cecal explant. Matrigel™ was allowed to solidify at 37°C for 30 minutes, then 250 μl of culture medium was gently added to each well and the explants were grown for 4 days at 37°C in a 5% CO2 incubator. For histology, the cultured cecal explants were fixed overnight in 4% PFA/PBS, embedded in OCT, and 6-um frozen sections were cut for H&E staining.
RESULTS
Fgf9 and Fgfr expression during cecal development
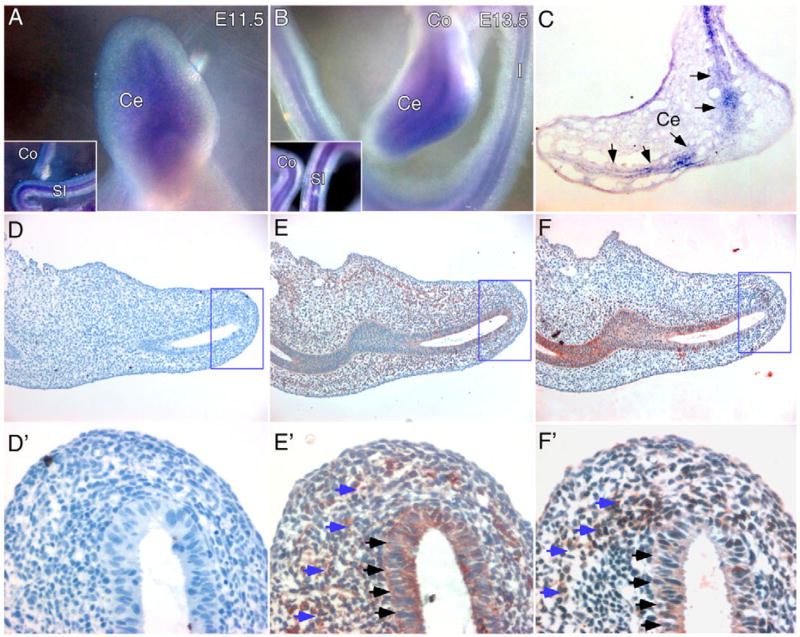
Previous studies have reported that Fgf9 is expressed in the mesothelial lining of the foregut, including the esophageal region, stomach and intestinal primordium at embryonic day 10.5 (E10.5) and in luminal epithelial cells at E14.5 (Colvin et al., 1999). To examine the spatial patterns of Fgf9 expression along the length of the GI tract, whole-mount in situ hybridization of dissected intestines was performed at E11.5–E14.5. At these developmental stages, Fgf9 was strongly and uniformly expressed throughout the small intestinal, cecal and colonic epithelium (Fig. 1A–C, data not shown). Expression of FGFR1 and FGFR2, potential receptors for FGF9, was localized to both cecal epithelium and mesenchyme (Fig. 1D–F′). In cecal mesenchyme, FGFR1 was broadly expressed, whereas FGFR2 showed higher expression on the caudal side of the cecal bud.
Fig. 1. Fgf9, FGFR1 and FGFR2 expression in the embryonic cecum.
(A,B) Whole-mount in situ hybridization was performed on isolated E11.5 (A) and E13.5 (B) wild-type GI tracts to detect Fgf9 expression. Insets show small intestine and colon. Fgf9 expression was uniformly expressed in the epithelium of the cecum, small intestine and colon. (C) Frozen section of an E13.5 whole-mount in situ hybridization showing Fgf9 expression in the cecal epithelium (arrows). (D–F) Immunohistochemical analysis of E12.5 cecal sections stained with an anti-FGFR1 (E) or anti-FGFR2 (F) polyclonal antibody. Control (D) staining, in which the primary antibody was absent, shows no signal. D′, E′ and F′ are higher magnification views of the boxes in D, E and F, respectively. FGFR1 (E′) and FGFR2 (F′) are expressed in both cecal epithelium (black arrows) and mesenchyme (blue arrows). Co, colon; I, Ileum; SI, small intestine; Ce, cecum.
Absent cecal development in Fgf9−/− and Fgf10−/− embryos
The development of the cecum in wild type control, Fgf9−/− and Fgf10−/− mouse embryos was examined at E12.5, E14.5 and E18.5. GI tracts from Fgf9+/− and Fgf10+/− animals were morphologically indistinguishable from their wild-type littermates at all stages examined (Fgf9, n=20; Fgf10, n=5, for each genotype at each stage). At E12.5, the developing cecum is readily identifiable as a mesothelial-covered, mesenchymal protrusion from the intestinal tract (Fig. 2A,D). This characteristic protrusion contains an epithelial bud and occurs at a distinctive bend in the gut tube at the junction of the distal small intestine (ileum) and colon. In Fgf9−/− intestines, this characteristic bend was present at this location, but the mesenchymal bud and its associated epithelial bud were absent (n=80/80 animals examined; e.g., Fig. 2A). By contrast, the ileo-cecal junction of Fgf10−/− mice maintained both the bend and mesenchymal bud, but lacked epithelial budding into cecal mesenchyme (n=20/20; e.g. Fig. 2D). In E14.5 wild-type embryos, both the epithelial and mesenchymal components of the cecum continued to elongate (Fig. 2B,E,G). At this same stage, all Fgf9−/− embryos examined showed no evidence of cecal development (Fig. 2B,H), whereas all Fgf10−/− embryos studied showed a continued mesenchymal bud with no epithelial budding (n=20; Fig. 2E,I). By E18.5, the wild type embryonic cecum was elongated and showed a mature curved morphology (Fig. 2C,F). Fgf9−/− embryos continued to show no cecal development (Fig. 2C), whereas the mesenchymal bud of Fgf10−/− embryos appeared to be degenerating when compared with earlier embryonic time points (Fig. 2F) (Burns et al., 2004).
Fig. 2. The development of the cecum is abnormal in both Fgf9−/− and Fgf10−/− embryos.
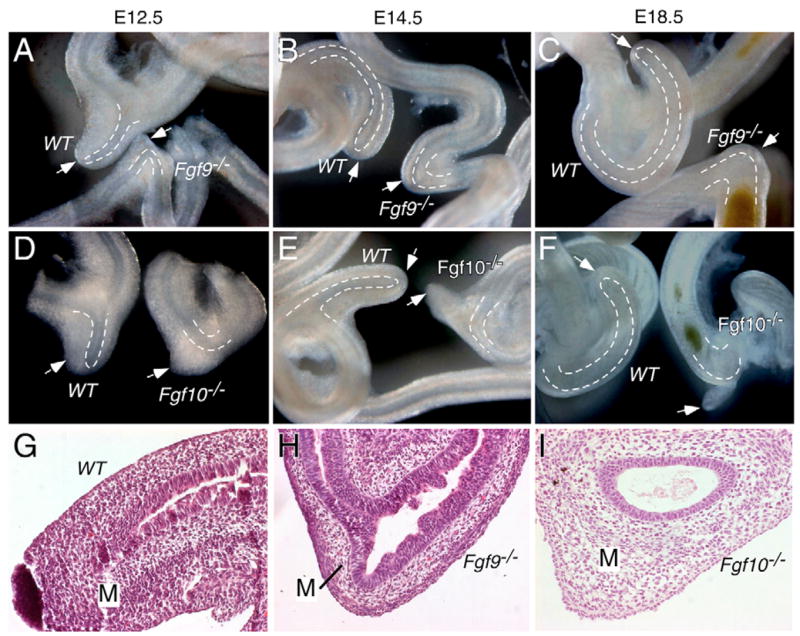
(A–F) Morphologic comparison of the development of the cecum in WT, Fgf9−/− and Fgf10−/− embryos at E12.5, E14.5 and E18.5. (G–I) Hematoxylin and Eosin (H&E) stained sections of E14.5 WT, Fgf9−/− and Fgf10−/− cecum. In WT embryos (A–G), the cecum is located at the ileo-colonic junction. (A–C,H) Fgf9−/− embryos show a normal appearing epithelial tube throughout the small intestine and colon, but no identifiable cecal bud. (D–F,I) Fgf10−/− embryos develop a mesenchymal bud at the ileo-colonic junction, but no epithelial budding or expansion into cecal mesenchyme was observed. (F) By E18.5, the cecal mesenchymal bud appeared to degenerate in Fgf10−/− tissue (arrow). Dashed lines indicate the border between the epithelium and mesenchyme in the cecal region. Arrows indicate the position of the cecal tip or the position of the ileo-colonic junction in the absence of a cecum. M, mesenchyme.
Cell proliferation in the Fgf9−/− embryonic cecum is reduced
Normally, cecal development is initiated at ~E11.5 as a mesenchymal bud at the ileo-colonic junction. By E12.5, epithelial budding was evident in control but not Fgf9−/− tissue (Fig. 3A,B). Therefore, we assessed mesenchymal and epithelial cell proliferation at E12.5 by BrdU incorporation. Compared with wild-type tissue, cell proliferation in Fgf9−/− cecum was reduced by 40% in epithelium and 47% in mesenchyme (Fig. 3, Table 1). Interestingly, mesenchymal proliferation in the cecal buds of wild-type mice was significantly greater than mesenchymal proliferation in the adjacent small intestine (Fig. 3, Table 1). By contrast, mesenchymal proliferation in Fgf9−/− cecal buds was significantly less than mesenchymal proliferation in the adjacent small intestine or colon (Table 1). Additionally, epithelial proliferation in Fgf9−/− cecal buds was less than epithelial proliferation in the small intestine or colon (Table 1). Comparable analysis of proliferation in the distal small intestine and colon revealed no significant difference between wild-type and Fgf9−/− mice in either mesenchyme or epithelium (Table 1). Together, these observations indicate that epithelial expression of FGF9 is required, either directly or indirectly, for the proper regulation of both mesenchymal and epithelial proliferation in the cecal region.
Fig. 3. Reduced cell proliferation rates in Fgf9−/− cecal mesenchyme and epithelium.
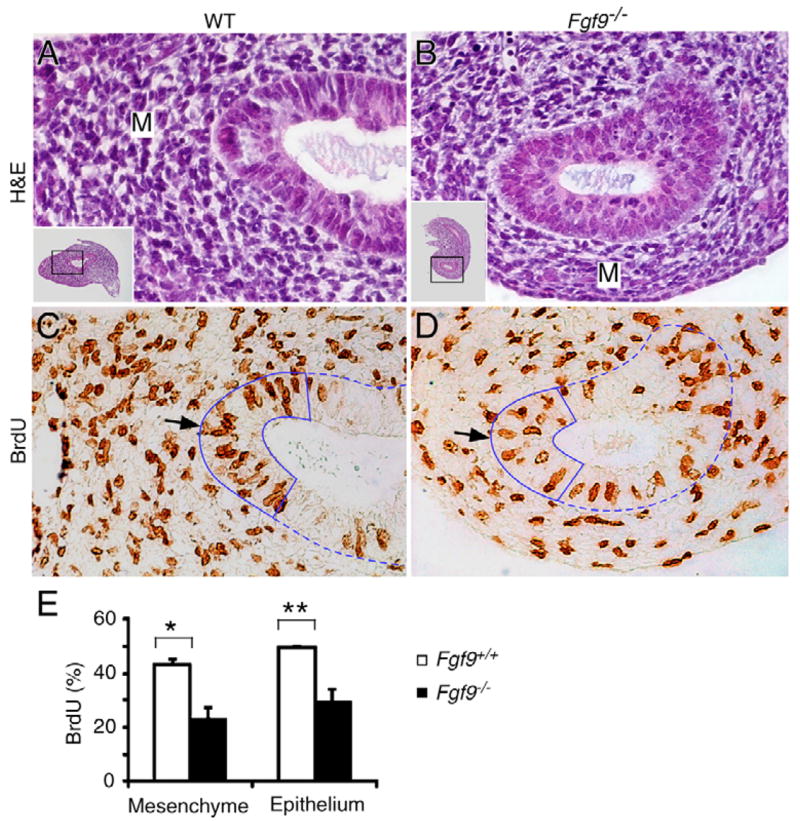
(A,B) H&E stained paraffin sections from E12.5 wild-type (A) and Fgf9−/− (B) cecum. The amount of mesenchyme (M) in Fgf9−/− cecal tissue is reduced compared with in wild-type tissue. The insets show a low-power view of the entire cecal section; the boxed region indicates the enlarged area shown. (C,D) BrdU labeling of adjacent cecal sections demonstrating reduced cell proliferation in both the mesenchyme and epithelium (arrow) in Fgf9−/− (D) tissue compared with in wild-type tissue (C). Dashed line indicates the border of mesenchyme and epithelium; the solid line indicates the apical cap of the cecal epithelium. (E) Quantification of percentage of BrdU-positive nuclei showing significantly decreased cell proliferation in both cecal mesenchyme (*P<0.001) and epithelium (**P<0.01) in Fgf9−/− tissue.
Table 1.
Cell proliferation in E12.5 control and Fgf9−/− GI tracts
| Epithelium
|
Mesenchyme
|
|||||
|---|---|---|---|---|---|---|
| Tissue | Genotype | n* | % BrdU±s.d. | P value† | % BrdU±s.d. | P value† |
| Cecum | +/+, +/− | 5 | 49±5 | 43±2‡ | ||
| −/− | 3 | 30±5§ | P<0.01 | 23±1§ | P<0.0001 | |
| Small intestine | +/+, +/− | 6 | 46±5 | 34±5‡ | ||
| −/− | 6 | 50±3§ | ns | 36±3§ | ns | |
| Colon | +/+, +/− | 6 | 50±2 | 42±5 | ||
| −/− | 6 | 51±3§ | ns | 41±4§ | ns | |
n, number of mice counted.
P value, two-tail P value from a two-sample t-test assuming equal variances for % BrdU labeled nuclei in Fgf9−/− tissue compared with wild-type or Fgf9+/− tissue.
P<0.001, comparison of the BrdU-labeling index in cecal and small intestinal mesenchyme from wild-type mice.
P<0.0001, comparison of the BrdU-labeling index in cecal and small intestine and colon epithelium and mesenchyme from Fgf9−/− mice.
s.d., standard deviation.
ns, non-significant difference.
Mesenchymal Fgf10 expression is downregulated in Fgf9−/− cecum
Decreased epithelial proliferation in Fgf9−/− cecum suggests that FGF9 must regulate additional factors that signal back to intestinal epithelium. One such factor is FGF10, given that previous studies reported that mesenchymal expression of Fgf10 is required for budding morphogenesis of the cecal epithelium (Burns et al., 2004). To examine the relationship between epithelial expression of Fgf9 and mesenchymal expression of Fgf10, Fgf10 expression was examined by whole-mount in situ hybridization at E11.5 and E12.5. In wild-type embryos, Fgf10 was expressed in caudal cecal mesenchyme (Fig. 4A,C). However, in Fgf9−/− embryos, Fgf10 expression was absent in mesenchyme at the ileo-colonic junction (n=6; e.g. Fig. 4B,D). These observations suggest that Fgf10 expression in cecal mesenchyme requires epithelial FGF9 signaling. Interestingly, in Fgf10−/− embryos, epithelial Fgf9 continued to be expressed in the ileo-colonic junction (Fig. 4E,F). This suggests that a reciprocal FGF10 signal is not required to maintain epithelial Fgf9 expression in the cecum, or in other regions of the GI tract where Fgf10 is not normally expressed. The possibility of other FGFs contributing to a reciprocal signal cannot be ruled out at this time.
Fig. 4. FGF9 signaling is required for Fgf10 expression.
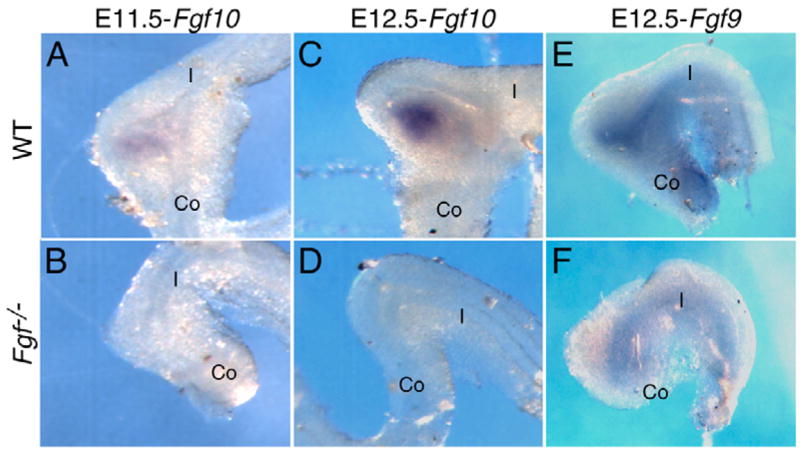
Whole-mount in situ hybridization was performed on E11.5 and E12.5 embryonic GI tracts from wild-type (WT), Fgf9−/− and Fgf10−/− embryos. Fgf10 was expressed in the cecum mesenchyme in WT tissue (A,C), but was completely absent in Fgf9−/− tissue (B,D). By contrast, Fgf9 was expressed in cecum epithelium from WT and Fgf10−/− tissue (E,F). Co, colon; I, terminal ileum.
Specificity of epithelial-mesenchymal signaling by FGF9 and FGF10
Based on the in vitro specificity of FGFs for alternatively spliced FGFRs, we predict that cecal FGF9 signals specifically to mesenchymal FGFRs, whereas FGF10 signals specifically to epithelial FGFRs (Ornitz and Itoh, 2001). An interesting exception to this rule is that FGF9 is able to activate the epithelial splice form of FGFR3 (FGFR3b) in vitro (Ornitz et al., 1996); however, this has not been demonstrated in vivo.
To examine the tissue-specific activity of FGF9 and FGF10 signaling in the developing cecum, intact cecal explants from wild-type E12.5 mice were cultured in Matrigel in the presence of recombinant BSA-, FGF9- or FGF10-soaked heparin beads (Fig. 5). In the presence of the control BSA-containing beads, explants did not grow or extend toward the bead over the 84-hour culture period (Fig. 5D,D′,G). In the presence of an FGF10-soaked bead, the apical cecal epithelium expanded and grew toward the FGF10 bead (Fig. 5E,E′,H), whereas cecal mesenchyme showed no additional growth. In the presence of an FGF9-soaked bead, expansion of both distal cecal mesenchyme and epithelium was observed (Fig. 5F,F′,I). These data suggest that FGF10 functions as both a mitogen and a chemoattractant for distal cecal epithelium, whereas FGF9 signals to cecal mesenchyme, and maintains or induces the expression of FGF10 to indirectly regulate cecal epithelial expansion. However, the possibility remains that FGF9 could signal to cecal epithelium through FGFR3b, as well as to cecal mesenchyme through c splice forms of other FGFRs.
Fig. 5. Effect of FGF9 and FGF10 on growth of cecal explants.
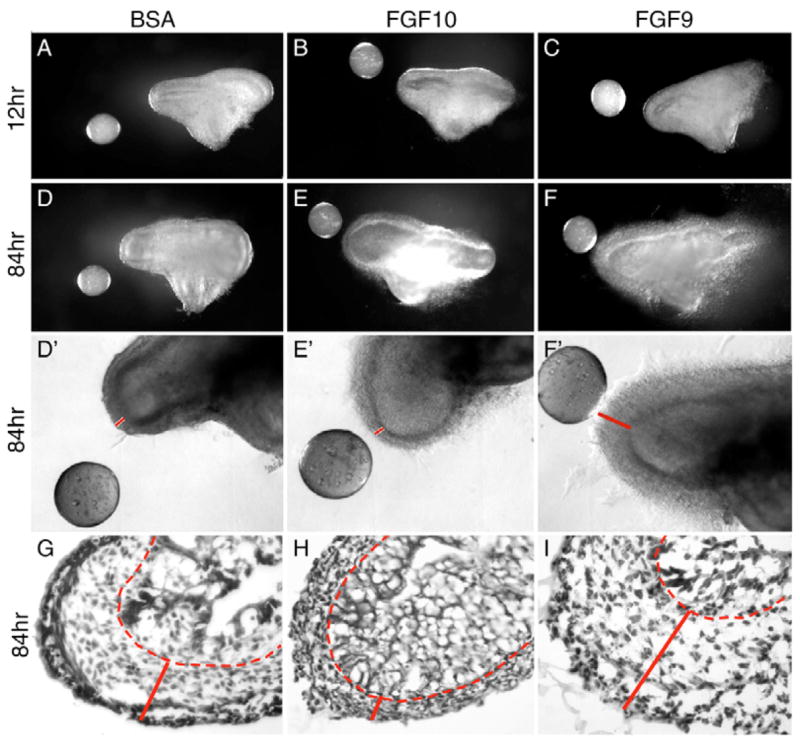
E12.5 embryonic wild-type cecums were removed and embedded in Matrigel. BSA-, FGF9- or FGF10-soaked heparin beads were placed one bead diameter from the cecal explant. (A–C) During the first 12 hours, the cecal epithelium and mesenchyme did not significantly extend toward either the FGF9- or FGF10-soaked bead compared with the BSA-soaked control bead. (D–F) After 84 hours, the cecal epithelium had expanded and extended toward the FGF10 bead (E,E′,H) compared to the BSA bead (D,D′,G); however, no mesenchymal growth was observed (red solid line in D′,E′,G,H). By contrast, cecal mesenchyme showed significant growth accompanied by epithelial expansion when placed adjacent to an FGF9 bead (F,F′,I). Note the significantly thickened mesenchymal regions in F′ and I (line). D′, E′ and F′ show higher magnification brightfield views of the explants in D, E and F, respectively. (G–I) H&E stained frozen sections showing the histology of cultured cecal explants at 84 hours. Red dashed lines indicate the border of mesenchyme and epithelium. The solid lines in D′–F′, G–I indicate the thickness of the cecal mesenchymal layer.
To directly test whether FGF9 could signal to intestinal epithelium, equal size pieces of mesenchyme-free distal intestinal epithelium were embedded in Matrigel and cultured with BSA-, FGF9- or FGF10-soaked heparin beads for varying periods of time (n=6 explants/condition). After 12 hours in culture, epithelial explants cultured with BSA beads were quiescent, whereas in the presence of either FGF9 or FGF10 beads, the explant expanded (Fig. 6E,I). Comparison of the FGF9- and FGF10-treated explants showed clear elongation of the FGF10-treated explant toward the bead over the 84-hour culture period (Fig. 6F–L). By 84 hours, the intestinal epithelium reached and began to surround the FGF10 bead (Fig. 6L). By contrast, epithelium placed near an FGF9 bead expanded but did not extend toward the bead (Fig. 6G,H). However, by 60 hours in culture, a mesenchymal halo was evident around the explant, suggesting that residual mesenchymal cells, possibly stimulated by FGF9, could account for the epithelial dilation by producing an FGF10-like activity. In contrast to FGF9- and FGF10-treated explants, no epithelial proliferation or expansion was observed when epithelial explants were exposed to a BSA bead over an 84-hour culture period (Fig. 6C,D).
Fig. 6. Distal small intestinal epithelium extends toward an FGF10, but not an FGF9, source.
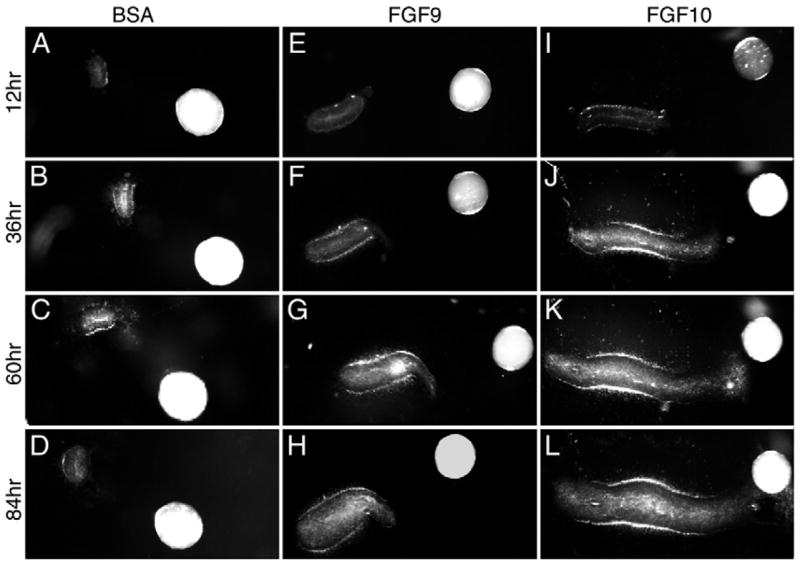
BSA-, FGF9- or FGF10-soaked beads were placed in Matrigel two-bead diameters from a wild-type distal intestinal epithelial explant in which mesenchymal tissue had been removed. (A–D) Intestinal epithelium did not grow when exposed to a BSA-soaked control bead over an 84-hour period. (E–H) In response to an FGF9 bead, intestinal epithelium showed a modest dilation but no directional extension towards the bead. (I–L) In response to an FGF10 bead, intestinal epithelium showed a significant dilation and extension towards the bead.
Gene regulation during Fgf9−/− and Fgf10−/− cecal development
Previous studies demonstrated that during early development of the mouse GI tract, epithelial expression of Shh and mesenchymal expression of Bmp4 are important regulators of epithelial-mesenchymal interactions (Bitgood and McMahon, 1995; Roberts et al., 1995; Roberts et al., 1998). Both SHH and BMP4 signals are known to interact with FGF signaling pathways in lung and limb development (Bellusci et al., 1997; Bellusci et al., 1996; Buckland et al., 1998; Khokha et al., 2003; Lebeche et al., 1999; Scherz et al., 2004; Weaver et al., 2000; Zuniga et al., 1999). To determine whether Shh or Bmp4 expression was affected by the loss of Fgf9 or Fgf10, we examined their expression at E12.5 and E13.5. In wild-type embryos, Bmp4 was strongly expressed in cecal mesenchyme (Fig. 7A,B). Its expression did not change in the absence of Fgf10 (Fig. 7C,D) (Burns et al., 2004). However, in Fgf9−/− gut, Bmp4 expression was absent in ileo-colonic junctional mesenchyme (Fig. 7E,F). In contrast to Bmp4, Shh expression was not changed in the cecum, small intestine or colonic epithelium in embryos lacking either Fgf9 or Fgf10 (Fig. 7G,H).
Fig. 7. Loss of Bmp4 but not Shh expression in the developing cecum of Fgf9−/− mice.
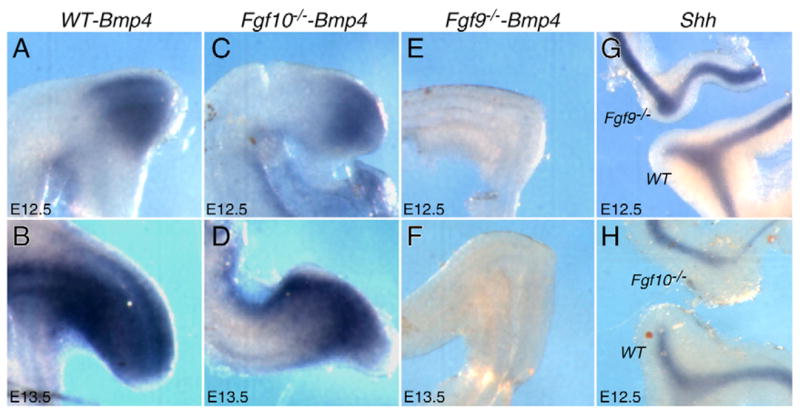
Whole-mount in situ hybridization was performed on E12.5 and E13.5 embryonic GI tracts from wild-type (WT), Fgf9−/− and Fgf10−/− embryos. (A–D) At E12.5 and E13.5, Bmp4 was expressed strongly in the cecal mesenchyme in WT tissue (A,B) and Fgf10−/− tissue (C,D). (E,F) At these same stages, Bmp4 expression was absent in Fgf9−/− cecum. (G,H) At E12.5, Shh was expressed in cecal epithelium in WT tissue, and its expression was not significantly affected by the absence of expression of either Fgf9 or Fgf10.
DISCUSSION
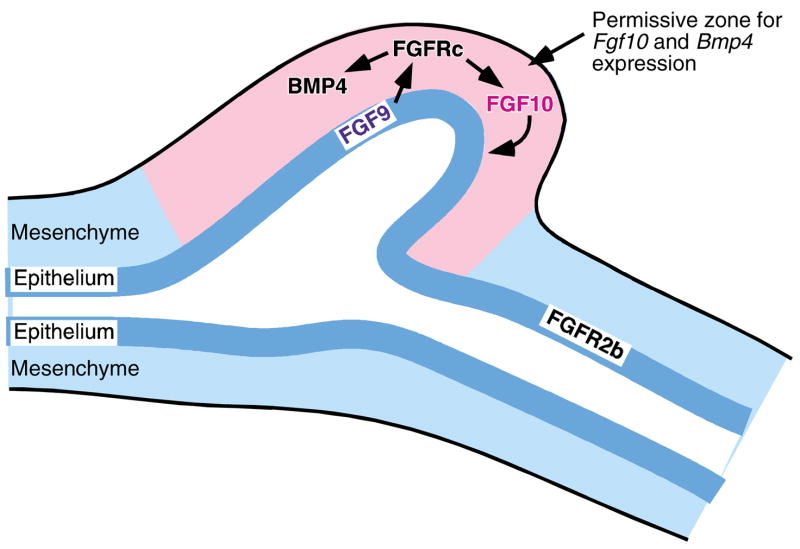
The cecum in adult mice and humans is an anatomically and functionally distinct unit. It is populated by a large and diverse population of indigenous microbes that play a key role in a number of biotransformations (Eckburg et al., 2005). These biotransformations include breakdown of plant polysaccharides that are delivered to the distal gut from the small intestine that otherwise could not be broken down because the mammalian host lacks the requisite glycoside hydrolases (Backhed et al., 2005). The factors that underlie proper cecal development have been poorly defined. Our data provide new evidence that epithelial-expressed FGF9 functions primarily to regulate cecal mesenchymal proliferation/differentiation, as well as mesenchymal FGF10 expression. Fig. 8 and the discussion below summarize our view about how FGF9 and FGF10 act in a cooperative and reciprocal manner, together with other factors, to generate a cecum from the developing gut tube.
Fig. 8. Model of reciprocal FGF signaling in cecum development.
During early cecal development, epithelial expression of FGF9 signals to mesenchymal FGFRs (FGFR1c and FGFR2c) to stimulate local mesenchymal proliferation and gene expression (Fgf10, Bmp4). Mesenchymal FGF10 signals to the epithelial FGFR2b to stimulate epithelial proliferation and extension of the epithelial bud into the expanded cecal mesenchyme. FGF9 is only able to induce mesenchymal proliferation and Fgf10 expression in the permissive junctional mesenchyme.
The GI tract consists of functionally distinct domains along the rostrocaudal axis (esophagus, stomach, small intestine and colon). A muscular sphincter that regulates the passage of intestinal contents from one region to the next separates each intestinal domain. Formation of these sphincters is controlled by regionally specific expression of transcription factors (HOX genes) and morphogens (Smith and Tabin, 1999; Zakany and Duboule, 1999). HOX genes, which are collinearly expressed along the length of the GI tract, can be spatially and temporally correlated with morphological specialization, suggesting that they may pattern the GI tract along the rostrocaudal axis (Kawazoe et al., 2002; Pitera et al., 1999; Roberts, 2000; Sekimoto et al., 1998; Yokouchi et al., 1995). In addition to regional specification by these early patterning genes, epithelial-mesenchymal interactions function to further guide gut morphogenesis.
Development of the cecum requires FGF9 and FGF10 reciprocal signals. However, these FGFs alone do not determine the location of the cecum or the establishment of the ileo-colonic junction. During GI tract development, Fgf9 is expressed uniformly in the epithelium along the entire length of the intestine (Fig. 1) (Colvin et al., 1999). Despite this fact, loss of FGF9 has region-specific effects along the rostrocaudal axis (i.e. a loss of cecal development). In contrast to Fgf9, Fgf10 is expressed at multiple, specific mesenchymal sites along the rostrocaudal axis, including the pyloric, ileo-colonic and ano-rectal junctions (Burns et al., 2004; Fairbanks et al., 2004). FGF10 signals to an epithelial receptor, FGFR2b, and both Fgf10 and Fgfr2b mutant mice fail to develop a cecal epithelial bud (Burns et al., 2004). This is consistent with (1) known roles for FGF10 in inducing epithelial branching in the lung and salivary glands (Min et al., 1998; Sekine et al., 1999; Steinberg et al., 2005), and (2) our observation that an FGF10 bead can induce the elongation of intestinal epithelium in the absence of intestinal mesenchyme.
The ileo-colonic junction is characterized by a distinct bend in the GI tract and the formation of the cecum as an intestinal appendage. This ileo-colonic bend is maintained in both Fgf9−/− and Fgf10−/− embryos. Furthermore, Bmp4 expression is absent in the Fgf9−/− ileo-colonic junction. Therefore, this signaling molecule is also not required to specify this boundary. Cecum development initiates as a mesenchymal bud that precedes the budding and elongation of the intestinal epithelium. Examination of the effects of both gain and loss of FGF9 activity on cecal development demonstrates that Fgf9 signals to GI mesenchyme at the ileo-colonic junction, where it induces mesenchymal proliferation. It is necessary for FGF10 expression, and through FGF10, induces epithelial budding and elongation (Fig. 8).
The expression pattern of Fgf9 and the phenotype of Fgf9−/− mice suggest that FGF9 acts as a signal that drives the progression of a predetermined developmental program. In the case of cecal development, FGF9 is necessary for mesenchymal expansion and Fgf10 expression, but FGF9 does not induce significant mesenchymal growth, or expression of Fgf10, in other regions of the small intestine. This suggests that permissive domains for mesenchymal growth and Fgf10 expression are determined by other factors, possibly HOX genes in combination with other signaling molecules (Fig. 8).
In several examples of organogenesis, epithelial and mesenchymal FGFs exhibit reciprocal signaling. However, the regulation of these reciprocal signals differs in each developmental situation. In limb bud development, mesenchymal FGF10 signals to the apical ectodermal ridge; epithelial FGFs 4, 8, 9 and 17, in turn, signal back to limb mesenchyme. This reciprocal FGF signaling is required for limb development (Martin, 1998). In the lung, epithelial FGF9 signals to its mesenchymal receptors, FGFR1c and FGFR2c, and is crucial for mesenchymal proliferation and Fgf10 expression (Colvin et al., 2001b) (A.C.W., unpublished). Mesenchymal FGF10, in turn, signals to epithelial FGFR2b to induce epithelial branching morphogenesis (Arman et al., 1999; Bellusci et al., 1997; Min et al., 1998; Park et al., 1998; Sekine et al., 1999). In the lung, FGF9 is necessary for FGF10 expression, but is not the primary determinant of the pattern of Fgf10 expression. Overexpression of FGF9 in the lung can induce the expression of FGF10 throughout lung mesenchyme. However, in the intestine, FGF9 can only induce FGF10 in cecal mesenchyme.
A reciprocal signal is also required to maintain cecal development, because in Fgf10−/− embryos, which lack cecal epithelial branching, the cecal mesenchymal bud degenerates by late gestation. It is possible that low-level Fgf9 expression in cecal epithelium or other FGF10-induced epithelial factors act as survival signals for cecal mesenchyme at later stages of development and that a cecal bud lacking epithelium has insufficient survival factors to maintain cecal mesenchyme. This reciprocal signaling between cecal epithelium and mesenchyme is similar to limb bud development; however, there are also important differences between cecal and limb bud development. In the limb, FGF10 is required to initiate formation of the apical ectodermal ridge, expression of epithelial FGFs and subsequent mesenchymal expansion. By contrast, in cecal development, Fgf10 expression appears to be secondary to epithelial FGF activity and mesenchymal growth.
Finally, understanding details of FGF-mediated signaling in the developing gut should prove useful in understanding and/or manipulating injury responses in the adult intestine. For example, exogenously introduced FGFs ameliorate damage from irradiation (Paris et al., 2001) or chemical injury (dextran sodium sulfate) (Chen et al., 2002; Jeffers et al., 2002). Further understanding of the cells that express specific FGFs and their receptors in the adult epithelium and underlying mesenchyme, particularly in the region of the crypt base where epithelial stem cells reside, could provide more specific therapeutic tools to prevent or repair damage to the intestinal mucosa.
Acknowledgments
We thank G. Schmid for animal husbandry and genotyping, and B. Hogan (Fgf10) and A. McMahon (Bmp4, Shh) for providing in situ probes. This work was supported by a grant from the March of Dimes (1FY02194) and the Washington University Digestive Disease Research Core (NIH P30-DK52574).
References
- Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci USA. 1999;96:11895–11899. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Buckland RA, Collinson JM, Graham E, Davidson DR, Hill RE. Antagonistic effects of FGF4 on BMP induction of apoptosis and chondrogenesis in the chick limb bud. Mech Dev. 1998;71:143–150. doi: 10.1016/s0925-4773(98)00008-2. [DOI] [PubMed] [Google Scholar]
- Burns RC, Fairbanks TJ, Sala F, De Langhe S, Mailleux A, Thiery JP, Dickson C, Itoh N, Warburton D, Anderson KD, et al. Requirement for fibroblast growth factor 10 or fibroblast growth factor receptor 2–IIIb signaling for cecal development in mouse. Dev Biol. 2004;265:61–74. doi: 10.1016/j.ydbio.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Calvert R, Pothier P. Migration of fetal intestinal intervillous cells in neonatal mice. Anat Rec. 1990;227:199–206. doi: 10.1002/ar.1092270208. [DOI] [PubMed] [Google Scholar]
- Cardoso WV. Molecular regulation of lung development. Annu Rev Physiol. 2001;63:471–494. doi: 10.1146/annurev.physiol.63.1.471. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci USA. 2002;99:14338–14343. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001a;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- Colvin JS, White A, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001b;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks TJ, Kanard R, Del Moral PM, Sala FG, De Langhe S, Warburton D, Anderson KD, Bellusci S, Burns RC. Fibroblast growth factor receptor 2 IIIb invalidation – a potential cause of familial duodenal atresia. J Pediatr Surg. 2004;39:872–874. doi: 10.1016/j.jpedsurg.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Melton DA. Endoderm development: from patterning to organogenesis. Trends Genet. 2000;16:124–130. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- Haffen K, Kedinger M, Simon-Assmann P. Mesenchyme-dependent differentiation of epithelial progenitor cells in the gut. J Pediatr Gastroenterol Nutr. 1987;6:14–23. doi: 10.1097/00005176-198701000-00005. [DOI] [PubMed] [Google Scholar]
- Jeffers M, McDonald WF, Chillakuru RA, Yang M, Nakase H, Deegler LL, Sylander ED, Rittman B, Bendele A, Sartor RB, et al. A novel human fibroblast growth factor treats experimental intestinal inflammation. Gastroenterology. 2002;123:1151–1162. doi: 10.1053/gast.2002.36041. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The Atlas of Mouse Development. San Diego (CA), London: Academic Press; 1992. [Google Scholar]
- Kawazoe Y, Sekimoto T, Araki M, Takagi K, Araki K, Yamamura K. Region-specific gastrointestinal Hox code during murine embryonal gut development. Dev Growth Differ. 2002;44:77–84. doi: 10.1046/j.1440-169x.2002.00623.x. [DOI] [PubMed] [Google Scholar]
- Kedinger M, Duluc I, Fritsch C, Lorentz O, Plateroti M, Freund JN. Intestinal epithelial-mesenchymal cell interactions. Ann New York Acad Sci. 1998;859:1–17. doi: 10.1111/j.1749-6632.1998.tb11107.x. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- Koike T, Yasugi S. In vitro analysis of mesenchymal influences on the differentiation of stomach epithelial cells of the chicken embryo. Differentiation. 1999;65:13–25. doi: 10.1046/j.1432-0436.1999.6510013.x. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lebeche D, Malpel S, Cardoso WV. Fibroblast growth factor interactions in the developing lung. Mech Dev. 1999;86:125–136. doi: 10.1016/s0925-4773(99)00124-0. [DOI] [PubMed] [Google Scholar]
- Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Paris F, Fuks Z, Kang A, Capodieci P, Juan G, Ehleiter D, Haimovitz-Friedman A, Cordon-Cardo C, Kolesnick R. Endothelial apoptosis as the primary lesion initiating intestinal radiation damage in mice. Science. 2001;293:293–297. doi: 10.1126/science.1060191. [DOI] [PubMed] [Google Scholar]
- Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- Pitera JE, Smith VV, Thorogood P, Milla PJ. Coordinated expression of 3′ hox genes during murine embryonal gut development: an enteric Hox code. Gastroenterology. 1999;117:1339–1351. doi: 10.1016/s0016-5085(99)70284-2. [DOI] [PubMed] [Google Scholar]
- Roberts DJ. Molecular mechanisms of development of the gastrointestinal tract. Dev Dyn. 2000;219:109–120. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1047>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Johnson RL, Burke AC, Nelson CE, Morgan BA, Tabin C. Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut. Development. 1995;121:3163–3174. doi: 10.1242/dev.121.10.3163. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Smith DM, Goff DJ, Tabin CJ. Epithelial-mesenchymal signaling during the regionalization of the chick gut. Development. 1998;125:2791–2801. doi: 10.1242/dev.125.15.2791. [DOI] [PubMed] [Google Scholar]
- Scherz PJ, Harfe BD, McMahon AP, Tabin CJ. The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science. 2004;305:396–399. doi: 10.1126/science.1096966. [DOI] [PubMed] [Google Scholar]
- Schmidt GH, Winton DJ, Ponder BA. Development of the pattern of cell renewal in the crypt-villus unit of chimaeric mouse small intestine. Development. 1988;103:785–790. doi: 10.1242/dev.103.4.785. [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Yoshinobu K, Yoshida M, Kuratani S, Fujimoto S, Araki M, Tajima N, Araki K, Yamamura K. Region-specific expression of murine Hox genes implies the Hox code-mediated patterning of the digestive tract. Genes Cells. 1998;3:51–64. doi: 10.1046/j.1365-2443.1998.00167.x. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Shannon JM, Hyatt BA. Epithelial-mesenchymal interactions in the developing lung. Annu Rev Physiol. 2004;66:625–645. doi: 10.1146/annurev.physiol.66.032102.135749. [DOI] [PubMed] [Google Scholar]
- Smith DM, Tabin CJ. BMP signalling specifies the pyloric sphincter. Nature. 1999;402:748–749. doi: 10.1038/45439. [DOI] [PubMed] [Google Scholar]
- Snell GD, Stevens LC. Biology of the laboratory mouse. In: Green EL, editor. Early embryology. New York: McGraw-Hill; 1966. pp. 205–245. [Google Scholar]
- Stappenbeck TS, Gordon JI. Rac1 mutations produce aberrant epithelial differentiation in the developing and adult mouse small intestine. Development. 2000;127:2629–2642. doi: 10.1242/dev.127.12.2629. [DOI] [PubMed] [Google Scholar]
- Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, Hoffman MP. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 2005;132:1223–1234. doi: 10.1242/dev.01690. [DOI] [PubMed] [Google Scholar]
- Tanaka EM, Gann AF. Limb development. The budding role of FGF. Curr Biol. 1995;5:594–597. doi: 10.1016/s0960-9822(95)00118-7. [DOI] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Sakiyama J, Kuroiwa A. Coordinated expression of Abd-B subfamily genes of the HoxA cluster in the developing digestive tract of chick embryo. Dev Biol. 1995;169:76–89. doi: 10.1006/dbio.1995.1128. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. Hox genes and the making of sphincters. Nature. 1999;401:761–762. doi: 10.1038/44511. [DOI] [PubMed] [Google Scholar]
- Zuniga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]