Abstract
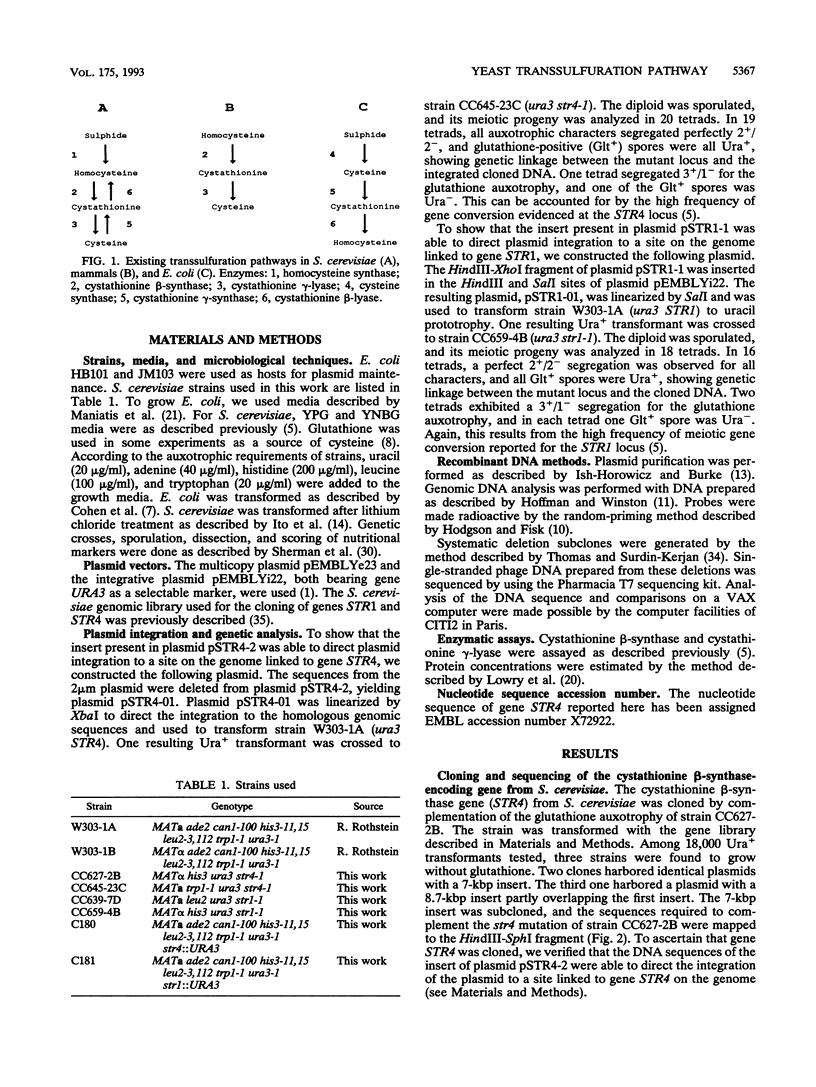
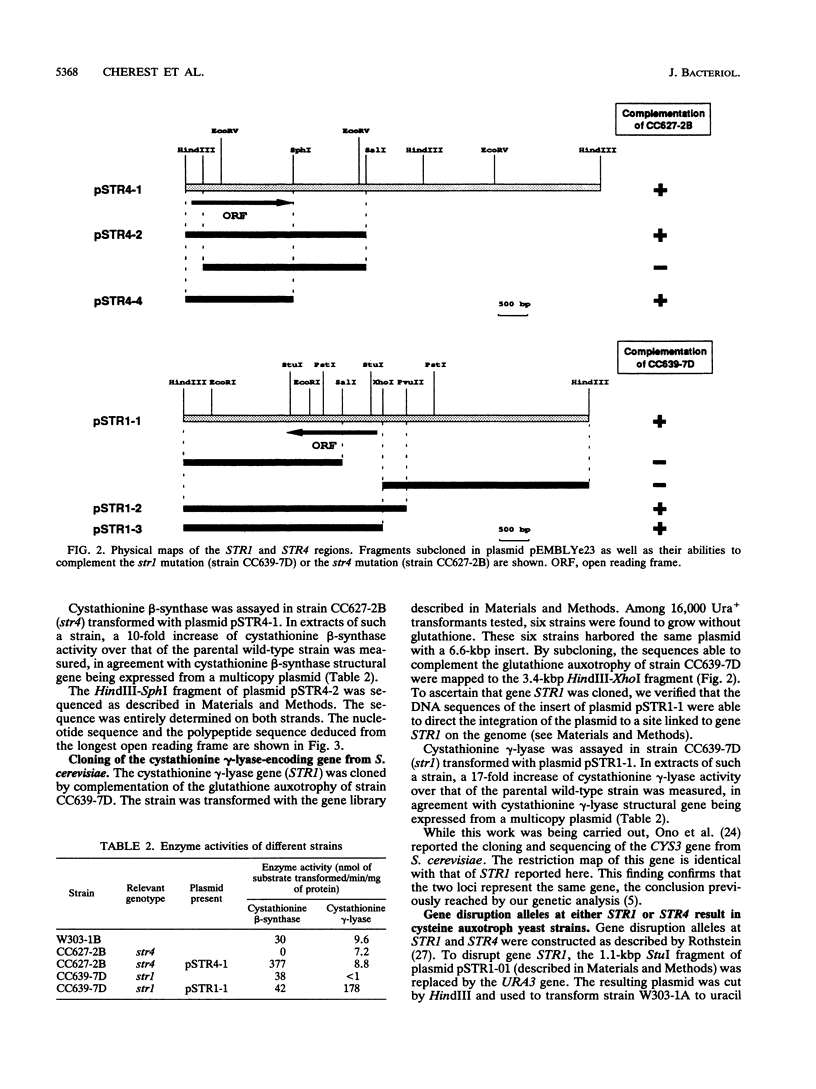
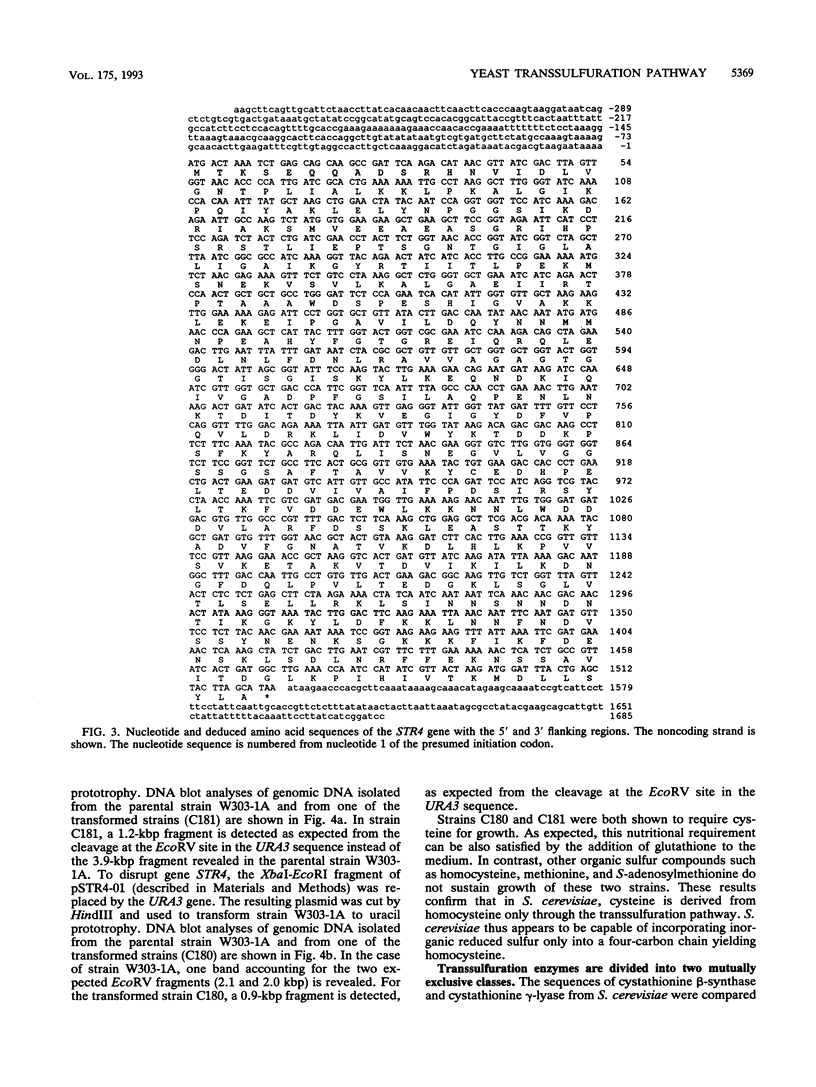
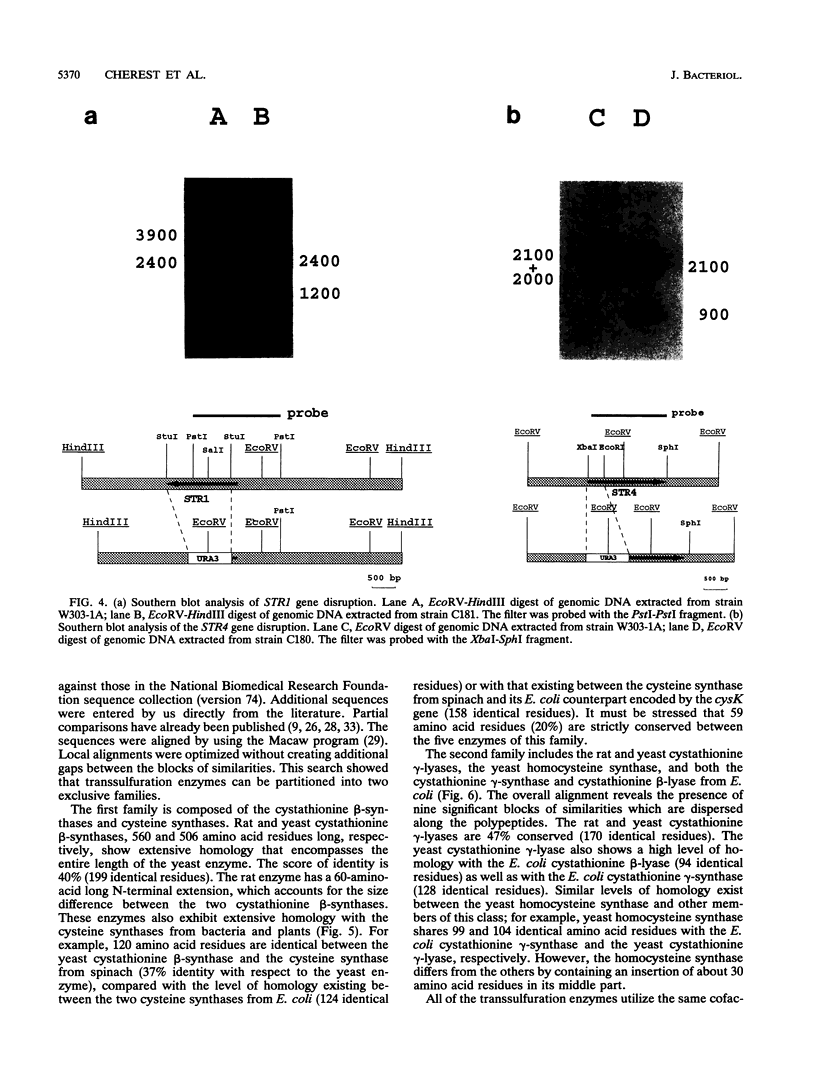
The transsulfuration pathways allow the interconversion of homocysteine and cysteine with the intermediary formation of cystathionine. The various organisms studied up to now incorporate reduced sulfur into a three- or a four-carbon chain and use differently the transsulfuration pathways to synthesize sulfur amino acids. In enteric bacteria, the synthesis of cysteine is the first step of organic sulfur metabolism and homocysteine is derived from cysteine. Fungi are capable of incorporating reduced sulfur into a four-carbon chain, and they possess two operating transsulfuration pathways. By contrast, synthesis of cysteine from homocysteine is the only existing transsulfuration pathway in mammals. In Saccharomyces cerevisiae, genetic, phenotypic, and enzymatic study of mutants has allowed us to demonstrate that homocysteine is the first sulfur amino acid to be synthesized and cysteine is derived only from homocysteine (H. Cherest and Y. Surdin-Kerjan, Genetics 130:51-58, 1992). We report here the cloning of genes STR4 and STR1, encoding cystathionine beta-synthase and cystathionine gamma-lyase, respectively. The only phenotypic consequence of the inactivation of STR1 or STR4 is cysteine auxotrophy. The sequencing of gene STR4 has allowed us to compare all of the known sequences of transsulfuration enzymes and enzymes catalyzing the incorporation of reduced sulfur in carbon chains. These comparisons reveal a partition into two families based on sequence motifs. This partition mainly correlates with similarities in the catalytic mechanisms of these enzymes.
Full text
PDF
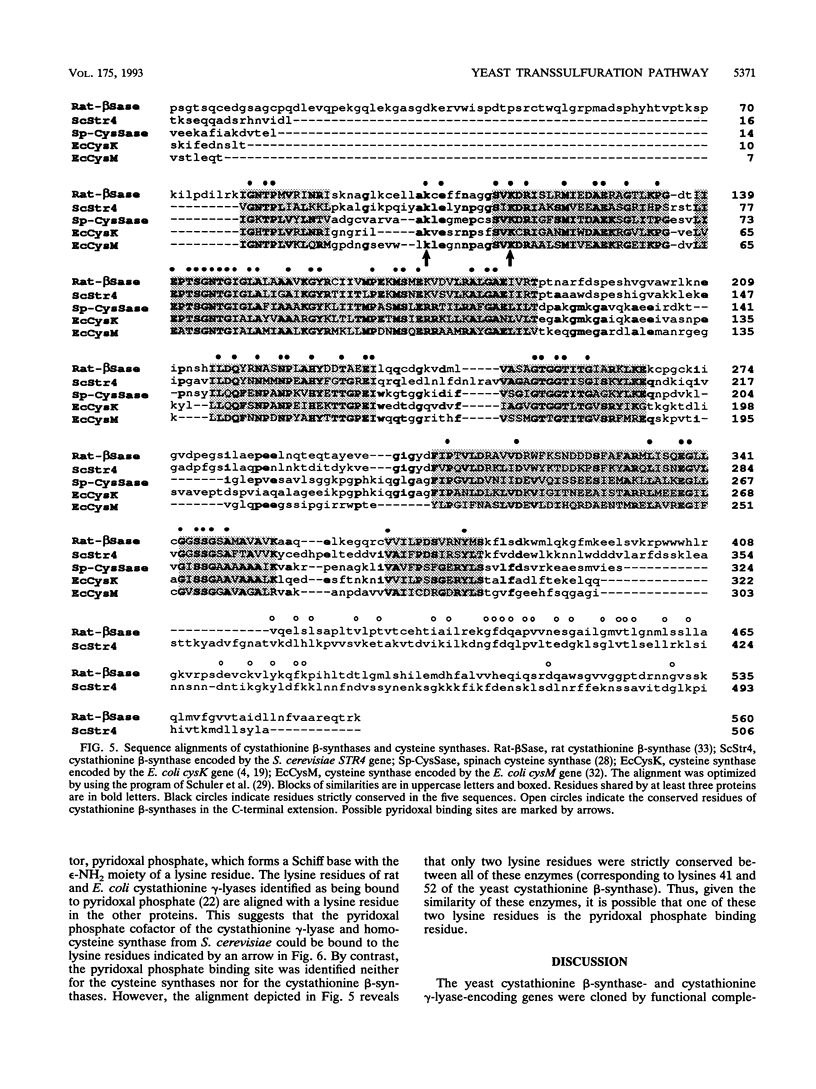



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldari C., Cesareni G. Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene. 1985;35(1-2):27–32. doi: 10.1016/0378-1119(85)90154-4. [DOI] [PubMed] [Google Scholar]
- Belfaiza J., Parsot C., Martel A., de la Tour C. B., Margarita D., Cohen G. N., Saint-Girons I. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci U S A. 1986 Feb;83(4):867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. A., D'Ari R., Newman E. B. A relationship between L-serine degradation and methionine biosynthesis in Escherichia coli K12. J Gen Microbiol. 1990 Jun;136(6):1017–1023. doi: 10.1099/00221287-136-6-1017. [DOI] [PubMed] [Google Scholar]
- Byrne C. R., Monroe R. S., Ward K. A., Kredich N. M. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J Bacteriol. 1988 Jul;170(7):3150–3157. doi: 10.1128/jb.170.7.3150-3157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H., Surdin-Kerjan Y. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics. 1992 Jan;130(1):51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H., Talbot G., Robichon-Szulmajster H. Role of homocysteine synthetase in an alternate route for methionine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1970 May;102(2):448–461. doi: 10.1128/jb.102.2.448-461.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elskens M. T., Jaspers C. J., Penninckx M. J. Glutathione as an endogenous sulphur source in the yeast Saccharomyces cerevisiae. J Gen Microbiol. 1991 Mar;137(3):637–644. doi: 10.1099/00221287-137-3-637. [DOI] [PubMed] [Google Scholar]
- Erickson P. F., Maxwell I. H., Su L. J., Baumann M., Glode L. M. Sequence of cDNA for rat cystathionine gamma-lyase and comparison of deduced amino acid sequence with related Escherichia coli enzymes. Biochem J. 1990 Jul 15;269(2):335–340. doi: 10.1042/bj2690335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson C. P., Fisk R. Z. Hybridization probe size control: optimized 'oligolabelling'. Nucleic Acids Res. 1987 Aug 11;15(15):6295–6295. doi: 10.1093/nar/15.15.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Horowitz N. H. On the Evolution of Biochemical Syntheses. Proc Natl Acad Sci U S A. 1945 Jun;31(6):153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. A. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Kerjan P., Cherest H., Surdin-Kerjan Y. Nucleotide sequence of the Saccharomyces cerevisiae MET25 gene. Nucleic Acids Res. 1986 Oct 24;14(20):7861–7871. doi: 10.1093/nar/14.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich N. M., Foote L. J., Hulanicka M. D. Studies on the mechanism of inhibition of Salmonella typhimurium by 1,2,4-triazole. J Biol Chem. 1975 Sep 25;250(18):7324–7331. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Levy S., Danchin A. Phylogeny of metabolic pathways: O-acetylserine sulphydrylase A is homologous to the tryptophan synthase beta subunit. Mol Microbiol. 1988 Nov;2(6):777–783. doi: 10.1111/j.1365-2958.1988.tb00089.x. [DOI] [PubMed] [Google Scholar]
- Martel A., Bouthier de la Tour C., Le Goffic F. Pyridoxal 5'phosphate binding site of Escherichia coli beta cystathionase and cystathionine gamma synthase comparison of their sequences. Biochem Biophys Res Commun. 1987 Sep 15;147(2):565–571. doi: 10.1016/0006-291x(87)90968-5. [DOI] [PubMed] [Google Scholar]
- Old I. G., Phillips S. E., Stockley P. G., Saint Girons I. Regulation of methionine biosynthesis in the Enterobacteriaceae. Prog Biophys Mol Biol. 1991;56(3):145–185. doi: 10.1016/0079-6107(91)90012-h. [DOI] [PubMed] [Google Scholar]
- Ono B., Tanaka K., Naito K., Heike C., Shinoda S., Yamamoto S., Ohmori S., Oshima T., Toh-e A. Cloning and characterization of the CYS3 (CYI1) gene of Saccharomyces cerevisiae. J Bacteriol. 1992 May;174(10):3339–3347. doi: 10.1128/jb.174.10.3339-3347.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsot C. Evolution of biosynthetic pathways: a common ancestor for threonine synthase, threonine dehydratase and D-serine dehydratase. EMBO J. 1986 Nov;5(11):3013–3019. doi: 10.1002/j.1460-2075.1986.tb04600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsot C., Saint-Girons I., Cohen G. N. Enzyme specialization during the evolution of amino acid biosynthetic pathways. Microbiol Sci. 1987 Sep;4(9):258, 260-2. [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saito K., Miura N., Yamazaki M., Hirano H., Murakoshi I. Molecular cloning and bacterial expression of cDNA encoding a plant cysteine synthase. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8078–8082. doi: 10.1073/pnas.89.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Simon M., Hong J. S. Direct homocysteine biosynthesis from O-succinylhomoserine in Escherichia coli: an alternate pathway that bypasses cystathionine. J Bacteriol. 1983 Jan;153(1):558–561. doi: 10.1128/jb.153.1.558-561.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko A., Hryniewicz M., Hulanicka D., Böck A. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J Bacteriol. 1990 Jun;172(6):3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop M., Bradley K., Ohura T., Tahara T., Roper M. D., Rosenberg L. E., Kraus J. P. Rat cystathionine beta-synthase. Gene organization and alternative splicing. J Biol Chem. 1992 Jun 5;267(16):11455–11461. [PubMed] [Google Scholar]
- Thomas D., Cherest H., Surdin-Kerjan Y. Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J. 1991 Mar;10(3):547–553. doi: 10.1002/j.1460-2075.1991.tb07981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Surdin-Kerjan Y. An improved strategy for generating a family of unidirectional deletions on large DNA fragments. Genet Anal Tech Appl. 1990 Jun;7(4):87–90. doi: 10.1016/0735-0651(90)90033-c. [DOI] [PubMed] [Google Scholar]
- Yamagata S. Low-molecular-weight O-acetylserine sulfhydrylase and serine sulfhydrylase of Saccharomyces cerevisiae are the same protein. J Bacteriol. 1981 Aug;147(2):688–690. doi: 10.1128/jb.147.2.688-690.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata S. Roles of O-acetyl-L-homoserine sulfhydrylases in micro-organisms. Biochimie. 1989 Nov-Dec;71(11-12):1125–1143. doi: 10.1016/0300-9084(89)90016-3. [DOI] [PubMed] [Google Scholar]
- Ycas M. On earlier states of the biochemical system. J Theor Biol. 1974 Mar;44(1):145–160. doi: 10.1016/s0022-5193(74)80035-4. [DOI] [PubMed] [Google Scholar]




