Abstract
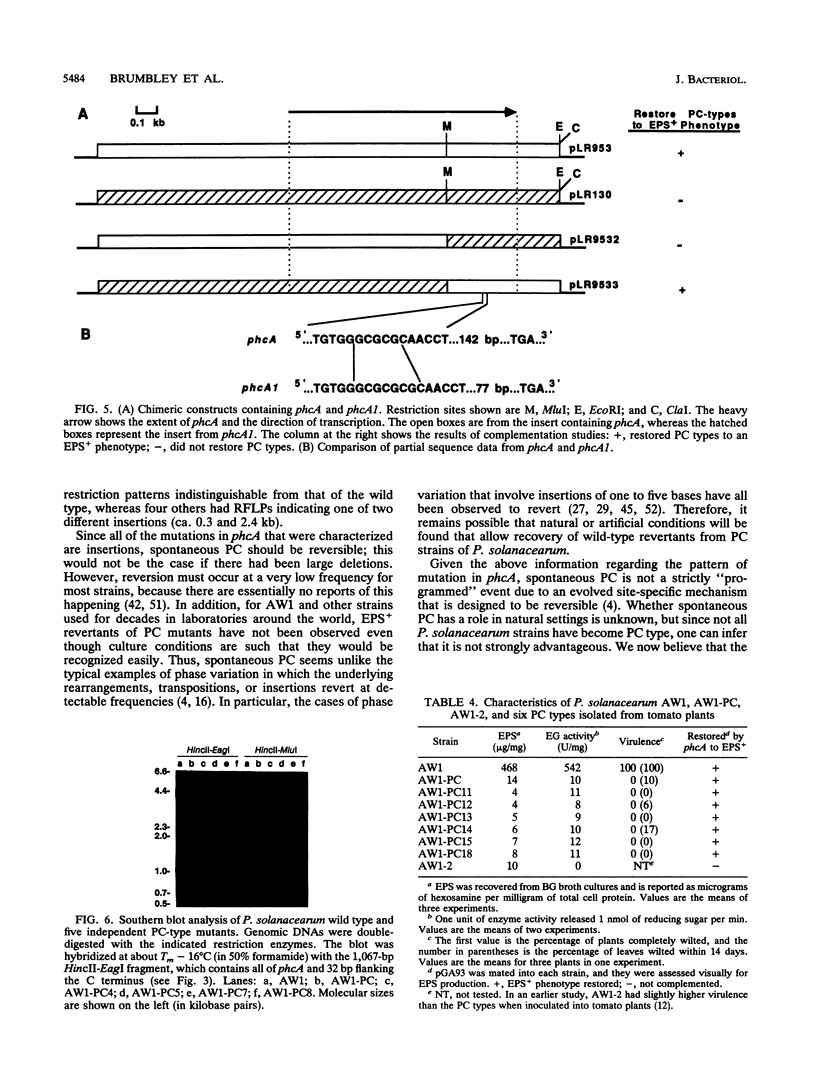
Phenotype conversion (PC) in Pseudomonas solanacearum is the coordinated change in production of extracellular polysaccharide and a variety of extracellular proteins, some of which contribute to virulence. Although PC is normally spontaneous, it is mimicked by transposon inactivation of the phcA locus (S. M. Brumbley and T. P. Denny, J. Bacteriol. 172:5677-5685, 1990). The DNA sequence of a 1.8-kb region from strain AW1 that contains phcA revealed one open reading frame that should encode a polypeptide of 38.6 kDa. The PhcA protein produced in Escherichia coli by using a T7 RNA polymerase expression system was of the predicted size. The deduced amino acid sequence of PhcA is similar to that of some members of the LysR transcriptional activator gene family, especially in the amino terminus, where a putative helix-turn-helix DNA-binding motif was identified. An analogous allele (phcA1) was cloned from the spontaneous PC mutant strain AW1-PC and found to be nonfunctional in complementation studies. When phcA1 was expressed in E. coli, the PhcA1 protein was 35.5 kDa, 3 kDa smaller than PhcA. Sequence analysis of phcA1 and chimeric constructs of phcA and phcA1 confirmed that PhcA1 is truncated by a 2-bp insertion 147 nucleotides upstream of the carboxyl terminus of PhcA. Southern blot analysis of 10 additional independently isolated PC mutants of strain AW1 revealed that two strains have larger insertions (0.2 and 1.0 kb) within phcA. These results suggest that phcA encodes a DNA-binding protein that regulates the transcription of one or more of the genes involved in P. solanacearum virulence and that spontaneous PC can be attributed to one of several different insertions within this locus.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appelbaum E. R., Thompson D. V., Idler K., Chartrain N. Rhizobium japonicum USDA 191 has two nodD genes that differ in primary structure and function. J Bacteriol. 1988 Jan;170(1):12–20. doi: 10.1128/jb.170.1.12-20.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Borst P., Greaves D. R. Programmed gene rearrangements altering gene expression. Science. 1987 Feb 6;235(4789):658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- Caldwell A. L., Gulig P. A. The Salmonella typhimurium virulence plasmid encodes a positive regulator of a plasmid-encoded virulence gene. J Bacteriol. 1991 Nov;173(22):7176–7185. doi: 10.1128/jb.173.22.7176-7185.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney B. F., Denny T. P. A cloned avirulence gene from Pseudomonas solanacearum determines incompatibility on Nicotiana tabacum at the host species level. J Bacteriol. 1990 Sep;172(9):4836–4843. doi: 10.1128/jb.172.9.4836-4843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRita V. J., Mekalanos J. J. Genetic regulation of bacterial virulence. Annu Rev Genet. 1989;23:455–482. doi: 10.1146/annurev.ge.23.120189.002323. [DOI] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Egelhoff T. T., Mulligan J. T., Long S. R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988 Mar;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Z., Schell M. A. In vivo interactions of the NahR transcriptional activator with its target sequences. Inducer-mediated changes resulting in transcription activation. J Biol Chem. 1991 Jun 15;266(17):10830–10838. [PubMed] [Google Scholar]
- Huang J. Z., Sukordhaman M., Schell M. A. Excretion of the egl gene product of Pseudomonas solanacearum. J Bacteriol. 1989 Jul;171(7):3767–3774. doi: 10.1128/jb.171.7.3767-3774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Sequeira L. Identification of a locus that regulates multiple functions in Pseudomonas solanacearum. J Bacteriol. 1990 Aug;172(8):4728–4731. doi: 10.1128/jb.172.8.4728-4731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C. C., Sequeira L. A gene cluster required for coordinated biosynthesis of lipopolysaccharide and extracellular polysaccharide also affects virulence of Pseudomonas solanacearum. J Bacteriol. 1991 Dec;173(24):7841–7847. doi: 10.1128/jb.173.24.7841-7847.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Maskell D., Moxon E. R. Genetics of Haemophilus influenzae lipopolysaccharide expression. Biochem Soc Trans. 1991 Aug;19(3):631–635. doi: 10.1042/bst0190631. [DOI] [PubMed] [Google Scholar]
- Maxon M. E., Redfield B., Cai X. Y., Shoeman R., Fujita K., Fisher W., Stauffer G., Weissbach H., Brot N. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):85–89. doi: 10.1073/pnas.86.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T. F., Gibbs C. P., Haas R. Variation and control of protein expression in Neisseria. Annu Rev Microbiol. 1990;44:451–477. doi: 10.1146/annurev.mi.44.100190.002315. [DOI] [PubMed] [Google Scholar]
- Ostrowski J., Jagura-Burdzy G., Kredich N. M. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J Biol Chem. 1987 May 5;262(13):5999–6005. [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Roberts D. P., Denny T. P., Schell M. A. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J Bacteriol. 1988 Apr;170(4):1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R. Frameshift mutations. Annu Rev Genet. 1974;8:319–346. doi: 10.1146/annurev.ge.08.120174.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Brown P. H., Raju S. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J Biol Chem. 1990 Mar 5;265(7):3844–3850. [PubMed] [Google Scholar]
- Schell M. A., Poser E. F. Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J Bacteriol. 1989 Feb;171(2):837–846. doi: 10.1128/jb.171.2.837-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Roberts D. P., Denny T. P. Analysis of the Pseudomonas solanacearum polygalacturonase encoded by pglA and its involvement in phytopathogenicity. J Bacteriol. 1988 Oct;170(10):4501–4508. doi: 10.1128/jb.170.10.4501-4508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaman H. R., Okker R. J., Lugtenberg B. J. Regulation of nodulation gene expression by NodD in rhizobia. J Bacteriol. 1992 Aug;174(16):5177–5182. doi: 10.1128/jb.174.16.5177-5182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibitz S., Aaronson W., Monack D., Falkow S. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature. 1989 Mar 16;338(6212):266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira S., Riikonen P., Saarilahti H., Sukupolvi S., Rhen M. The mkaC virulence gene of the Salmonella serovar typhimurium 96 kb plasmid encodes a transcriptional activator. Mol Gen Genet. 1991 Sep;228(3):381–384. doi: 10.1007/BF00260630. [DOI] [PubMed] [Google Scholar]
- Viale A. M., Kobayashi H., Akazawa T., Henikoff S. rbcR [correction of rcbR], a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J Bacteriol. 1991 Aug;173(16):5224–5229. doi: 10.1128/jb.173.16.5224-5229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev D., Rosengarten R., Watson-McKown R., Wise K. S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5' regulatory sequences. EMBO J. 1991 Dec;10(13):4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]





