Abstract
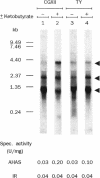
Acetohydroxy acid synthase (AHAS) and isomeroreductase (IR) catalyze subsequent reactions in the flux of metabolites towards isoleucine, valine, leucine, and pantothenate. A 4,705-bp DNA fragment from Corynebacterium glutamicum known to code for AHAS and IR was sequenced and analyzed by Northern (RNA blot) analysis. As in other bacteria, the AHAS of this gram-positive organism is encoded by two genes, ilvB and ilvN. Gene disruption verified that these genes encode the single AHAS activity in C. glutamicum. The start of ilvB was determined by amino-terminal sequencing of a fusion peptide. By Northern analysis of the ilvBNC cluster, three in vivo transcripts of 3.9, 2.3, and 1.1 kb were identified, corresponding to ilvBNC, ilvNC, and ilvC messages, respectively. The ilvC transcript (encoding IR) was by far the most abundant one. With a clone from which the ilvB upstream regions had been deleted, only the ilvNC and ilvC transcripts were synthesized, and with a clone from which the ilvN upstream regions had been deleted, only the smallest ilvC transcript was formed. It is therefore concluded that in the ilv operon of C. glutamicum, three promoters are active. The amounts of the ilvBNC and ilvNC transcripts increased in response to the addition of alpha-ketobutyrate to the growth medium. This was correlated to an increase in specific AHAS activity, whereas IR activity was not increased because of the relatively large amount of the ilvC transcript present under all conditions assayed. Therefore, the steady-state level of the ilvBNC and ilvNC messages contributes significantly to the total activity of the single AHAS. The ilvC transcript of this operon, however, is regulated independently and present in a large excess, which is in accord with the constant IR activities determined.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barak Z., Chipman D. M., Gollop N. Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. J Bacteriol. 1987 Aug;169(8):3750–3756. doi: 10.1128/jb.169.8.3750-3756.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börmann E. R., Eikmanns B. J., Sahm H. Molecular analysis of the Corynebacterium glutamicum gdh gene encoding glutamate dehydrogenase. Mol Microbiol. 1992 Feb;6(3):317–326. doi: 10.1111/j.1365-2958.1992.tb01474.x. [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Cronan J. E., Jr Common ancestry of Escherichia coli pyruvate oxidase and the acetohydroxy acid synthases of the branched-chain amino acid biosynthetic pathway. J Bacteriol. 1988 Sep;170(9):3937–3945. doi: 10.1128/jb.170.9.3937-3945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes C., Möckel B., Eggeling L., Sahm H. Cloning, organization and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene. 1992 Mar 1;112(1):113–116. doi: 10.1016/0378-1119(92)90311-c. [DOI] [PubMed] [Google Scholar]
- Friden P., Tsui P., Okamoto K., Freundlich M. Interaction of cyclic AMP receptor protein with the ilvB biosynthetic operon in E. coli. Nucleic Acids Res. 1984 Nov 12;12(21):8145–8160. doi: 10.1093/nar/12.21.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Chopin M. C., Ehrlich S. D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandoni J. A., Zahler S. A., Calvo J. M. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J Bacteriol. 1992 May;174(10):3212–3219. doi: 10.1128/jb.174.10.3212-3219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Coppola G., Calhoun D. H. Multiple transcripts encoded by the ilvGMEDA gene cluster of Escherichia coli K-12. J Bacteriol. 1992 Aug;174(15):4871–4877. doi: 10.1128/jb.174.15.4871-4877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey C. J., Warburg R. J., Halvorson H. O., Zahler S. A. Genetic and physical analysis of the ilvBC-leu region in Bacillus subtilis. Gene. 1984 Dec;32(1-2):49–56. doi: 10.1016/0378-1119(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Menkel E., Thierbach G., Eggeling L., Sahm H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl Environ Microbiol. 1989 Mar;55(3):684–688. doi: 10.1128/aem.55.3.684-688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrachko G. T., Chunduru S. K., Calvo K. C. The pH dependence of the kinetic parameters of ketol acid reductoisomerase indicates a proton shuttle mechanism for alkyl migration. Arch Biochem Biophys. 1992 May 1;294(2):446–453. doi: 10.1016/0003-9861(92)90710-e. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nilsson B., Abrahmsén L. Fusions to staphylococcal protein A. Methods Enzymol. 1990;185:144–161. doi: 10.1016/0076-6879(90)85015-g. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrumpf B., Schwarzer A., Kalinowski J., Pühler A., Eggeling L., Sahm H. A functionally split pathway for lysine synthesis in Corynebacterium glutamicium. J Bacteriol. 1991 Jul;173(14):4510–4516. doi: 10.1128/jb.173.14.4510-4516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer A., Pühler A. Manipulation of Corynebacterium glutamicum by gene disruption and replacement. Biotechnology (N Y) 1991 Jan;9(1):84–87. doi: 10.1038/nbt0191-84. [DOI] [PubMed] [Google Scholar]
- Schwinde J. W., Thum-Schmitz N., Eikmanns B. J., Sahm H. Transcriptional analysis of the gap-pgk-tpi-ppc gene cluster of Corynebacterium glutamicum. J Bacteriol. 1993 Jun;175(12):3905–3908. doi: 10.1128/jb.175.12.3905-3908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Kalinowski J., Simon R., Seep-Feldhaus A. H., Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990 Mar;172(3):1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Guss B., Nilsson B., Gatenbeck S., Philipson L., Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984 Feb 10;259(3):1695–1702. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weinstock O., Sella C., Chipman D. M., Barak Z. Properties of subcloned subunits of bacterial acetohydroxy acid synthases. J Bacteriol. 1992 Sep;174(17):5560–5566. doi: 10.1128/jb.174.17.5560-5566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Willins D. A., Ryan C. W., Platko J. V., Calvo J. M. Characterization of Lrp, and Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991 Jun 15;266(17):10768–10774. [PubMed] [Google Scholar]