Abstract
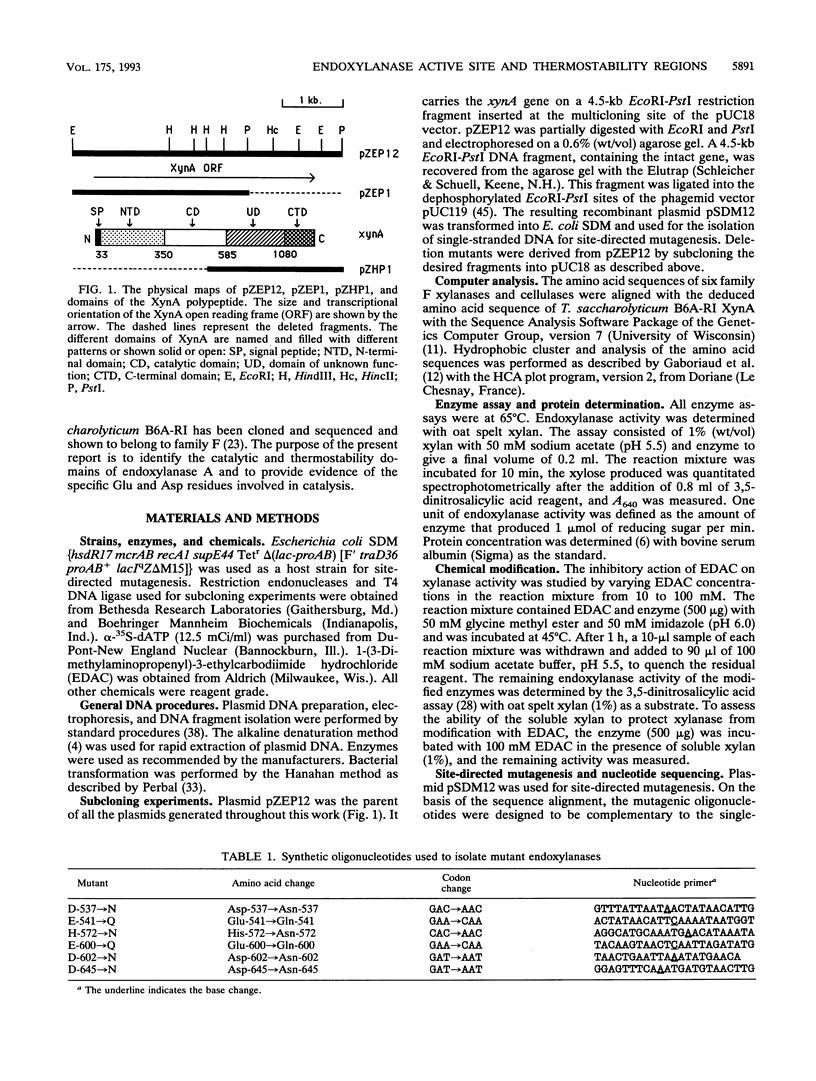
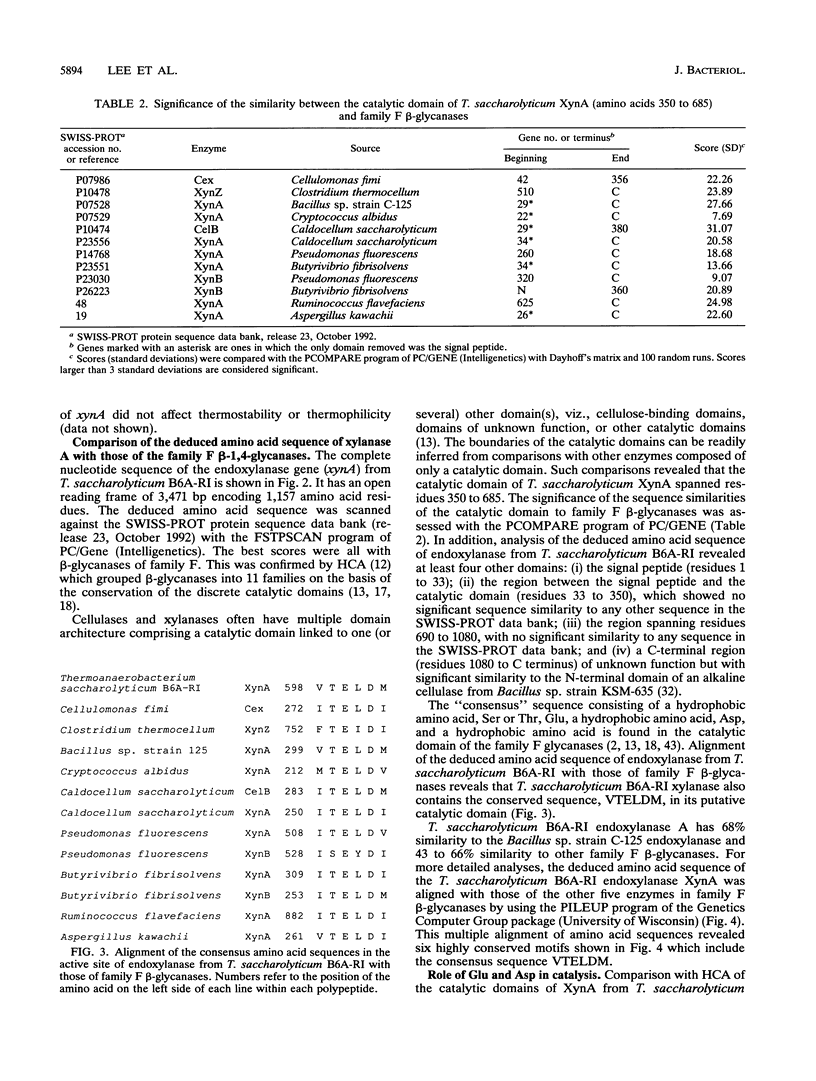
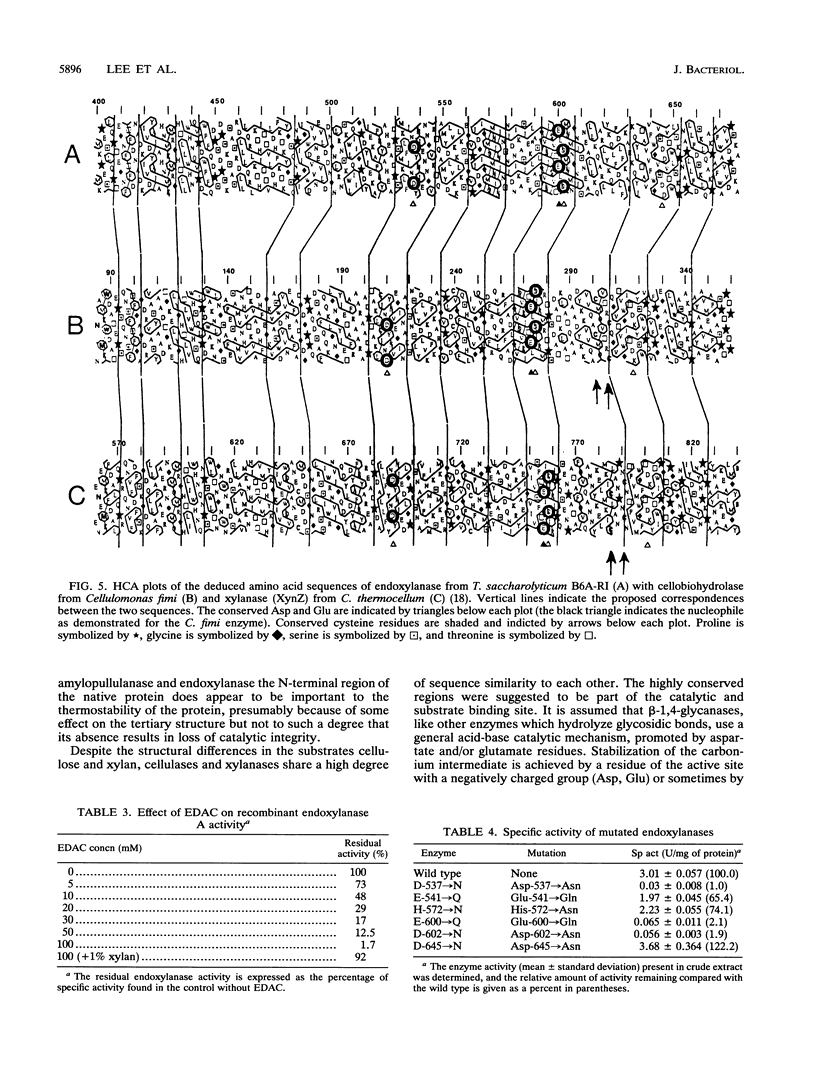
Deletion mutants were constructed from pZEP12, which contained the intact Thermoanaerobacterium saccharolyticum endoxylanase gene (xynA). Deletion of 1.75 kb from the N-terminal end of xynA resulted in a mutant enzyme that retained activity but lost thermostability. Deletion of 1.05 kb from the C terminus did not alter thermostability or activity. The deduced amino acid sequence of T. saccharolyticum B6A-RI endoxylanase XynA was aligned with five other family F beta-glycanases by using the PILEUP program of the Genetics Computer Group package. This multiple alignment of amino acid sequences revealed six highly conserved motifs which included the consensus sequence consisting of a hydrophobic amino acid, Ser or Thr, Glu, a hydrophobic amino acid, Asp, and a hydrophobic amino acid in the catalytic domain. Endoxylanase was inhibited by EDAC [1-(3-dimethylamino propenyl)-3-ethylcarbodiimide hydrochloride], suggesting that Asp and/or Glu was involved in catalysis. Three aspartic acids, two glutamic acids, and one histidine were conserved in all six enzymes aligned. Hydrophobic cluster analysis revealed that two Asp and one Glu occur in the same hydrophobic clusters in T. saccharolyticum B6A-RI endoxylanase and two other enzymes belonging to family F beta-glycanases and suggests their involvement in a catalytic triad. These two Asp and one Glu in XynA from T. saccharolyticum were targeted for analysis by site-specific mutagenesis. Substitution of Asp-537 and Asp-602 by Asn and Glu-600 by Gln completely destroyed endoxylanase activity. These results suggest that these three amino acids form a catalytic triad that functions in a general acid catalysis mechanism.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baird S. D., Hefford M. A., Johnson D. A., Sung W. L., Yaguchi M., Seligy V. L. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-beta-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1035–1039. doi: 10.1016/0006-291x(90)91998-8. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher F., Morosoli R., Durand S. Complete nucleotide sequence of the xylanase gene from the yeast Cryptococcus albidus. Nucleic Acids Res. 1988 Oct 25;16(20):9874–9874. doi: 10.1093/nar/16.20.9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bray M. R., Clarke A. J. Essential carboxy groups in xylanase A. Biochem J. 1990 Aug 15;270(1):91–96. doi: 10.1042/bj2700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Chauvaux S., Béguin P., Aubert J. P. Site-directed mutagenesis of essential carboxylic residues in Clostridium thermocellum endoglucanase CelD. J Biol Chem. 1992 Mar 5;267(7):4472–4478. [PubMed] [Google Scholar]
- Chipman D. M., Sharon N. Mechanism of lysozyme action. Science. 1969 Aug 1;165(3892):454–465. doi: 10.1126/science.165.3892.454. [DOI] [PubMed] [Google Scholar]
- Clarke A. J., Yaguchi M. The role of carboxyl groups in the function of endo-beta-1,4-glucanase from Schizophyllum commune. Eur J Biochem. 1985 Jun 3;149(2):233–238. doi: 10.1111/j.1432-1033.1985.tb08917.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grépinet O., Chebrou M. C., Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988 Oct;170(10):4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Huskisson N. S., Durrant A. J., Gilbert H. J. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol Microbiol. 1989 Sep;3(9):1211–1219. doi: 10.1111/j.1365-2958.1989.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Ito K., Ikemasu T., Ishikawa T. Cloning and sequencing of the xynA gene encoding xylanase A of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992 Jun;56(6):906–912. doi: 10.1271/bbb.56.906. [DOI] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. E., Lowe S. E., Zeikus J. G. Gene cloning, sequencing, and biochemical characterization of endoxylanase from Thermoanaerobacterium saccharolyticum B6A-RI. Appl Environ Microbiol. 1993 Sep;59(9):3134–3137. doi: 10.1128/aem.59.9.3134-3137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. E., Lowe S. E., Zeikus J. G. Regulation and Characterization of Xylanolytic Enzymes of Thermoanaerobacterium saccharolyticum B6A-RI. Appl Environ Microbiol. 1993 Mar;59(3):763–771. doi: 10.1128/aem.59.3.763-771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. L., Thomson J. A. Cloning, sequencing and expression of a gene encoding a 73 kDa xylanase enzyme from the rumen anaerobe Butyrivibrio fibrisolvens H17c. Mol Gen Genet. 1991 Aug;228(1-2):55–61. doi: 10.1007/BF00282447. [DOI] [PubMed] [Google Scholar]
- Lüthi E., Love D. R., McAnulty J., Wallace C., Caughey P. A., Saul D., Bergquist P. L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Apr;56(4):1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannarelli B. M., Evans S., Lee D. Cloning, sequencing, and expression of a xylanase gene from the anaerobic ruminal bacterium Butyrivibrio fibrisolvens. J Bacteriol. 1990 Aug;172(8):4247–4254. doi: 10.1128/jb.172.8.4247-4254.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Hata Y., Yamaguchi H., Sato M., Shinmyo A., Tanaka N., Okada H., Katsube Y. Crystallization and preliminary X-ray studies of Bacillus pumilus IPO xylanase. J Mol Biol. 1987 Jan 5;193(1):237–238. doi: 10.1016/0022-2836(87)90644-9. [DOI] [PubMed] [Google Scholar]
- O'Neill G., Goh S. H., Warren R. A., Kilburn D. G., Miller R. C., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44(2-3):325–330. doi: 10.1016/0378-1119(86)90197-6. [DOI] [PubMed] [Google Scholar]
- Ozaki K., Shikata S., Kawai S., Ito S., Okamoto K. Molecular cloning and nucleotide sequence of a gene for alkaline cellulase from Bacillus sp. KSM-635. J Gen Microbiol. 1990 Jul;136(7):1327–1334. doi: 10.1099/00221287-136-7-1327. [DOI] [PubMed] [Google Scholar]
- Podkovyrov S. M., Burdette D., Zeikus J. G. Analysis of the catalytic center of cyclomaltodextrinase from Thermoanaerobacter ethanolicus 39E. FEBS Lett. 1993 Feb 15;317(3):259–262. doi: 10.1016/0014-5793(93)81288-b. [DOI] [PubMed] [Google Scholar]
- Py B., Bortoli-German I., Haiech J., Chippaux M., Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng. 1991 Feb;4(3):325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- Rose D. R., Birnbaum G. I., Tan L. U., Saddler J. N. Crystallization and preliminary X-ray diffraction study of a xylanase from Trichoderma harzianum. J Mol Biol. 1987 Apr 20;194(4):755–756. doi: 10.1016/0022-2836(87)90254-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul D. J., Williams L. C., Love D. R., Chamley L. W., Bergquist P. L. Nucleotide sequence of a gene from Caldocellum saccharolyticum encoding for exocellulase and endocellulase activity. Nucleic Acids Res. 1989 Jan 11;17(1):439–439. doi: 10.1093/nar/17.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull D., Withers S. G., Gilkes N. R., Kilburn D. G., Warren R. A., Aebersold R. Glutamic acid 274 is the nucleophile in the active site of a "retaining" exoglucanase from Cellulomonas fimi. J Biol Chem. 1991 Aug 25;266(24):15621–15625. [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Ward O. P., Moo-Young M. Enzymatic degradation of cell wall and related plant polysaccharides. Crit Rev Biotechnol. 1989;8(4):237–274. doi: 10.3109/07388558909148194. [DOI] [PubMed] [Google Scholar]
- Wong K. K., Tan L. U., Saddler J. N. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. X., Flint H. J. A bifunctional xylanase encoded by the xynA gene of the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 comprises two dissimilar domains linked by an asparagine/glutamine-rich sequence. Mol Microbiol. 1992 Apr;6(8):1013–1023. doi: 10.1111/j.1365-2958.1992.tb02167.x. [DOI] [PubMed] [Google Scholar]


