Abstract
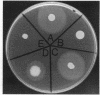
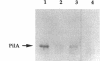
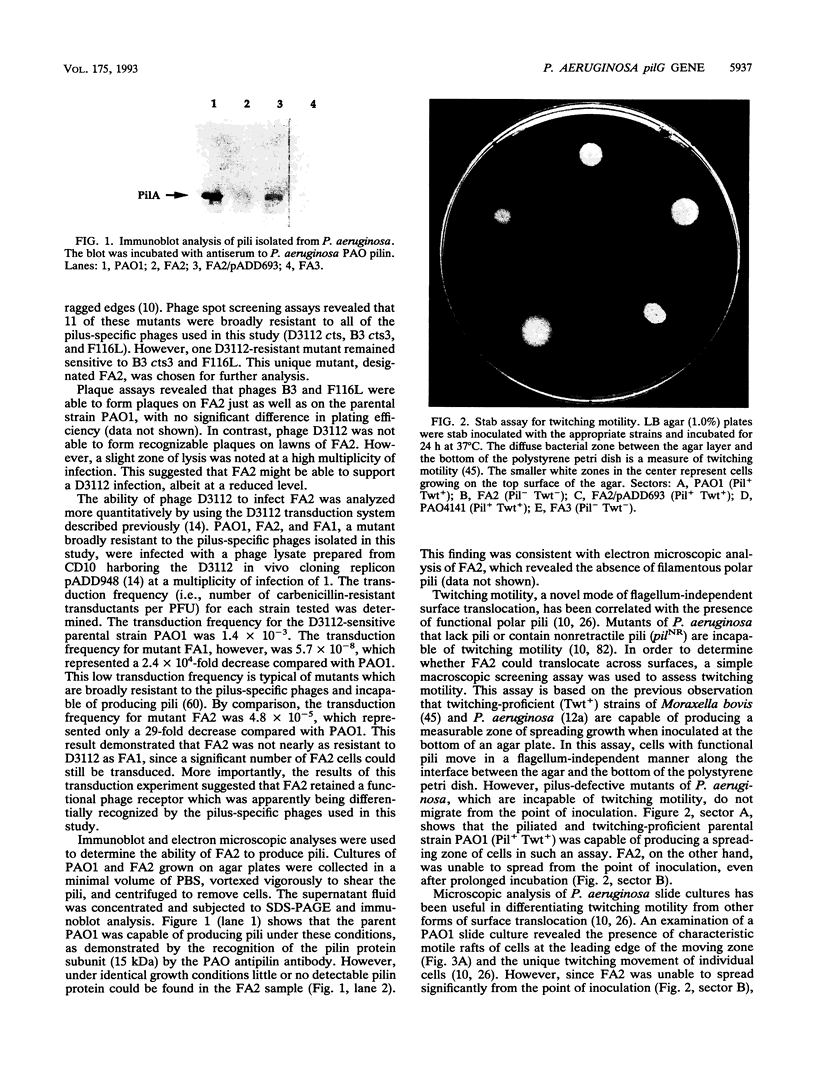
The Pseudomonas aeruginosa pilG gene, encoding a protein which is involved in pilus production, was cloned by phenotypic complementation of a unique, pilus-defective mutant of strain PAO1. This mutant, designated FA2, although resistant to the pilus-specific phage D3112 was sensitive to the pilus-specific phages B3 and F116L. In spite of the unusual phage sensitivity pattern, FA2 lacked the ability to produce functional polar pili (pil) and was incapable of twitching motility (twt). Genetic analysis revealed that the FA2 pil mutation, designated pilG1, mapped near the met-28 marker located at 20 min and was distinct from the previously described pilT mutation. This map location was confirmed by localization of a 6.2-kb EcoRI fragment that complemented FA2 on the SpeI and DpnI physical map of the P. aeruginosa PAO1 chromosome. A 700-bp region encompassing the pilG gene was sequenced, and a 405-bp open reading frame, with characteristic P. aeruginosa codon bias, was identified. The molecular weight of the protein predicted from the amino acid sequence of PilG, which was determined to be 14,717, corresponded very closely to that of a polypeptide with the apparent molecular weight of 15,000 detected after expression of pilG from the T7 promoter in Escherichia coli. Moreover, the predicted amino acid sequence of PilG showed significant homology to that of the enteric CheY protein, a single-domain response regulator. A chromosomal pilG insertion mutant, constructed by allele replacement of the wild-type gene, was not capable of pilus production or twitching motility but displayed normal flagellum-mediated motility. These results, therefore, suggest that PilG may be an important part of the signal transduction system involved in the elaboration of P. aeruginosa pili.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright L. M., Huala E., Ausubel F. M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- BRAMMAR W. J., CLARKE P. H. INDUCTION AND REPRESSION OF PSEUDOMONAS AERUGINOSA AMIDASE. J Gen Microbiol. 1964 Dec;37:307–319. doi: 10.1099/00221287-37-3-307. [DOI] [PubMed] [Google Scholar]
- Berg C. M., Vartak N. B., Wang G., Xu X., Liu L., MacNeil D. J., Gewain K. M., Wiater L. A., Berg D. E. The m gamma delta-1 element, a small gamma delta (Tn1000) derivative useful for plasmid mutagenesis, allele replacement and DNA sequencing. Gene. 1992 Apr 1;113(1):9–16. doi: 10.1016/0378-1119(92)90664-b. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980 Feb;26(2):146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Basic characterization of a Pseudomonas aeruginosa pilus-dependent bacteriophage with a long noncontractile tail. J Virol. 1973 Nov;12(5):1139–1148. doi: 10.1128/jvi.12.5.1139-1148.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E., Pitt T. L. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J Gen Virol. 1974 Jul;24(1):1–15. doi: 10.1099/0022-1317-24-1-1. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology. 1974 Mar;58(1):149–163. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. In vivo cloning of Pseudomonas aeruginosa genes with mini-D3112 transposable bacteriophage. J Bacteriol. 1989 Jul;171(7):3917–3925. doi: 10.1128/jb.171.7.3917-3925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J Bacteriol. 1989 Jul;171(7):3909–3916. doi: 10.1128/jb.171.7.3909-3916.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
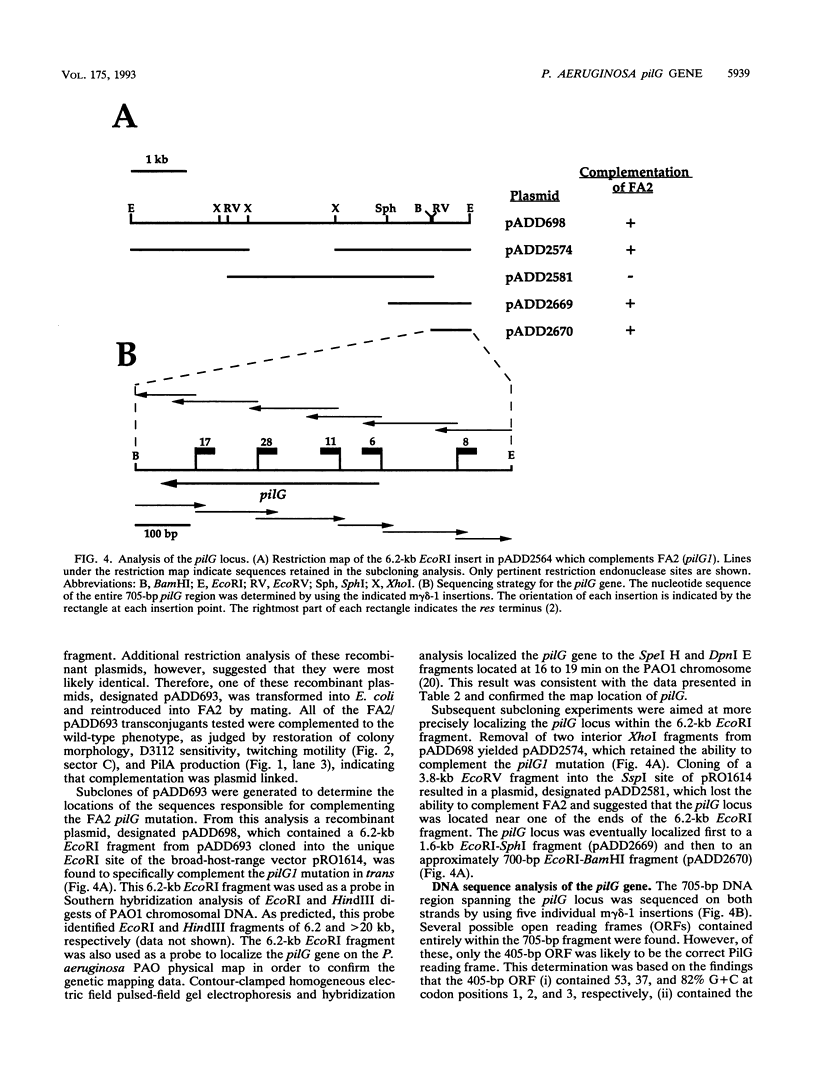
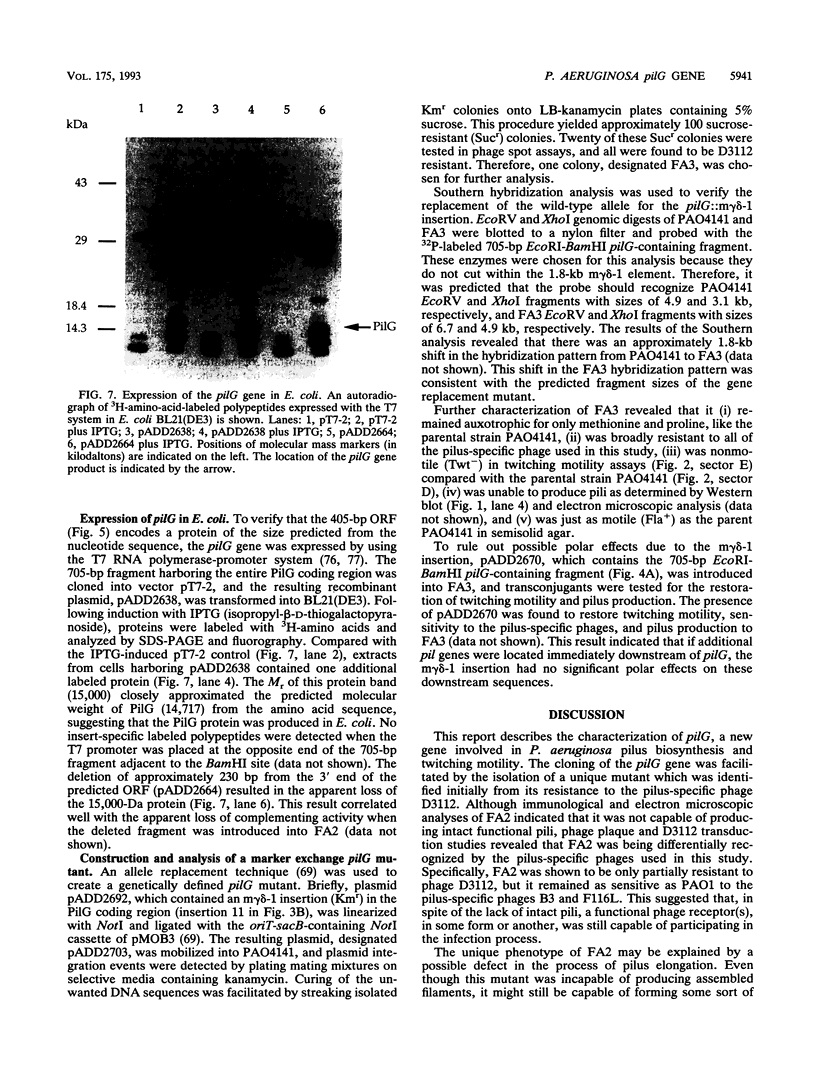
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Wang S. K., Vanags R. I., Chakrabarty A. M. Clustering of mutations affecting alginic acid biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1985 Nov;164(2):516–524. doi: 10.1128/jb.164.2.516-524.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig P., Todd T., Sastry P. A., Lee K. K., Hodges R. S., Paranchych W., Irvin R. T. Role of pili in adhesion of Pseudomonas aeruginosa to human respiratory epithelial cells. Infect Immun. 1988 Jun;56(6):1641–1646. doi: 10.1128/iai.56.6.1641-1646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Adam S. A., Choi Y. D. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984 Mar;4(3):415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha M. A., Ronald S. L., Kropinski A. M., Paranchych W. Localization of the virulence-associated genes pilA, pilR, rpoN, fliA, fliC, ent, and fbp on the physical map of Pseudomonas aeruginosa PAO1 by pulsed-field electrophoresis. Infect Immun. 1993 Apr;61(4):1571–1575. doi: 10.1128/iai.61.4.1571-1575.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Pagratis N., Casadaban M. J. Genome mapping and protein coding region identification using bacteriophage Mu. Gene. 1991 Mar 1;99(1):1–7. doi: 10.1016/0378-1119(91)90026-8. [DOI] [PubMed] [Google Scholar]
- HOLLOWAY B. W., EGAN J. B., MONK M. Lysogeny in Pseudomonas aeruginosa. Aust J Exp Biol Med Sci. 1960 Aug;38:321–329. doi: 10.1038/icb.1960.34. [DOI] [PubMed] [Google Scholar]
- HOLLOWAY B. W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955 Dec;13(3):572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- Haas D. Genetic aspects of biodegradation by pseudomonads. Experientia. 1983 Nov 15;39(11):1199–1213. doi: 10.1007/BF01990357. [DOI] [PubMed] [Google Scholar]
- Hasan N., Szybalski W. Control of cloned gene expression by promoter inversion in vivo: construction of improved vectors with a multiple cloning site and the Ptac promoter. Gene. 1987;56(1):145–151. doi: 10.1016/0378-1119(87)90167-3. [DOI] [PubMed] [Google Scholar]
- Henrichsen J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Morgan A. F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979 Mar;43(1):73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ishimoto K. S., Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto K. S., Lory S. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J Bacteriol. 1992 Jun;174(11):3514–3521. doi: 10.1128/jb.174.11.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson W. G., Jr, Higuchi J. H., Chaudhuri T. R., Woods D. E. Bacterial adherence to epithelial cells in bacillary colonization of the respiratory tract. Am Rev Respir Dis. 1980 Jan;121(1):55–63. doi: 10.1164/arrd.1980.121.1.55. [DOI] [PubMed] [Google Scholar]
- Johnson K., Lory S. Characterization of Pseudomonas aeruginosa mutants with altered piliation. J Bacteriol. 1987 Dec;169(12):5663–5667. doi: 10.1128/jb.169.12.5663-5667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnapillai V. A novel transducing phage. Its role in recognition of a possible new host-controlled modification system in Pseudomonas aeruginosa. Mol Gen Genet. 1972;114(2):134–143. doi: 10.1007/BF00332784. [DOI] [PubMed] [Google Scholar]
- Kupersztoch Y. M., Helinski D. R. A catenated DNA molecule as an intermediate in the replication of the resistance transfer factor R6K in Escherichia coli. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1451–1459. doi: 10.1016/0006-291x(73)91149-2. [DOI] [PubMed] [Google Scholar]
- Lukat G. S., Lee B. H., Mottonen J. M., Stock A. M., Stock J. B. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991 May 5;266(13):8348–8354. [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Manning P. A., Morelli G., Achtman M. traG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7487–7491. doi: 10.1073/pnas.78.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H., Nakazawa T., Ohta S., Terawaki Y. Chromosomal locations of catA, pobA, dcu and chu genes in Pseudomonas aeruginosa. Genet Res. 1981 Dec;38(3):251–266. doi: 10.1017/s0016672300020590. [DOI] [PubMed] [Google Scholar]
- Matsumura P., Rydel J. J., Linzmeier R., Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984 Oct;160(1):36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael J. C. Bacterial differentiation within Moraxella bovis colonies growing at the interface of the agar medium with the Petri dish. J Gen Microbiol. 1992 Dec;138(12):2687–2695. doi: 10.1099/00221287-138-12-2687. [DOI] [PubMed] [Google Scholar]
- Miller R. V., Ku C. M. Characterization of Pseudomonas aeruginosa mutants deficient in the establishment of lysogeny. J Bacteriol. 1978 Jun;134(3):875–883. doi: 10.1128/jb.134.3.875-883.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. A. Transformation and preservation of competent bacterial cells by freezing. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- Mutoh N., Simon M. I. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986 Jan;165(1):161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lory S. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):47–51. doi: 10.1073/pnas.89.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D., Bergman S., Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990 Jun;172(6):2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hoy K., Krishnapillai V. Recalibration of the Pseudomonas aeruginosa strain PAO chromosome map in time units using high-frequency-of-recombination donors. Genetics. 1987 Apr;115(4):611–618. doi: 10.1093/genetics/115.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranchych W., Frost L. S. The physiology and biochemistry of pili. Adv Microb Physiol. 1988;29:53–114. doi: 10.1016/s0065-2911(08)60346-x. [DOI] [PubMed] [Google Scholar]
- Pemberton J. M. F116: a DNA bacteriophage specific for the pili of Pseudomonas aeruginosa strain PAO. Virology. 1973 Oct;55(2):558–560. doi: 10.1016/0042-6822(73)90203-1. [DOI] [PubMed] [Google Scholar]
- Ramphal R., Pyle M. Evidence for mucins and sialic acid as receptors for Pseudomonas aeruginosa in the lower respiratory tract. Infect Immun. 1983 Jul;41(1):339–344. doi: 10.1128/iai.41.1.339-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramphal R., Sadoff J. C., Pyle M., Silipigni J. D. Role of pili in the adherence of Pseudomonas aeruginosa to injured tracheal epithelium. Infect Immun. 1984 Apr;44(1):38–40. doi: 10.1128/iai.44.1.38-40.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmat S., Shapiro J. A. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol Gen Genet. 1983;192(3):416–423. doi: 10.1007/BF00392184. [DOI] [PubMed] [Google Scholar]
- Roman S. J., Meyers M., Volz K., Matsumura P. A chemotactic signaling surface on CheY defined by suppressors of flagellar switch mutations. J Bacteriol. 1992 Oct;174(19):6247–6255. doi: 10.1128/jb.174.19.6247-6255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Darzins A., Casadaban M. J. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J Bacteriol. 1990 Apr;172(4):1899–1904. doi: 10.1128/jb.172.4.1899-1904.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle P. L., Matsumoto H., Holloway B. W. Genetic circularity of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1981 Jan;145(1):145–155. doi: 10.1128/jb.145.1.145-155.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. R., Tecklenburg M., Betz J. L. Plasmids containing many tandem copies of a synthetic lactose operator. Gene. 1980 Feb;8(3):279–300. doi: 10.1016/0378-1119(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry P. A., Finlay B. B., Pasloske B. L., Paranchych W., Pearlstone J. R., Smillie L. B. Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J Bacteriol. 1985 Nov;164(2):571–577. doi: 10.1128/jb.164.2.571-577.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Okinaga K., Saito H. Role of pili in the pathogenesis of Pseudomonas aeruginosa burn infection. Microbiol Immunol. 1988;32(2):131–139. doi: 10.1111/j.1348-0421.1988.tb01372.x. [DOI] [PubMed] [Google Scholar]
- Schandel K. A., Maneewannakul S., Ippen-Ihler K., Webster R. E. A traC mutant that retains sensitivity to f1 bacteriophage but lacks F pili. J Bacteriol. 1987 Jul;169(7):3151–3159. doi: 10.1128/jb.169.7.3151-3159.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandel K. A., Maneewannakul S., Vonder Haar R. A., Ippen-Ihler K., Webster R. E. Nucleotide sequence of the F plasmid gene, traC, and identification of its product. Gene. 1990 Nov 30;96(1):137–140. doi: 10.1016/0378-1119(90)90354-t. [DOI] [PubMed] [Google Scholar]
- Schandel K. A., Muller M. M., Webster R. E. Localization of TraC, a protein involved in assembly of the F conjugative pilus. J Bacteriol. 1992 Jun;174(11):3800–3806. doi: 10.1128/jb.174.11.3800-3806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer H. P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992 May;6(9):1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- Starnbach M. N., Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992 Feb;6(4):459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- Stock A., Koshland D. E., Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann M., Hamilton B. A., Mayeda C. A., Simon M. I., Meyerowitz E. M., Palazzolo M. J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S., Nunn D., Lory S. Multiple roles of the pilus biogenesis protein pilD: involvement of pilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991 Feb;173(3):1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Hobbs M., Livingston S. P., Krishnapillai V., Mattick J. S. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991 May 15;101(1):33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- Woods D. E., Straus D. C., Johanson W. G., Jr, Berry V. K., Bass J. A. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun. 1980 Sep;29(3):1146–1151. doi: 10.1128/iai.29.3.1146-1151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]