Abstract
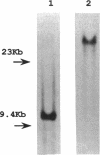
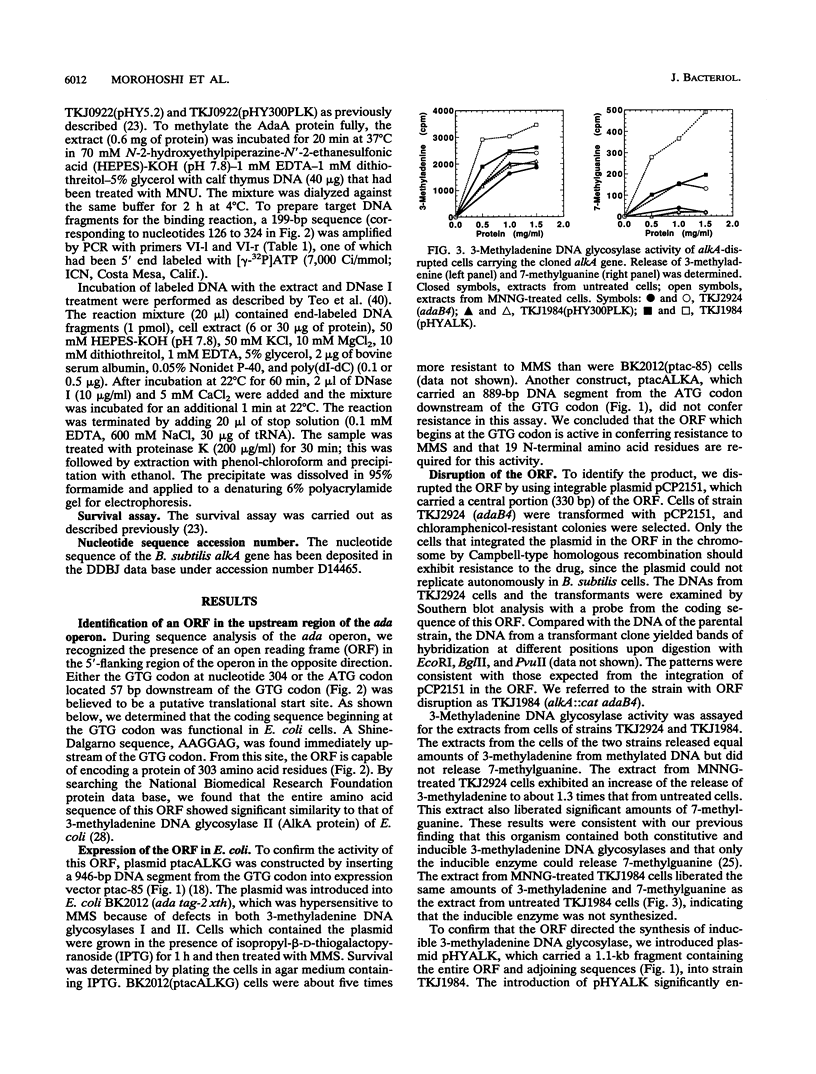
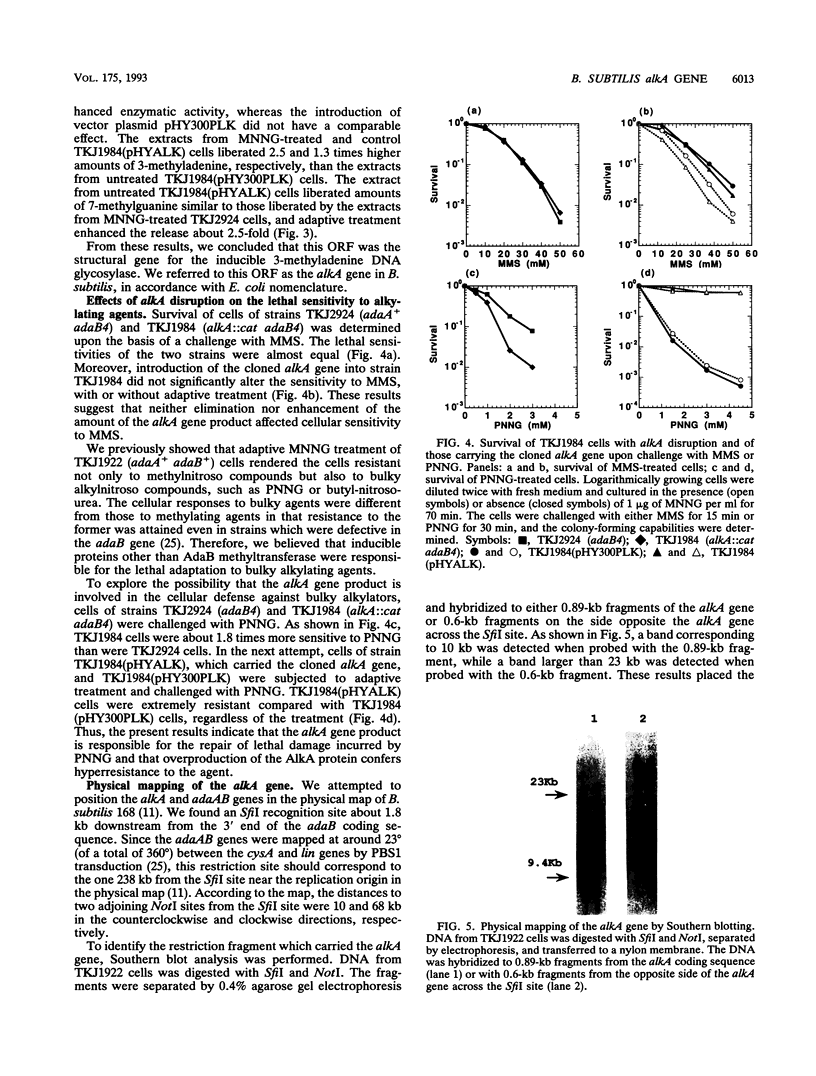
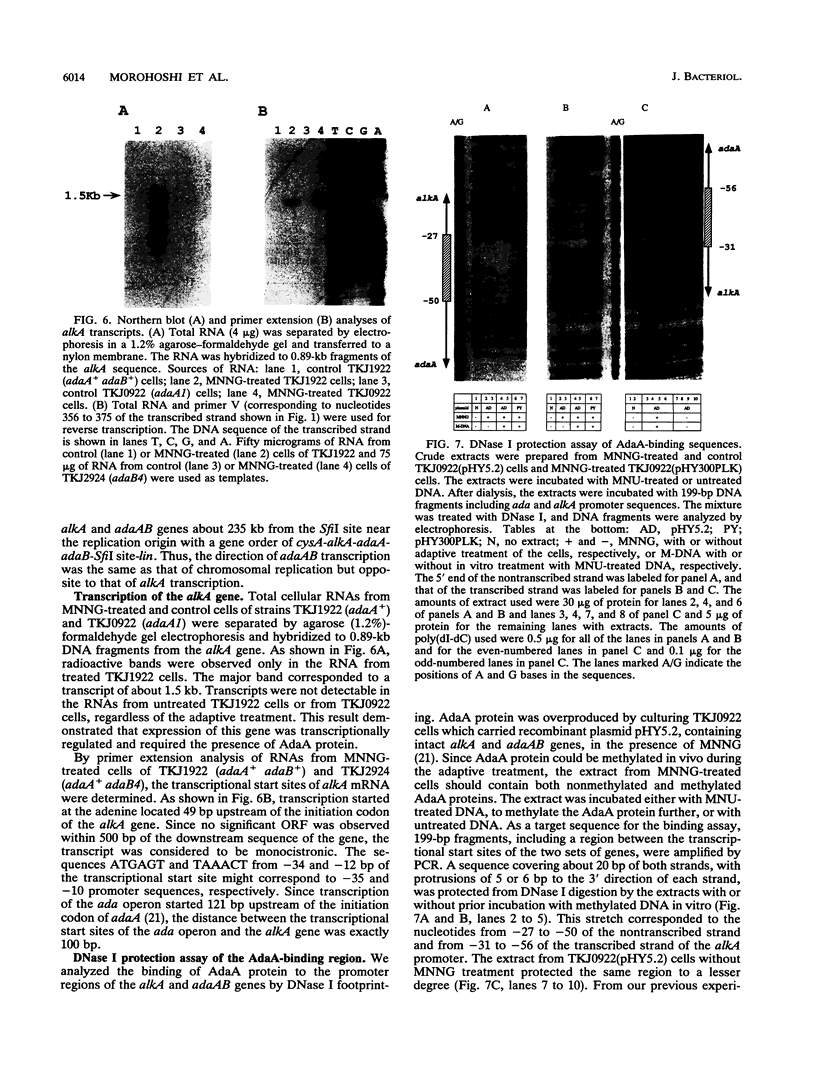
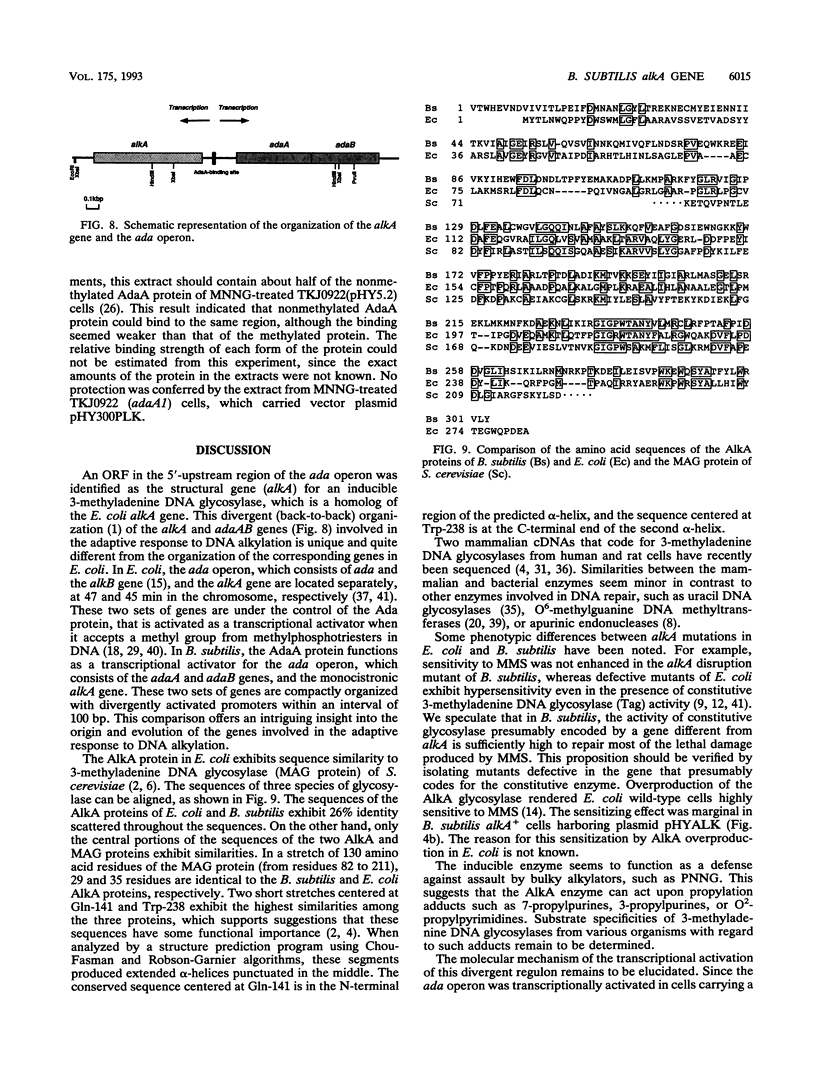
In Bacillus subtilis, the adaptive response to DNA alkylation depends on the ada operon, which consists of the adaA and adaB genes, which encode methylphosphotriester DNA methyltransferase (AdaA protein) and O6-methylguanine DNA methyltransferase (AdaB protein), respectively. A structural gene (alkA) that encodes 3-methyladenine DNA glycosylase was found upstream of the ada operon, but in the opposite orientation. This cluster of genes was mapped at about 235 kb from the SfiI recognition site near the origin of replication in the physical map of the B. subtilis chromosome. Disruption of the alkA gene sensitized cells to N-propyl-N'-nitro-N-nitrosoguanidine, while its overproduction rendered cells highly resistant to N-propyl-N'-nitro-N-nitrosoguanidine, indicating that lethal DNA damage produced by bulky alkylating agents was effectively counteracted by AlkA glycosylase. Transcription of the alkA gene was induced by treating adaA+ cells with methylating agents concurrent with transcription of the ada operon. This was accomplished by using methylated AdaA protein bound to a 30-bp segment in the middle of the 100-bp sequence between the transcriptional start sites of the alkA gene and ada operon. Thus, in this organism, the adaptive response to DNA alkylation is achieved by autologous activation of a divergent regulon composed of the genes for a DNA glycosylase and two species of DNA alkyltransferase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck C. F., Warren R. A. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988 Sep;52(3):318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdal K. G., Bjørås M., Bjelland S., Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990 Dec;9(13):4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent T. P. Partial purification and characterization of a human 3-methyladenine-DNA glycosylase. Biochemistry. 1979 Mar 6;18(5):911–916. doi: 10.1021/bi00572a028. [DOI] [PubMed] [Google Scholar]
- Chakravarti D., Ibeanu G. C., Tano K., Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991 Aug 25;266(24):15710–15715. [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke N. D., Kvaal M., Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197(3):368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- Demple B., Herman T., Chen D. S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Itaya M., Tanaka T. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J Mol Biol. 1991 Aug 5;220(3):631–648. doi: 10.1016/0022-2836(91)90106-g. [DOI] [PubMed] [Google Scholar]
- Kaasen I., Evensen G., Seeberg E. Amplified expression of the tag+ and alkA+ genes in Escherichia coli: identification of gene products and effects on alkylation resistance. J Bacteriol. 1986 Nov;168(2):642–647. doi: 10.1128/jb.168.2.642-647.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Kondo H., Nakabeppu Y., Kataoka H., Kuhara S., Kawabata S., Sekiguchi M. Structure and expression of the alkB gene of Escherichia coli related to the repair of alkylated DNA. J Biol Chem. 1986 Nov 25;261(33):15772–15777. [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976 Jan 1;259(5538):64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Marsh P. Ptac-85, an E. coli vector for expression of non-fusion proteins. Nucleic Acids Res. 1986 Apr 25;14(8):3603–3603. doi: 10.1093/nar/14.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Bacillus subtilis ada operon encodes two DNA alkyltransferases. Nucleic Acids Res. 1990 Sep 25;18(18):5473–5480. doi: 10.1093/nar/18.18.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Bacillus subtilis gene coding for constitutive O6-methylguanine-DNA alkyltransferase. Nucleic Acids Res. 1989 Aug 25;17(16):6531–6543. doi: 10.1093/nar/17.16.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Molecular analysis of Bacillus subtilis ada mutants deficient in the adaptive response to simple alkylating agents. J Bacteriol. 1991 Dec;173(24):7834–7840. doi: 10.1128/jb.173.24.7834-7840.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Munakata N. Bacillus subtilis mutants deficient in the adaptive response to simple alkylating agents. J Bacteriol. 1985 Mar;161(3):825–830. doi: 10.1128/jb.161.3.825-830.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Munakata N. Multiple species of Bacillus subtilis DNA alkyltransferase involved in the adaptive response to simple alkylating agents. J Bacteriol. 1987 Feb;169(2):587–592. doi: 10.1128/jb.169.2.587-592.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Munakata N. Two classes of Bacillus subtilis mutants deficient in the adaptive response to simple alkylating agents. Mol Gen Genet. 1986 Feb;202(2):200–206. doi: 10.1007/BF00331637. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984 Nov 25;259(22):13730–13736. [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tokumoto Y., Sakumi K., Koike G., Nakabeppu Y., Sekiguchi M. Expression of the ada gene of Escherichia coli in response to alkylating agents. Identification of transcriptional regulatory elements. J Mol Biol. 1988 Aug 5;202(3):483–494. doi: 10.1016/0022-2836(88)90280-x. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990 Oct;9(10):3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. W., Gitt M. A., Doi R. H. Isolation and physical mapping of the gene encoding the major sigma factor of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4074–4078. doi: 10.1073/pnas.80.13.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Sakumi K., Nakabeppu Y., Yamamoto Y., Kawabata S., Iwanaga S., Sekiguchi M. Purification and structure of 3-methyladenine-DNA glycosylase I of Escherichia coli. J Biol Chem. 1986 Nov 25;261(33):15761–15766. [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Samson L., Derfler B., Boosalis M., Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B. Genetic mapping of ada and adc mutations affecting the adaptive response of Escherichia coli to alkylating agents. J Bacteriol. 1982 May;150(2):984–988. doi: 10.1128/jb.150.2.984-988.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog Nucleic Acid Res Mol Biol. 1975;15(0):219–284. [PubMed] [Google Scholar]
- Tano K., Shiota S., Collier J., Foote R. S., Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci U S A. 1990 Jan;87(2):686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]