Abstract
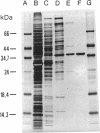
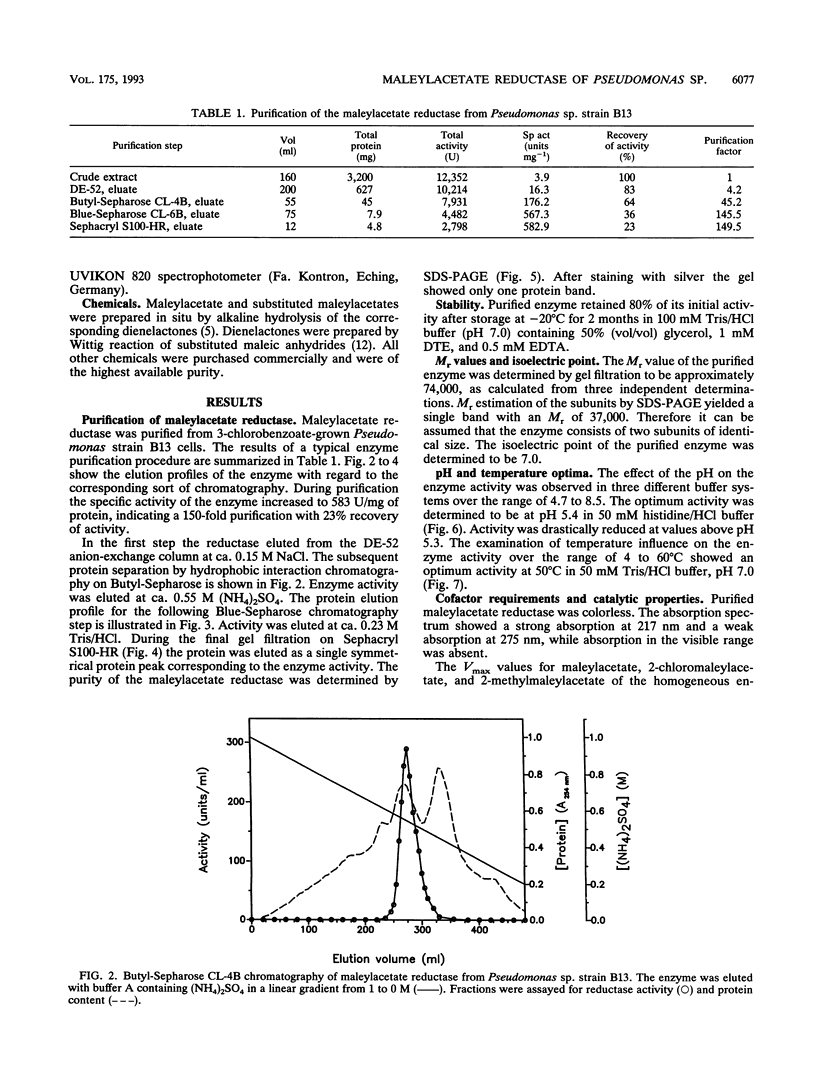
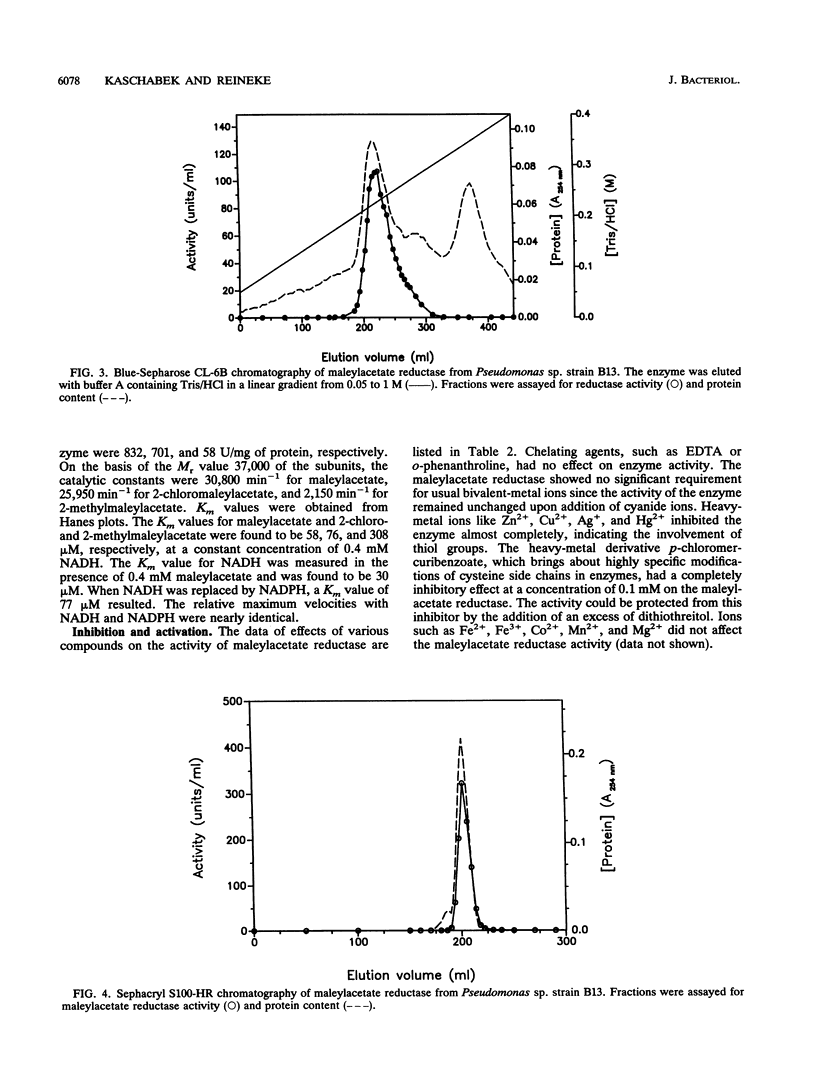
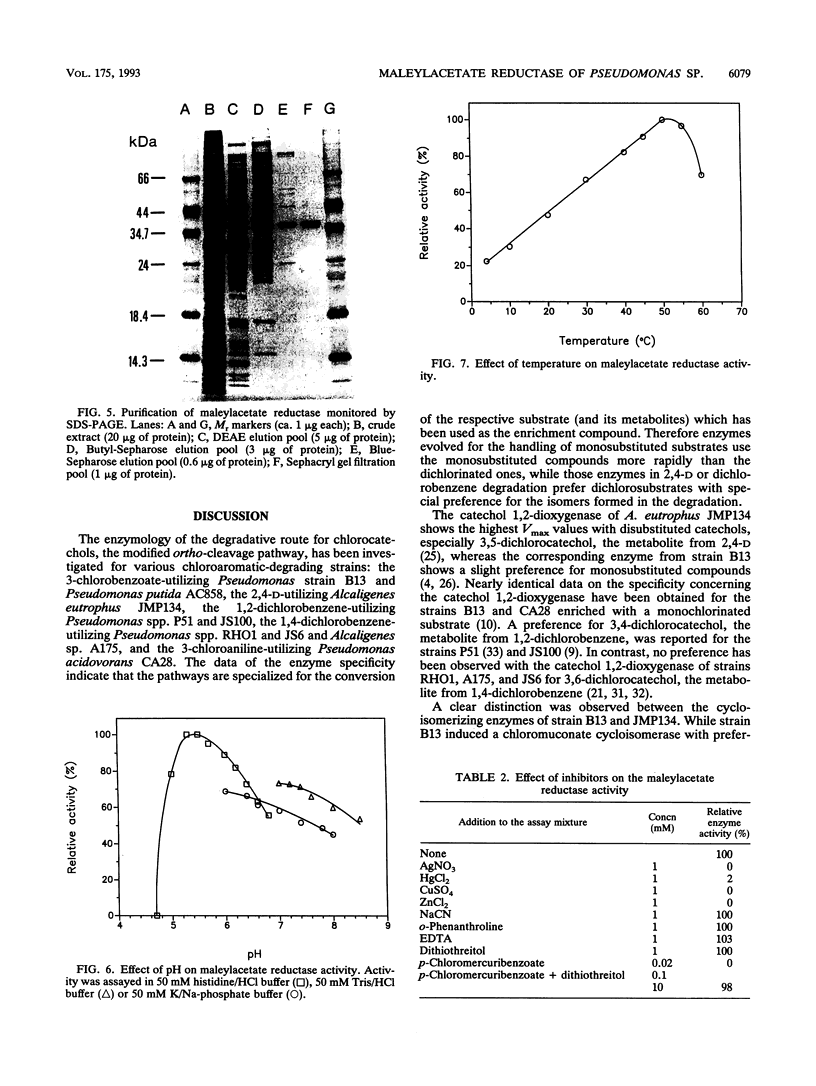
Maleylacetate reductase of Pseudomonas sp. strain B13 was purified to homogeneity by chromatography on DEAE-cellulose, Butyl-Sepharose, Blue-Sepharose, and Sephacryl S100. The final preparation gave a single band by polyacrylamide gel electrophoresis under denaturing conditions and a single symmetrical peak by gel filtration under nondenaturing conditions. The subunit M(r) value was 37,000 (determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Estimation of the native M(r) value by gel filtration gave a value of 74,000 with a Superose 6 column, indicating that the enzyme is dimeric. The pH and temperature optima were 5.4 and 50 degrees C, respectively. The pI of the enzyme was estimated to be 7.0. The apparent Km values for maleylacetate and NADH were 58 and 30 microM, respectively, and the maximum velocity was 832 U/mg of protein for maleylacetate. Maleylacetate and various substituted maleylacetates, such as 2-chloro- and 2-methyl-maleylacetate, were reduced at significant rates. NADPH was also used as a cofactor instead of NADH with nearly the same Vmax value, but its Km value was estimated to be 77 microM. Reductase activity was inhibited by a range of thiol-blocking reagents. The absorption spectrum indicated that there was no bound cofactor or prosthetic group in the enzyme.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broderick J. B., O'Halloran T. V. Overproduction, purification, and characterization of chlorocatechol dioxygenase, a non-heme iron dioxygenase with broad substrate tolerance. Biochemistry. 1991 Jul 23;30(29):7349–7358. doi: 10.1021/bi00243a040. [DOI] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Dorn E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978 Jul 15;174(1):85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. C., Smith B. S., Moss P., Fernley H. N. Bacterial metabolism of 4-chlorophenoxyacetate. Biochem J. 1971 May;122(4):509–517. doi: 10.1042/bj1220509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Chakrabarty A. M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Ngai K. L., Chatterjee D. K., Ornston L. N., Chakrabarty A. M. Nucleotide sequence and expression of clcD, a plasmid-borne dienelactone hydrolase gene from Pseudomonas sp. strain B13. J Bacteriol. 1987 Feb;169(2):704–709. doi: 10.1128/jb.169.2.704-709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal A. B., Neujahr H. Y. Maleylacetate reductase from Trichosporon cutaneum. Biochem J. 1980 Mar 1;185(3):783–786. doi: 10.1042/bj1850783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler B. E., Nishino S. F., Spain J. C. Degradation of 1,2-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1988 Feb;54(2):294–301. doi: 10.1128/aem.54.2.294-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinteregger C., Loidl M., Streichsbier F. Characterization of isofunctional ring-cleaving enzymes in aniline and 3-chloroaniline degradation by Pseudomonas acidovorans CA28. FEMS Microbiol Lett. 1992 Oct 15;76(3):261–266. doi: 10.1016/0378-1097(92)90346-p. [DOI] [PubMed] [Google Scholar]
- Kaschabek S. R., Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch Microbiol. 1992;158(6):412–417. doi: 10.1007/BF00276301. [DOI] [PubMed] [Google Scholar]
- Kuhm A. E., Schlömann M., Knackmuss H. J., Pieper D. H. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP 134. Biochem J. 1990 Mar 15;266(3):877–883. [PMC free article] [PubMed] [Google Scholar]
- Kukor J. J., Olsen R. H., Siak J. S. Recruitment of a chromosomally encoded maleylacetate reductase for degradation of 2,4-dichlorophenoxyacetic acid by plasmid pJP4. J Bacteriol. 1989 Jun;171(6):3385–3390. doi: 10.1128/jb.171.6.3385-3390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Ngai K. L., Schlömann M., Knackmuss H. J., Ornston L. N. Dienelactone hydrolase from Pseudomonas sp. strain B13. J Bacteriol. 1987 Feb;169(2):699–703. doi: 10.1128/jb.169.2.699-703.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Ngai K. L. Crystallization and preliminary x-ray crystallographic data of dienelactone hydrolase from Pseudomonas sp. B13. J Biol Chem. 1985 Aug 15;260(17):9818–9819. [PubMed] [Google Scholar]
- Pathak D., Ngai K. L., Ollis D. X-ray crystallographic structure of dienelactone hydrolase at 2.8 A. J Mol Biol. 1988 Nov 20;204(2):435–445. doi: 10.1016/0022-2836(88)90587-6. [DOI] [PubMed] [Google Scholar]
- Pathak D., Ollis D. Refined structure of dienelactone hydrolase at 1.8 A. J Mol Biol. 1990 Jul 20;214(2):497–525. doi: 10.1016/0022-2836(90)90196-s. [DOI] [PubMed] [Google Scholar]
- Perkins E. J., Gordon M. P., Caceres O., Lurquin P. F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990 May;172(5):2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Hybrid pathway for chlorobenzoate metabolism in Pseudomonas sp. B13 derivatives. J Bacteriol. 1980 May;142(2):467–473. doi: 10.1128/jb.142.2.467-473.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- Schmidt E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem J. 1980 Oct 15;192(1):339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraa G., Boone M. L., Jetten M. S., van Neerven A. R., Colberg P. J., Zehnder A. J. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986 Dec;52(6):1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain J. C., Nishino S. F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987 May;53(5):1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer M. D., Stadler-Fritzsche K., Schlömann M. Conversion of 2-chloromaleylacetate in Alcaligenes eutrophus JMP134. Arch Microbiol. 1993;159(2):182–188. doi: 10.1007/BF00250280. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Eggen R. I., Zehnder A. J., de Vos W. M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991 Apr;173(8):2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]