Abstract
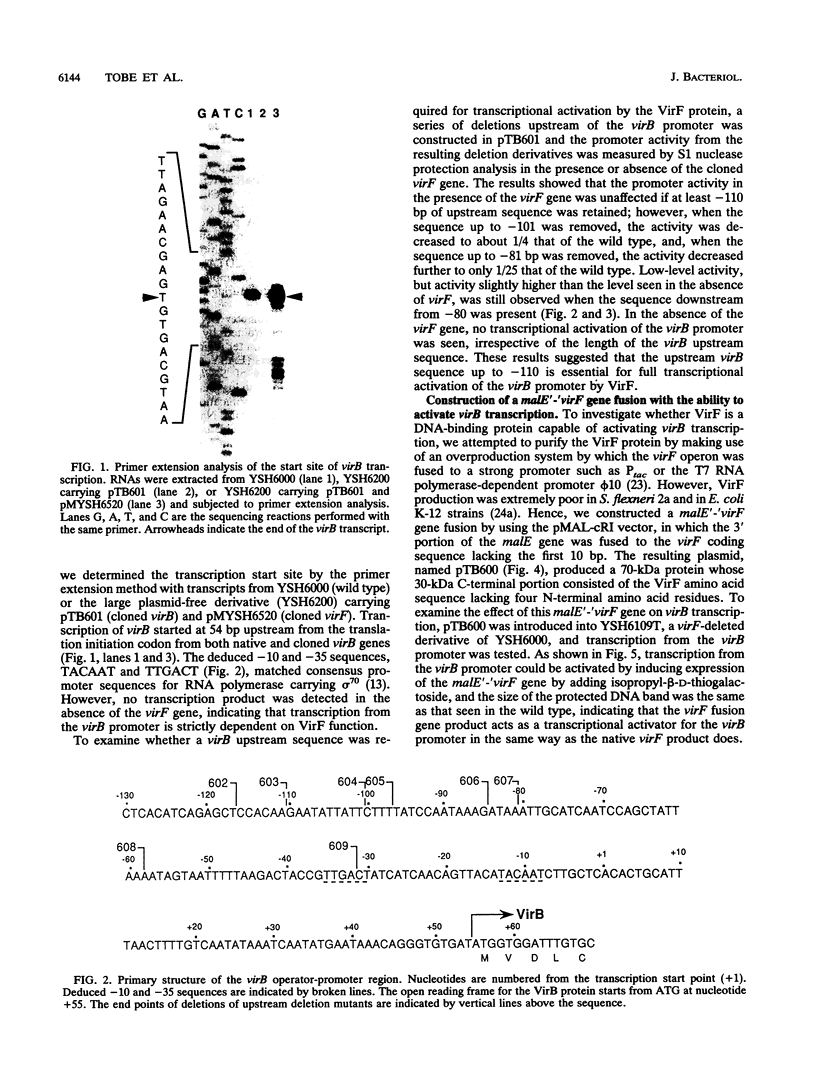
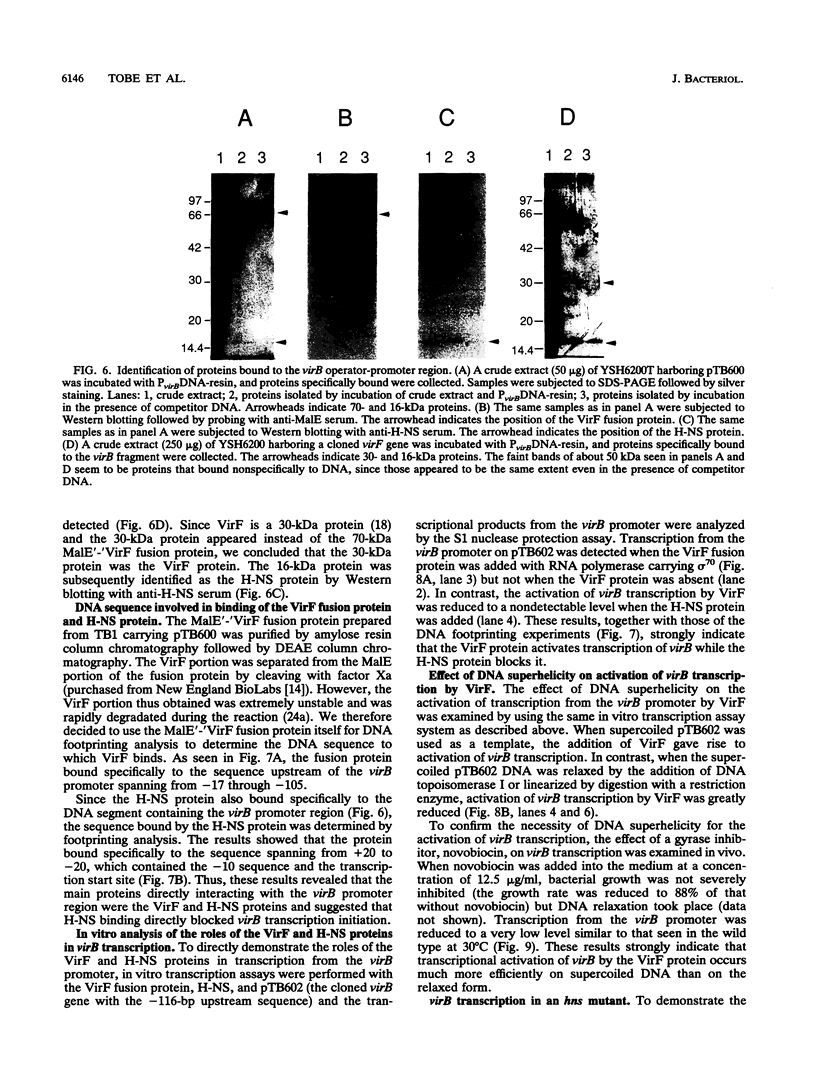
Expression of invasion genes encoded by the large 230-kb plasmid of Shigella flexneri is controlled by the virB gene, which is itself activated by another regulator, virF. Transcription of the invasion genes is temperature regulated, since they are activated in bacteria grown at 37 but not at 30 degrees C. Recently, we have shown that the thermoregulated expression of invasion genes is mediated by thermal activation of virB transcription (T. Tobe, S. Nagai, B. Adler, M. Yoshikawa, and C. Sasakawa, Mol. Microbiol. 5:887-893, 1991). It has also been shown that a mutation that inactivates H-NS, the product of virR (hns), derepresses transcription of virB. To elucidate the molecular mechanisms underlying virB activation, we determined the location of the transcription start site and found it to be 54 bp upstream of the 5' end of the virB coding sequence. Deletion analysis revealed that transcriptional activation by virF requires a DNA segment of 110 bp extending upstream of the transcription start site. By using a protein binding assay with crude extracts of S. flexneri harboring the malE'-'virF fusion gene, which was able to activate virB transcription, two protein species, one of 70 kDa (MalE'-'VirF fusion) and another of 16 kDa (H-NS), were shown to bind specifically to the virB promoter region. DNA footprinting analysis indicated that the VirF fusion and H-NS proteins bound to the upstream sequence spanning from -17 to -117 and to the sequence from -20 to +20, in which virB transcription starts, respectively. In an vitro transcription assay, the VirF fusion protein was shown to activate virB transcription while the H-NS protein blocked it. virB activation was seen only when negatively supercoiled DNA was used as a template. In in vivo studies, virB transcription was significantly decreased by adding novobiocin, a gyrase inhibitor, into the culture medium while virB transcription was increased by mutating hns. These in vitro and in vivo studies indicated that transcription of virB is activated through VirF binding to the upstream sequence of the virB promoter in a DNA-topology-dependent manner and is directly repressed by H-NS binding to the virB transcription start site.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler B., Sasakawa C., Tobe T., Makino S., Komatsu K., Yoshikawa M. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989 May;3(5):627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- Dagberg B., Uhlin B. E. Regulation of virulence-associated plasmid genes in enteroinvasive Escherichia coli. J Bacteriol. 1992 Dec;174(23):7606–7612. doi: 10.1128/jb.174.23.7606-7612.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Ni Bhriain N., Higgins C. F. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature. 1990 Apr 19;344(6268):789–792. doi: 10.1038/344789a0. [DOI] [PubMed] [Google Scholar]
- Göransson M., Forsman P., Nilsson P., Uhlin B. E. Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol Microbiol. 1989 Nov;3(11):1557–1565. doi: 10.1111/j.1365-2958.1989.tb00141.x. [DOI] [PubMed] [Google Scholar]
- Göransson M., Sondén B., Nilsson P., Dagberg B., Forsman K., Emanuelsson K., Uhlin B. E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990 Apr 12;344(6267):682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hinton J. C., Hulton C. S., Owen-Hughes T., Pavitt G. D., Seirafi A. Protein H1: a role for chromatin structure in the regulation of bacterial gene expression and virulence? Mol Microbiol. 1990 Dec;4(12):2007–2012. doi: 10.1111/j.1365-2958.1990.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Hromockyj A. E., Tucker S. C., Maurelli A. T. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1(Tyr)). Mol Microbiol. 1992 Aug;6(15):2113–2124. doi: 10.1111/j.1365-2958.1992.tb01385.x. [DOI] [PubMed] [Google Scholar]
- Hulton C. S., Seirafi A., Hinton J. C., Sidebotham J. M., Waddell L., Pavitt G. D., Owen-Hughes T., Spassky A., Buc H., Higgins C. F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990 Nov 2;63(3):631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- Jordi B. J., Dagberg B., de Haan L. A., Hamers A. M., van der Zeijst B. A., Gaastra W., Uhlin B. E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992 Jul;11(7):2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrec E. H., Schneider H., Magnani T. J., Formal S. B. EPITHELIAL CELL PENETRATION AS AN ESSENTIAL STEP IN THE PATHOGENESIS OF BACILLARY DYSENTERY. J Bacteriol. 1964 Nov;88(5):1503–1518. doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli A. T., Blackmon B., Curtiss R., 3rd Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun. 1984 Jan;43(1):397–401. doi: 10.1128/iai.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli A. T., Sansonetti P. J. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2820–2824. doi: 10.1073/pnas.85.8.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Thøgersen H. C. Generation of beta-globin by sequence-specific proteolysis of a hybrid protein produced in Escherichia coli. 1984 Jun 28-Jul 4Nature. 309(5971):810–812. doi: 10.1038/309810a0. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T. A., Pavitt G. D., Santos D. S., Sidebotham J. M., Hulton C. S., Hinton J. C., Higgins C. F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992 Oct 16;71(2):255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- Rimsky S., Spassky A. Sequence determinants for H1 binding on Escherichia coli lac and gal promoters. Biochemistry. 1990 Apr 17;29(15):3765–3771. doi: 10.1021/bi00467a024. [DOI] [PubMed] [Google Scholar]
- Sakai T., Sasakawa C., Makino S., Kamata K., Yoshikawa M. Molecular cloning of a genetic determinant for Congo red binding ability which is essential for the virulence of Shigella flexneri. Infect Immun. 1986 Feb;51(2):476–482. doi: 10.1128/iai.51.2.476-482.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Sasakawa C., Makino S., Yoshikawa M. DNA sequence and product analysis of the virF locus responsible for congo red binding and cell invasion in Shigella flexneri 2a. Infect Immun. 1986 Nov;54(2):395–402. doi: 10.1128/iai.54.2.395-402.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Sasakawa C., Yoshikawa M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton virF protein. Mol Microbiol. 1988 Sep;2(5):589–597. doi: 10.1111/j.1365-2958.1988.tb00067.x. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Kamata K., Sakai T., Makino S., Yamada M., Okada N., Yoshikawa M. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J Bacteriol. 1988 Jun;170(6):2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa C., Kamata K., Sakai T., Murayama S. Y., Makino S., Yoshikawa M. Molecular alteration of the 140-megadalton plasmid associated with loss of virulence and Congo red binding activity in Shigella flexneri. Infect Immun. 1986 Feb;51(2):470–475. doi: 10.1128/iai.51.2.470-475.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelkoul P. H., Willshaw G. A., McConnell M. M., Smith H. R., Hamers A. M., van der Zeijst B. A., Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990 Feb;8(2):91–99. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe T., Nagai S., Okada N., Adler B., Yoshikawa M., Sasakawa C. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol Microbiol. 1991 Apr;5(4):887–893. doi: 10.1111/j.1365-2958.1991.tb00762.x. [DOI] [PubMed] [Google Scholar]
- Yamada H., Muramatsu S., Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990 Sep;108(3):420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- Yamada H., Yoshida T., Tanaka K., Sasakawa C., Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991 Nov;230(1-2):332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]