Abstract
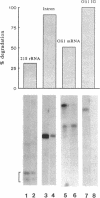
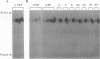
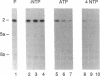
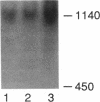
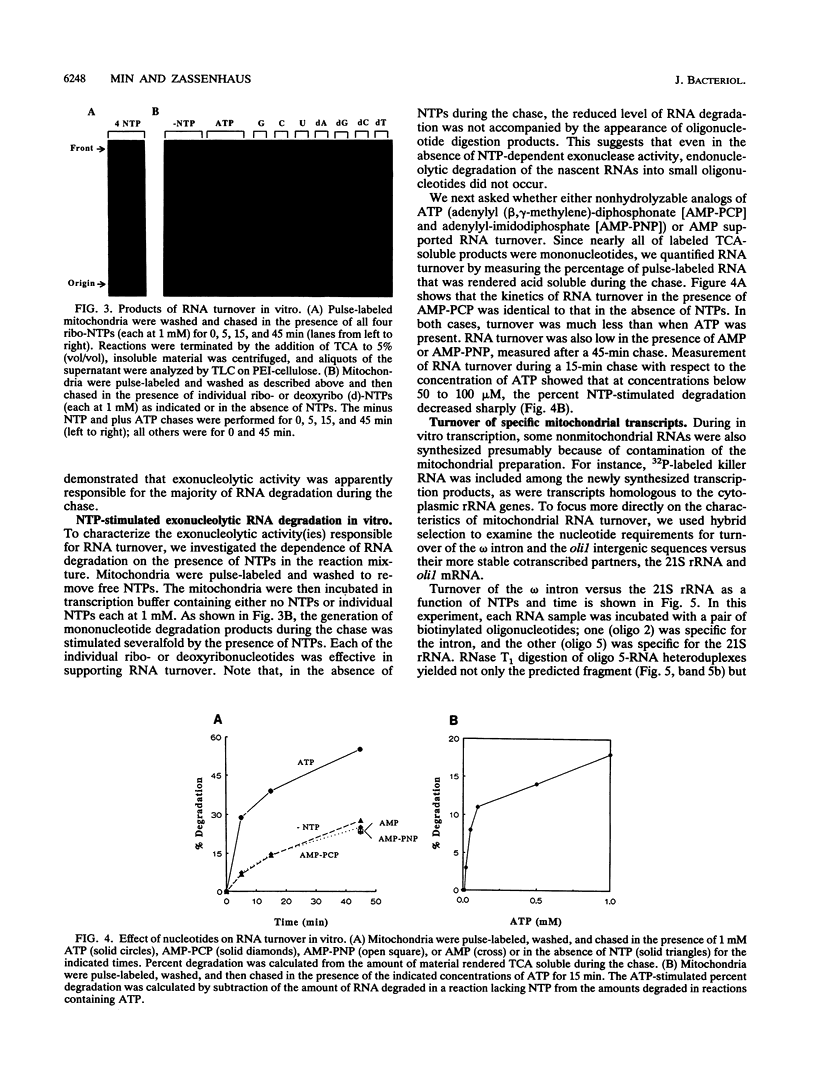
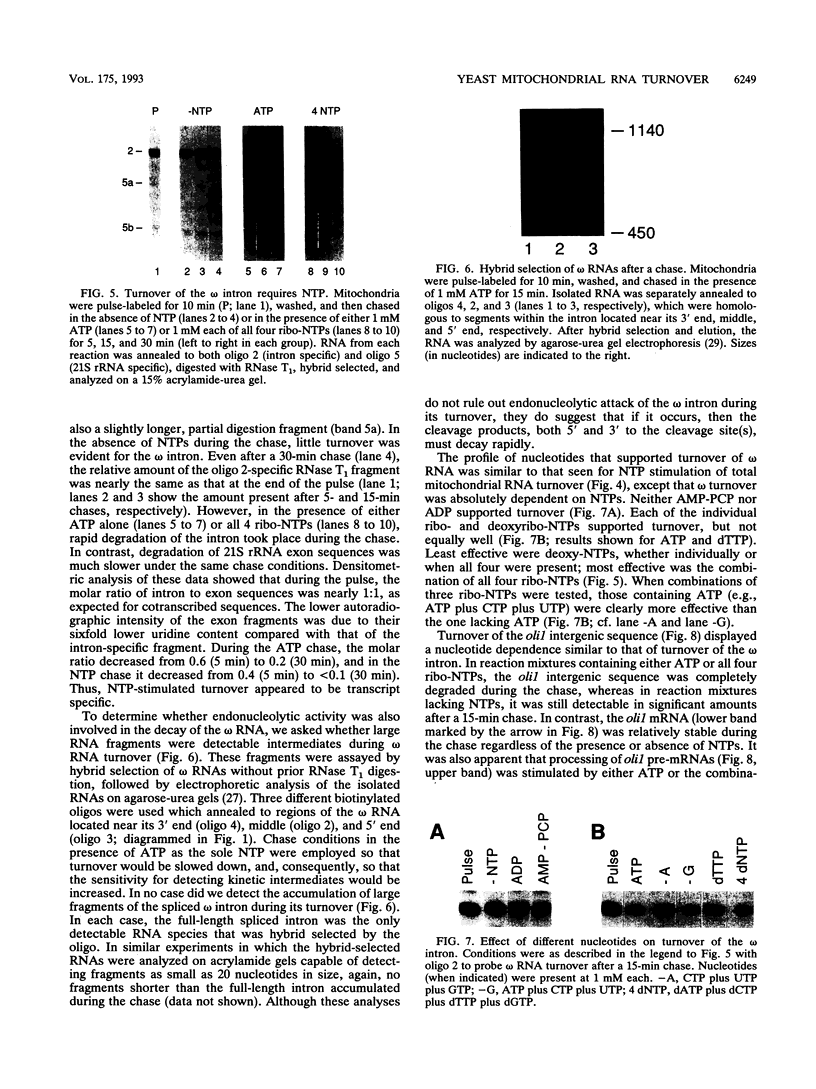
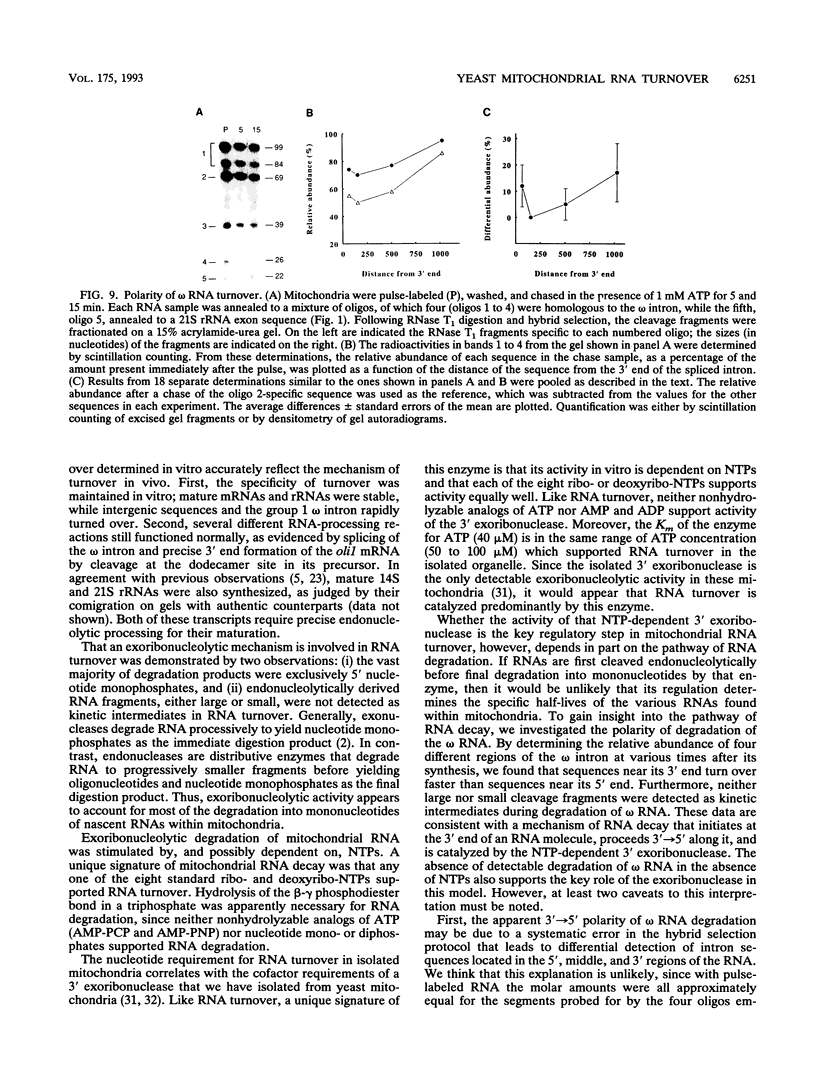
We have employed cell-free transcription reactions with mitochondria isolated from Saccharomyces cerevisiae to study the mechanism of RNA turnover. The specificity of RNA turnover was preserved in these preparations, as were other RNA-processing reactions, including splicing, 3' end formation of mRNAs, and maturation of rRNAs. Turnover of nascent RNAs was found to occur exonucleolytically; endonucleolytic cleavage products were not detected during turnover of the omega intron RNA, which was studied in detail. However, these experiments still leave open the possibility that endonucleolytic cleavage products with very short half-lives are kinetic intermediates in the decay of omega RNA. Exonucleolytic turnover was regulated by nucleotide triphosphates and required their hydrolysis. A unique signature of this regulation was that any one of the eight standard ribo- or deoxyribonucleotide triphosphates supported RNA turnover. A novel hybrid selection protocol was used to determine the turnover rates of the 5', middle, and 3' portions of one mitochondrial transcript, the omega intron RNA. The results suggested that degradation along that transcript occurred with a 3'-->5' polarity. The similarity between features of mitochondrial RNA turnover and the properties of a nucleotide triphosphate-dependent 3' exoribonuclease that has been purified from yeast mitochondria suggests that this single enzyme is a key activity whose regulation is involved in the specificity of mitochondrial RNA turnover.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Beutler E., Gelbart T., Han J. H., Koziol J. A., Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc Natl Acad Sci U S A. 1989 Jan;86(1):192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder R., Hwang S. P., Ratnasabapathy R., Williams D. L. Degradation of apolipoprotein II mRNA occurs via endonucleolytic cleavage at 5'-AAU-3'/5'-UAA-3' elements in single-stranded loop domains of the 3'-noncoding region. J Biol Chem. 1989 Oct 5;264(28):16910–16918. [PubMed] [Google Scholar]
- Boerner P., Mason T. L., Fox T. D. Synthesis and processing of ribosomal RNA in isolated yeast mitochondria. Nucleic Acids Res. 1981 Dec 11;9(23):6379–6390. doi: 10.1093/nar/9.23.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Homison G., Thalenfeld B. E., Tzagoloff A., Nobrega F. G. Assembly of the mitochondrial membrane system. Processing of the apocytochrome b precursor RNAs in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1982 Jun 10;257(11):6268–6274. [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J., Purvis I. J., Santiago T. C., Bettany A. J., Loughlin L., Moore J. Messenger RNA degradation in Saccharomyces cerevisiae. Gene. 1988 Dec 10;72(1-2):151–160. doi: 10.1016/0378-1119(88)90137-0. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur J Biochem. 1989 May 1;181(2):363–370. doi: 10.1111/j.1432-1033.1989.tb14733.x. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Capasso O., Bleecker G. C., Heintz N. Sequences controlling histone H4 mRNA abundance. EMBO J. 1987 Jun;6(6):1825–1831. doi: 10.1002/j.1460-2075.1987.tb02437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Belasco J. G. Degradation of pufLMX mRNA in Rhodobacter capsulatus is initiated by nonrandom endonucleolytic cleavage. J Bacteriol. 1990 Aug;172(8):4578–4586. doi: 10.1128/jb.172.8.4578-4586.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Edwards J. C., Mueller D. M., Rabinowitz M. Identification of a single transcriptional initiation site for the glutamic tRNA and COB genes in yeast mitochondria. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5564–5568. doi: 10.1073/pnas.80.18.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad-Webb H., Perlman P. S., Zhu H., Butow R. A. The nuclear SUV3-1 mutation affects a variety of post-transcriptional processes in yeast mitochondria. Nucleic Acids Res. 1990 Mar 25;18(6):1369–1376. doi: 10.1093/nar/18.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dake E., Hofmann T. J., McIntire S., Hudson A., Zassenhaus H. P. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1988 Jun 5;263(16):7691–7702. [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Dieckmann C. L., Mittelmeier T. M. Nuclearly-encoded CBP1 interacts with the 5' end of mitochondrial cytochrome b pre-mRNA. Curr Genet. 1987;12(6):391–397. doi: 10.1007/BF00434815. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Pape L. K., Tzagoloff A. Identification and cloning of a yeast nuclear gene (CBP1) involved in expression of mitochondrial cytochrome b. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1805–1809. doi: 10.1073/pnas.79.6.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Groot G. S., van Harten-Loosbroek N., van Ommen G. J., Pijst H. L. RNA synthesis in isolated yeast mitochondria. Nucleic Acids Res. 1981 Dec 11;9(23):6369–6377. doi: 10.1093/nar/9.23.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug G., Cohen S. N. Combined actions of multiple hairpin loop structures and sites of rate-limiting endonucleolytic cleavage determine differential degradation rates of individual segments within polycistronic puf operon mRNA. J Bacteriol. 1990 Sep;172(9):5140–5146. doi: 10.1128/jb.172.9.5140-5146.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker J. Analytical and preparative electrophoresis of RNA in agarose-urea. Anal Biochem. 1979 Oct 1;98(2):358–367. doi: 10.1016/0003-2697(79)90154-4. [DOI] [PubMed] [Google Scholar]
- Locker J., Rabinowitz M. Transcription in yeast mitochondria: analysis of the 21 S rRNA region and its transcripts. Plasmid. 1981 Nov;6(3):302–314. doi: 10.1016/0147-619x(81)90038-x. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J. J., Zassenhaus H. P. Characterization of a novel NTP-dependent 3' exoribonuclease from yeast mitochondria. SAAS Bull Biochem Biotechnol. 1991 Jan;4:1–5. [PubMed] [Google Scholar]
- Min J., Heuertz R. M., Zassenhaus H. P. Isolation and characterization of an NTP-dependent 3'-exoribonuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1993 Apr 5;268(10):7350–7357. [PubMed] [Google Scholar]
- Min J., Zassenhaus H. P. Identification of a protein complex that binds to a dodecamer sequence found at the 3' ends of yeast mitochondrial mRNAs. Mol Cell Biol. 1993 Jul;13(7):4167–4173. doi: 10.1128/mcb.13.7.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Zassenhaus H. P. Quantitation of radioactively labeled RNA by hybrid selection using biotinylated oligonucleotides. Biotechniques. 1992 Dec;13(6):870–874. [PubMed] [Google Scholar]
- Mittelmeier T. M., Dieckmann C. L. CBP1 function is required for stability of a hybrid cob-oli1 transcript in yeast mitochondria. Curr Genet. 1990 Dec;18(5):421–428. doi: 10.1007/BF00309911. [DOI] [PubMed] [Google Scholar]
- Mueller D. M., Getz G. S. Steady state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisiae under catabolite repression and derepression. J Biol Chem. 1986 Sep 5;261(25):11816–11822. [PubMed] [Google Scholar]
- Mueller D. M., Getz G. S. Transcriptional regulation of the mitochondrial genome of yeast Saccharomyces cerevisiae. J Biol Chem. 1986 Sep 5;261(25):11756–11764. [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Osinga K. A., De Vries E., Van der Horst G., Tabak H. F. Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 1984 Apr;3(4):829–834. doi: 10.1002/j.1460-2075.1984.tb01892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Brewer G., Bernstein P., Hart P. A., Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1(2):99–126. [PubMed] [Google Scholar]
- Santiago T. C., Purvis I. J., Bettany A. J., Brown A. J. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Nov 11;14(21):8347–8360. doi: 10.1093/nar/14.21.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Stepien P. P., Margossian S. P., Landsman D., Butow R. A. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6813–6817. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartwout S. G., Kinniburgh A. J. c-myc RNA degradation in growing and differentiating cells: possible alternate pathways. Mol Cell Biol. 1989 Jan;9(1):288–295. doi: 10.1128/mcb.9.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H. F., Van der Horst G., Osinga K. A., Arnberg A. C. Splicing of large ribosomal precursor RNA and processing of intron RNA in yeast mitochondria. Cell. 1984 Dec;39(3 Pt 2):623–629. doi: 10.1016/0092-8674(84)90469-0. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Xing Y. Y., Worcel A. A 3' exonuclease activity degrades the pseudogene 5S RNA transcript and processes the major oocyte 5S RNA transcript in Xenopus oocytes. Genes Dev. 1989 Jul;3(7):1008–1018. doi: 10.1101/gad.3.7.1008. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Hofmann T. J., Uthayashanker R., Vincent R. D., Zona M. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 1988 Apr 25;16(8):3283–3296. doi: 10.1093/nar/16.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zassenhaus H. P., Martin N. C., Butow R. A. Origins of transcripts of the yeast mitochondrial var 1 gene. J Biol Chem. 1984 May 10;259(9):6019–6027. [PubMed] [Google Scholar]
- Zhu H., Conrad-Webb H., Liao X. S., Perlman P. S., Butow R. A. Functional expression of a yeast mitochondrial intron-encoded protein requires RNA processing at a conserved dodecamer sequence at the 3' end of the gene. Mol Cell Biol. 1989 Apr;9(4):1507–1512. doi: 10.1128/mcb.9.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The AT spacers and the var1 genes from the mitochondrial genomes of Saccharomyces cerevisiae and Torulopsis glabrata: evolutionary origin and mechanism of formation. Gene. 1987;54(1):1–22. doi: 10.1016/0378-1119(87)90342-8. [DOI] [PubMed] [Google Scholar]