Abstract
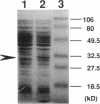
In Pseudomonas paucimobilis UT26, gamma-hexachlorocyclohexane (gamma-HCH) is converted by two steps of dehydrochlorination to a chemically unstable intermediate, 1,3,4,6-tetrachloro-1,4-cyclohexadiene (1,4-TCDN), which is then metabolized to 2,5-dichloro-2,5-cyclohexadiene-1,4-diol (2,5-DDOL) by two steps of hydrolytic dehalogenation via the chemically unstable intermediate 2,4,5-trichloro-2,5-cyclohexadiene-1-ol (2,4,5-DNOL). To clone a gene encoding the enzyme responsible for the conversion of the chemically unstable intermediates 1,4-TCDN and 2,4,5-DNOL, a genomic library of P. paucimobilis UT26 was constructed in Pseudomonas putida PpY101LA into which the linA gene had been introduced by Tn5. An 8-kb BglII fragment from one of the cosmid clones, which could convert gamma-HCH to 2,5-DDOL, was subcloned, and subsequent deletion analyses revealed that a ca. 1.1-kb region was responsible for the activity. Nucleotide sequence analysis revealed an open reading frame (designated the linB gene) of 885 bp within the region. The deduced amino acid sequence of LinB showed significant similarity to hydrolytic dehalogenase, DhlA (D. B. Janssen, F. Pries, J. van der Ploeg, B. Kazemier, P. Terpstra, and B. Witholt, J. Bacteriol. 171:6791-6799, 1989). The protein product of the linB gene was 32 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Not only 1-chlorobutane but also 1-chlorodecane (C10) and 2-chlorobutane, which are poor substrates for other dehalogenases, were good substrates for LinB, suggesting that LinB may be a member of haloalkane dehalogenases with broad-range specificity for substrates.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann A., Walet P., Wijnen P., de Bruin W., Huntjens J. L., Roelofsen W., Zehnder A. J. Biodegradation of alpha- and beta-hexachlorocyclohexane in a soil slurry under different redox conditions. Appl Environ Microbiol. 1988 Jan;54(1):143–149. doi: 10.1128/aem.54.1.143-149.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunya S. P., Jena G. B. Genotoxic potential of the organochlorine insecticide lindane (gamma-BHC): an in vivo study in chicks. Mutat Res. 1992 Oct;272(2):175–181. doi: 10.1016/0165-1161(92)90045-n. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken S. M., Rozeboom H. J., Kalk K. H., Dijkstra B. W. Crystal structure of haloalkane dehalogenase: an enzyme to detoxify halogenated alkanes. EMBO J. 1991 Jun;10(6):1297–1302. doi: 10.1002/j.1460-2075.1991.tb07647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Ngai K. L., Ornston L. N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989 Nov;171(11):6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman D. J. Biotransformation of halogenated compounds. Crit Rev Biotechnol. 1991;11(1):1–40. doi: 10.3109/07388559109069182. [DOI] [PubMed] [Google Scholar]
- Imai R., Nagata Y., Fukuda M., Takagi M., Yano K. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from gamma-hexachlorocyclohexane. J Bacteriol. 1991 Nov;173(21):6811–6819. doi: 10.1128/jb.173.21.6811-6819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen D. B., Gerritse J., Brackman J., Kalk C., Jager D., Witholt B. Purification and characterization of a bacterial dehalogenase with activity toward halogenated alkanes, alcohols and ethers. Eur J Biochem. 1988 Jan 15;171(1-2):67–72. doi: 10.1111/j.1432-1033.1988.tb13759.x. [DOI] [PubMed] [Google Scholar]
- Janssen D. B., Pries F., van der Ploeg J., Kazemier B., Terpstra P., Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol. 1989 Dec;171(12):6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuning S., Janssen D. B., Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985 Aug;163(2):635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbara K., Hashimoto T., Fukuda M., Koana T., Takagi M., Oishi M., Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989 May;171(5):2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marks T. S., Allpress J. D., Maule A. Dehalogenation of lindane by a variety of porphyrins and corrins. Appl Environ Microbiol. 1989 May;55(5):1258–1261. doi: 10.1128/aem.55.5.1258-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menn F. M., Zylstra G. J., Gibson D. T. Location and sequence of the todF gene encoding 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase in Pseudomonas putida F1. Gene. 1991 Jul 31;104(1):91–94. doi: 10.1016/0378-1119(91)90470-v. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Imai R., Sakai A., Fukuda M., Yano K., Takagi M. Isolation and characterization of Tn5-induced mutants of Pseudomonas paucimobilis UT26 defective in gamma-hexachlorocyclohexane dehydrochlorinase (LinA). Biosci Biotechnol Biochem. 1993 May;57(5):703–709. doi: 10.1271/bbb.57.703. [DOI] [PubMed] [Google Scholar]
- Nordlund I., Shingler V. Nucleotide sequences of the meta-cleavage pathway enzymes 2-hydroxymuconic semialdehyde dehydrogenase and 2-hydroxymuconic semialdehyde hydrolase from Pseudomonas CF600. Biochim Biophys Acta. 1990 Jun 21;1049(2):227–230. doi: 10.1016/0167-4781(90)90046-5. [DOI] [PubMed] [Google Scholar]
- Pathak D., Ngai K. L., Ollis D. X-ray crystallographic structure of dienelactone hydrolase at 2.8 A. J Mol Biol. 1988 Nov 20;204(2):435–445. doi: 10.1016/0022-2836(88)90587-6. [DOI] [PubMed] [Google Scholar]
- Pathak D., Ollis D. Refined structure of dienelactone hydrolase at 1.8 A. J Mol Biol. 1990 Jul 20;214(2):497–525. doi: 10.1016/0022-2836(90)90196-s. [DOI] [PubMed] [Google Scholar]
- Powlowski J., Sahlman L., Shingler V. Purification and properties of the physically associated meta-cleavage pathway enzymes 4-hydroxy-2-ketovalerate aldolase and aldehyde dehydrogenase (acylating) from Pseudomonas sp. strain CF600. J Bacteriol. 1993 Jan;175(2):377–385. doi: 10.1128/jb.175.2.377-385.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozeboom H. J., Kingma J., Janssen D. B., Dijkstra B. W. Crystallization of haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Mol Biol. 1988 Apr 5;200(3):611–612. doi: 10.1016/0022-2836(88)90548-7. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Tardif G., Greer C. W., Labbé D., Lau P. C. Involvement of a large plasmid in the degradation of 1,2-dichloroethane by Xanthobacter autotrophicus. Appl Environ Microbiol. 1991 Jun;57(6):1853–1857. doi: 10.1128/aem.57.6.1853-1857.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada I., Kwon S. T., Miyata Y., Matsuzawa H., Ohta T. Unique precursor structure of an extracellular protease, aqualysin I, with NH2- and COOH-terminal pro-sequences and its processing in Escherichia coli. J Biol Chem. 1990 Apr 25;265(12):6576–6581. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yabuuchi E., Yano I., Oyaizu H., Hashimoto Y., Ezaki T., Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34(2):99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]